Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culture Conditions
2.2. Construction of Plasmids
2.3. Analytical Methods
2.4. Transcriptome Analysis
2.5. Real-Time Quantitative PCR (RT-qPCR)
3. Results and Discussion
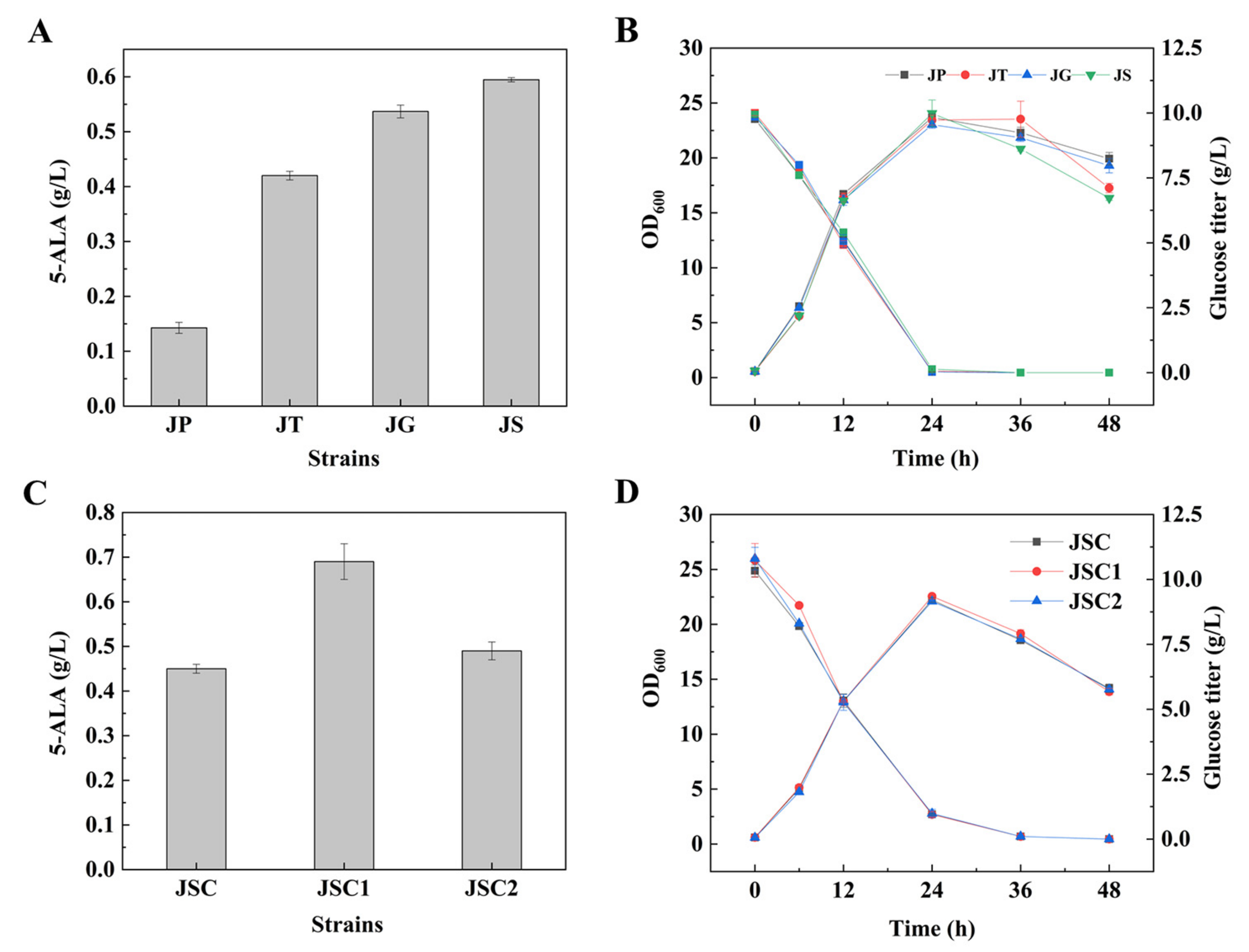
3.1. Enhancement of 5-ALA Synthesis by Overexpressing NCgl0580
3.2. Unexpected Effect of NCgl0580 Deletion on 5-ALA Biosynthesis
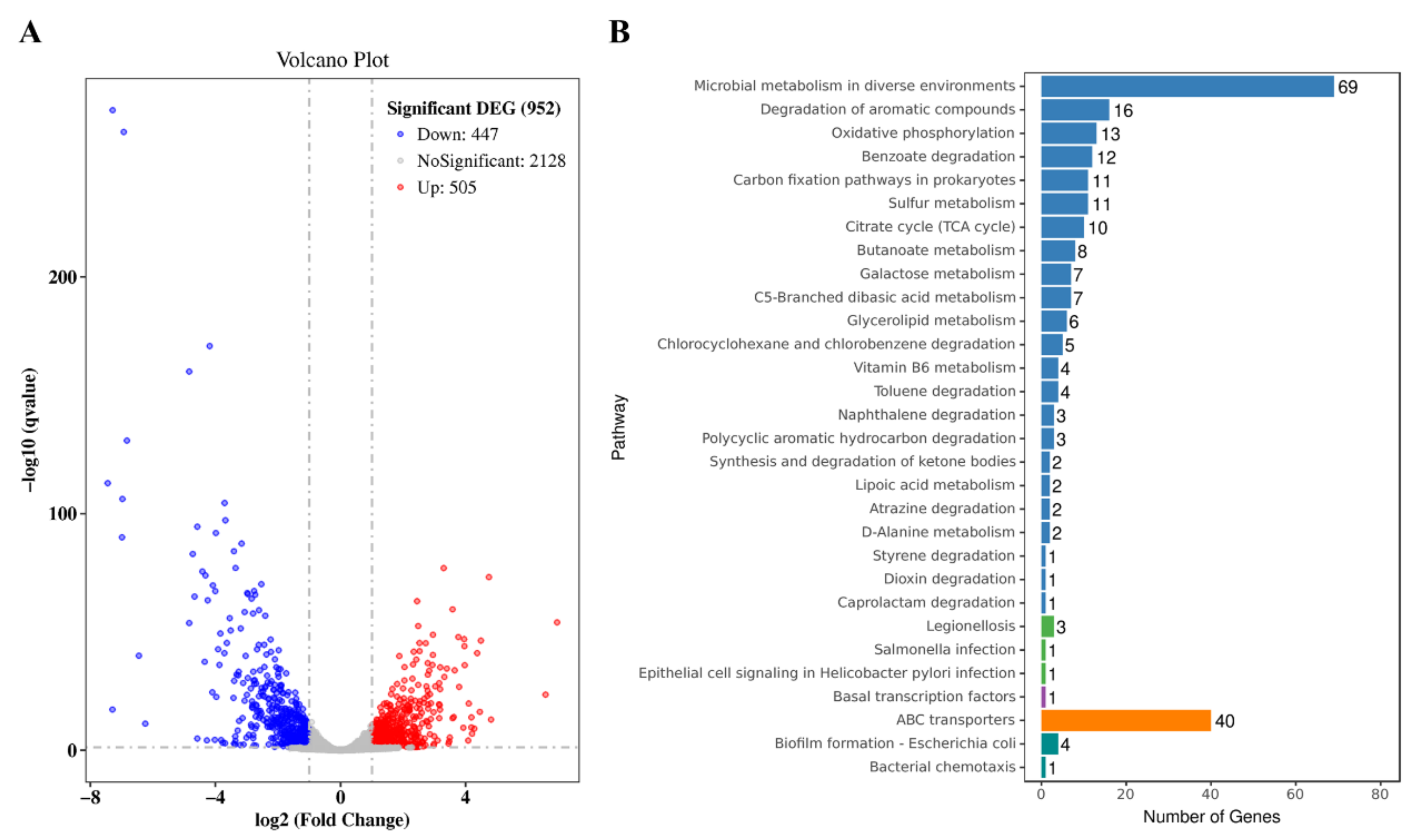
3.3. Transcriptomic Analysis of the Effect of NCgl0580 Knockout on 5-ALA Synthesis
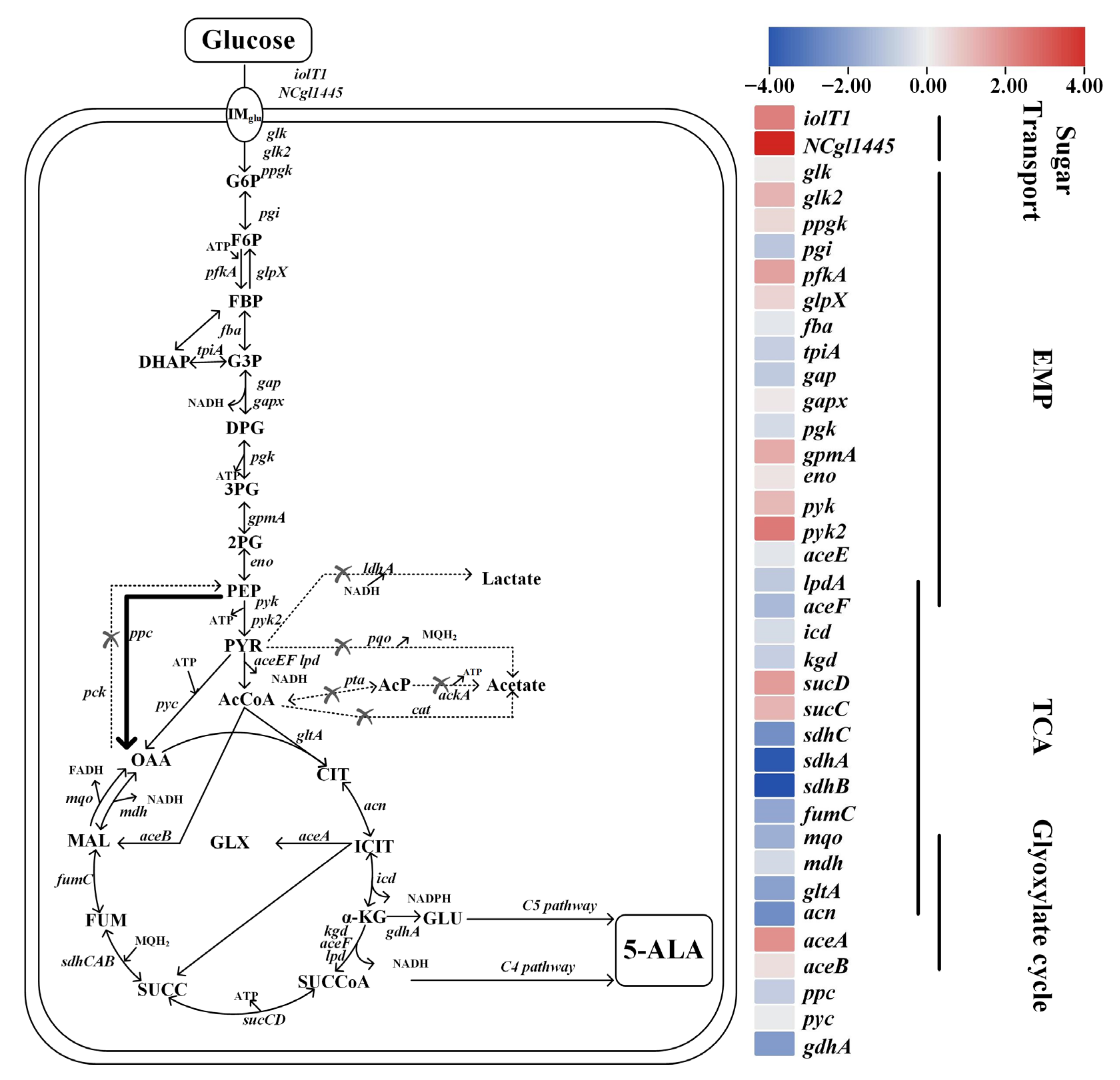
3.4. Redistribution of Central Carbon Fluxes toward Succinyl-CoA-Enhanced 5-ALA Biosynthesis
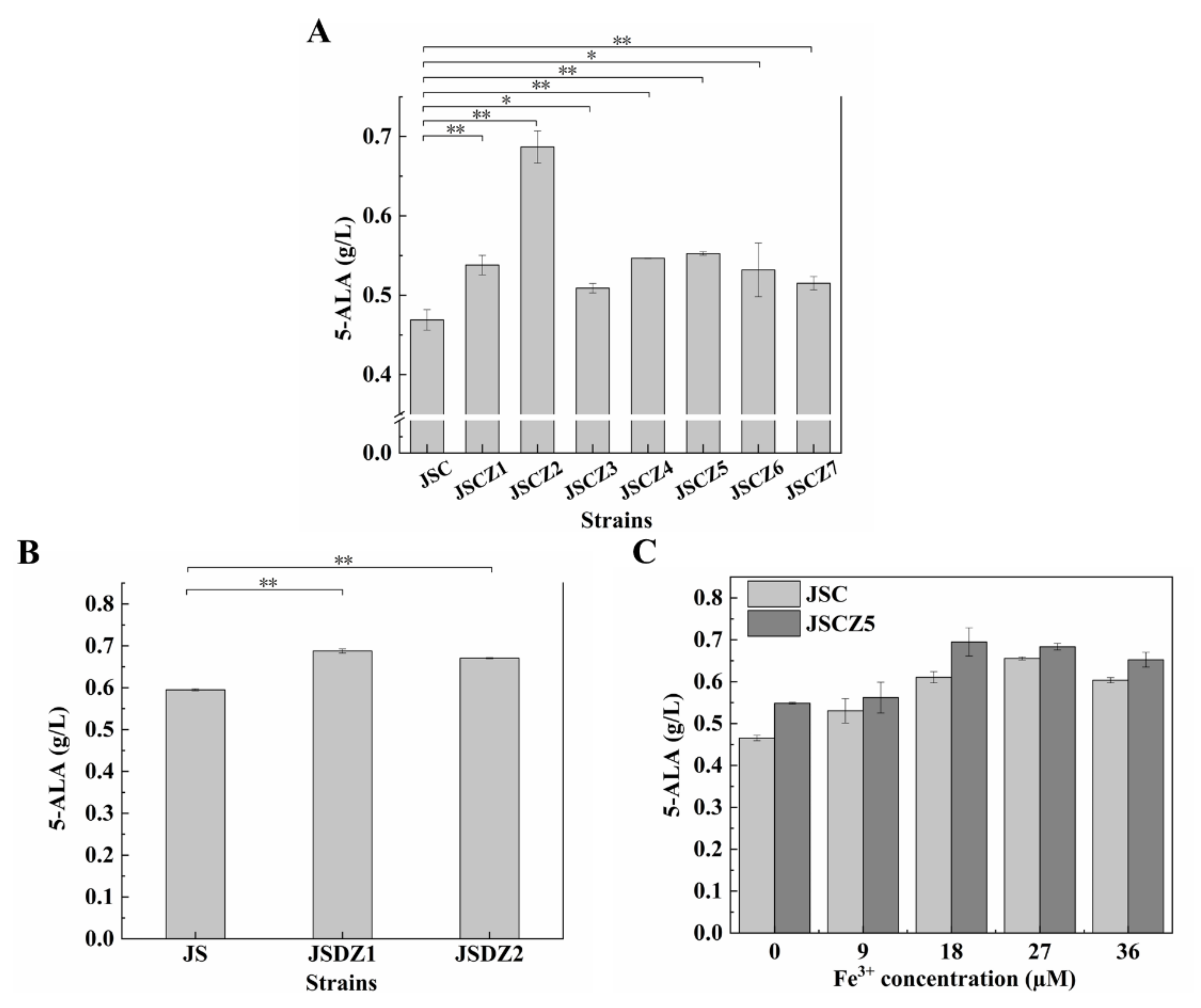
3.5. Enhancement of Iron and Phosphate Uptake Improves 5-ALA Synthesis
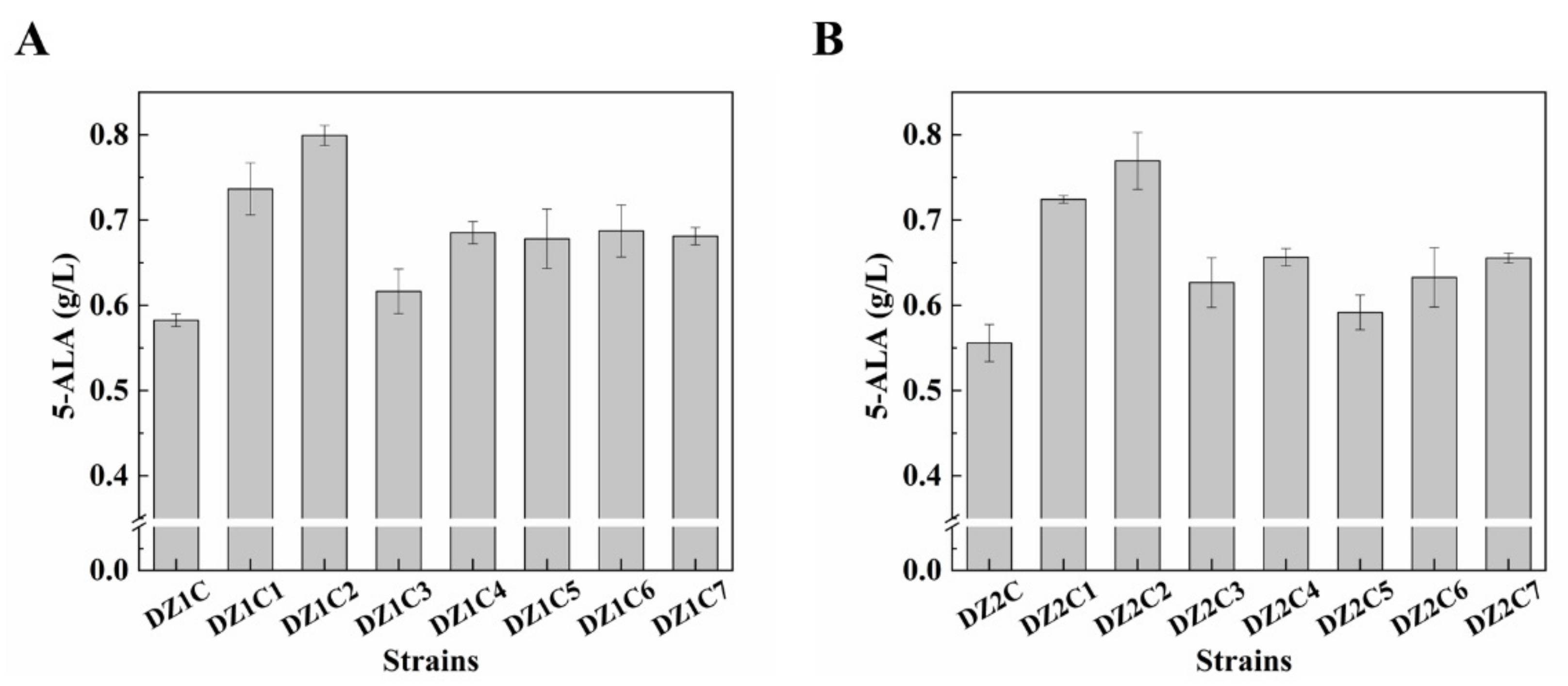
3.6. Effect of Multiple Gene Expression Perturbation on 5-ALA Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kang, Z.; Zhang, J.; Zhou, J.; Qi, Q.; Du, G.; Chen, J. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol. Adv. 2012, 30, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhao, M.; Wang, W.; Hong, L.; Wu, Z.; Luo, G.; Lu, S.; Tang, Y.; Li, J.; Wang, J.; et al. 5-ALA mediated photodynamic therapy with combined treatment improves anti-tumor efficacy of immunotherapy through boosting immunogenic cell death. Cancer Lett. 2022, 554, 216032. [Google Scholar] [CrossRef] [PubMed]
- McCracken, D.J.; Schupper, A.J.; Lakomkin, N.; Malcolm, J.; Painton Bray, D.; Hadjipanayis, C.G. Turning on the light for brain tumor surgery: A 5-aminolevulinic acid story. Neuro. Oncol. 2022, 24, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Suero Molina, E.; Kaneko, S.; Black, D.; Stummer, W. 5-Aminolevulinic acid-induced porphyrin contents in various brain tumors: Implications regarding imaging device design and their validation. Neurosurgery 2021, 89, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, Y.; Liao, X.; Tang, Y.; Ni, Q.; Sun, J.; Zhao, Y.; Zhang, J.; Teng, Z.; Lu, G. Enhancing selective photosensitizer accumulation and oxygen supply for high-efficacy photodynamic therapy toward glioma by 5-aminolevulinic acid loaded nanoplatform. J. Colloid Interf. Sci. 2020, 565, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Ngwe Tun, M.M.; Sakura, T.; Sakurai, Y.; Kurosaki, Y.; Inaoka, D.K.; Shioda, N.; Yasuda, J.; Kita, K.; Morita, K. Antiviral activity of 5-aminolevulinic acid against variants of severe acute respiratory syndrome coronavirus 2. Trop. Med. Health 2022, 50, 6. [Google Scholar] [CrossRef]
- Sakurai, Y.; Ngwe Tun, M.M.; Kurosaki, Y.; Sakura, T.; Inaoka, D.K.; Fujine, K.; Kita, K.; Morita, K.; Yasuda, J. 5-amino levulinic acid inhibits SARS-CoV-2 infection in vitro. Biochem. Biophys. Res. Commun. 2021, 545, 203–207. [Google Scholar] [CrossRef]
- Yang, L.; Wu, Y.; Wang, X.; Lv, J.; Tang, Z.; Hu, L.; Luo, S.; Wang, R.; Ali, B.; Yu, J. Physiological mechanism of exogenous 5-aminolevulinic acid improved the tolerance of chinese cabbage (Brassica pekinensis L.) to cadmium stress. Front. Plant Sci. 2022, 13, 427. [Google Scholar] [CrossRef]
- Dayan, F.E.; Duke, S.O. Natural compounds as next-generation herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef]
- Global 5-aminolevulinic Acid Hydrochloride Market Growth 2022–2028. Available online: https://www.360researchreports.com/global-5-aminolevulinic-acid-hydrochloride-market-19884300 (accessed on 30 December 2022).
- Zhang, J.; Cui, Z.; Zhu, Y.; Zhu, Z.; Qi, Q.; Wang, Q. Recent advances in microbial production of high-value compounds in the tetrapyrrole biosynthesis pathway. Biotechnol. Adv. 2022, 55, 107904. [Google Scholar] [CrossRef]
- Shemin, D.; Russell, C.S. δ-aminolevulinic acid, its role in the biosynthesis of porphyrins and purines1. J. Am. Chem. Soc. 1953, 75, 4873–4874. [Google Scholar] [CrossRef]
- Wang, L.Y.; Brown, L.; Elliott, M.; Elliott, T. Regulation of heme biosynthesis in Salmonella typhimurium: Activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism which increases abundance of the protein. J. Bacteriol. 1997, 179, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Woodard, S.I.; Dailey, H.A. Regulation of heme biosynthesis in Escherichia coli. Arch. Biochem. Biophys. 1995, 316, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Hong, K.; Mao, Y.; Ma, H.; Chen, T.; Wang, Z. Natural 5-aminolevulinic acid: Sources, biosynthesis, detection and applications. Front. Bioeng Biotechnol. 2022, 10, 841443. [Google Scholar] [CrossRef]
- Jørgensen, M.G.; Nielsen, J.S.; Boysen, A.; Franch, T.; Møller-Jensen, J.; Valentin-Hansen, P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012, 84, 36–50. [Google Scholar] [CrossRef]
- Livshits, V.A.; Zakataeva, N.P.; Aleshin, V.V.; Vitushkina, M.V. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 2003, 154, 123–135. [Google Scholar] [CrossRef]
- Kang, Z.; Wang, Y.; Gu, P.; Wang, Q.; Qi, Q. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab. Eng. 2011, 13, 492–498. [Google Scholar] [CrossRef]
- Tan, S.-I.; You, S.-C.; Shih, I.T.; Ng, I.S. Quantification, regulation and production of 5-aminolevulinic acid by green fluorescent protein in recombinant Escherichia coli. J. Biosci. Bioeng. 2020, 129, 387–394. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Zhu, F.; Li, Z.; Lu, N.; Li, Y.; Xu, Q.; Chen, N. Metabolic engineering of an auto-regulated Corynebacterium glutamicum chassis for biosynthesis of 5-aminolevulinic acid. Bioresour. Technol. 2020, 318, 124064. [Google Scholar] [CrossRef]
- Feng, L.L.; Zhang, Y.; Fu, J.; Mao, Y.F.; Chen, T.; Zhao, X.M.; Wang, Z.W. Metabolic engineering of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid. Biotechnol. Bioeng. 2016, 113, 1284–1293. [Google Scholar] [CrossRef]
- Laneelle, M.A.; Tropis, M.; Daffe, M. Current knowledge on mycolic acids in Corynebacterium glutamicum and their relevance for biotechnological processes. Appl. Microbiol. Biot. 2013, 97, 9923–9930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Gao, Y.J.; Chen, Z.W.; Xu, G.Q.; Zhang, X.J.; Li, H.; Shi, J.S.; Koffas, M.A.G.; Xu, Z.H. High-yield production of l-serine through a novel identified exporter combined with synthetic pathway in Corynebacterium glutamicum. Microb. Cell Fact 2020, 19, 115. [Google Scholar] [CrossRef] [PubMed]
- Kishino, M.; Kondoh, M.; Hirasawa, T. Enhanced L-cysteine production by overexpressing potential L-cysteine exporter genes in an L-cysteine-producing recombinant strain of Corynebacterium glutamicum. Biosci. Biotech. Bioch. 2019, 83, 2390–2393. [Google Scholar] [CrossRef] [PubMed]
- Ghiffary, M.R.; Prabowo, C.P.S.; Adidjaja, J.J.; Lee, S.Y.; Kim, H.U. Systems metabolic engineering of Corynebacterium glutamicum for the efficient production of β-alanine. Metab. Eng. 2022, 74, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Mao, Y.; Luo, J.; Liu, P.; Jiang, M.; He, G.; Zhang, Z.; Cao, Q.; Shen, J.; Ma, H.; et al. Global cellular metabolic rewiring adapts Corynebacterium glutamicum to efficient nonnatural xylose utilization. Appl. Environ. Microbiol. 2022, 88, e0151822. [Google Scholar] [CrossRef]
- Sinclair, P.R.; Gorman, N.; Jacobs, J.M. Measurement of heme concentration. Curr. Protoc. Toxicol. 1999, 1, 831–837. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Zheng, P.; Zhao, J.; Tan, Z.J.; Chen, J.Z.; Rao, D.M.; Sun, J.B.; Ma, Y.H. 5-aminolevulinic Acid (ALA) Synthetase Mutant and Host Cells and Application Thereof. China Patent CN108251396B, 1 April 2022. Available online: https://worldwide.espacenet.com/publicationDetails/biblio?CC=CN&NR=108251396B&KC=B&FT=D (accessed on 30 December 2022).
- Shang, X.L.; Chai, X.; Lu, X.M.; Li, Y.; Zhang, Y.; Wang, G.Q.; Zhang, C.; Liu, S.W.; Zhang, Y.; Ma, J.Y.; et al. Native promoters of Corynebacterium glutamicum and its application in l-lysine production. Biotechnol. Lett. 2018, 40, 383–391. [Google Scholar] [CrossRef]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; de Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb. Cell Fact 2015, 14, 201. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.M.; Xu, G.Q.; Zhang, X.J.; Shi, J.S.; Xu, Z.H. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve L-serine yield in Corynebacterium glutamicum. Appl. Microbiol. Biot. 2018, 102, 5939–5951. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Zakataeva, N.P.; Kutukova, E.A.; Gronskiy, S.V.; Troshin, P.V.; Livshits, V.A.; Aleshin, V.V. Export of metabolites by the proteins of the DMT and RhtB families and its possible role in intercellular communication. Microbiology 2006, 75, 438–448. [Google Scholar] [CrossRef]
- Zavilgelsky, G.B.; Manukhov, I.V. Quorum Sensing, or How Bacteria “Talk” to Each Other. Mol. Biol. 2001, 35, 224–232. [Google Scholar] [CrossRef]
- Banerjee, D.; Eng, T.; Sasaki, Y.; Srinivasan, A.; Oka, A.; Herbert, R.A.; Trinh, J.; Singan, V.R.; Sun, N.; Putnam, D.; et al. Genomics characterization of an engineered Corynebacterium glutamicum in bioreactor cultivation under ionic liquid stress. Front. Bioeng. Biotechnol. 2021, 9, 766674. [Google Scholar] [CrossRef] [PubMed]
- Krings, E.; Krumbach, K.; Bathe, B.; Kelle, R.; Wendisch, V.F.; Sahm, H.; Eggeling, L. Characterization of myo-inositol utilization by Corynebacterium glutamicum: The stimulon, identification of transporters, and influence on L-lysine formation. J. Bacteriol. 2006, 188, 8054–8061. [Google Scholar] [CrossRef]
- Lindner, S.N.; Seibold, G.M.; Henrich, A.; Kramer, R.; Wendisch, V.F. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl. Environ. Microb. 2011, 77, 3571–3581. [Google Scholar] [CrossRef] [PubMed]
- Man, Z.W.; Xu, M.J.; Rao, Z.M.; Guo, J.; Yang, T.W.; Zhang, X.; Xu, Z.H. Systems pathway engineering of Corynebacterium crenatum for improved L-arginine production. Sci. Rep. 2016, 6, 28629. [Google Scholar] [CrossRef]
- Xu, J.Z.; Wu, Z.H.; Gao, S.J.; Zhang, W.G. Rational modification of tricarboxylic acid cycle for improving L-lysine production in Corynebacterium glutamicum. Microb Cell Fact 2018, 17, 105. [Google Scholar] [CrossRef]
- Zhang, X.M.; Lai, L.H.; Xu, G.Q.; Zhang, X.J.; Shi, J.S.; Koffas, M.A.G.; Xu, Z.H. Rewiring the central metabolic pathway for high-yield l-serine production in Corynebacterium glutamicum by using glucose. Biotechnol. J. 2019, 14, 1800497. [Google Scholar] [CrossRef]
- Ghiffary, M.R.; Prabowo, C.P.S.; Sharma, K.; Yan, Y.; Lee, S.Y.; Kim, H.U. High-level production of the natural blue pigment indigoidine from metabolically engineered Corynebacterium glutamicum for sustainable fabric dyes. ACS Sustain. Chem. Eng. 2021, 9, 6613–6622. [Google Scholar] [CrossRef]
- Litsanov, B.; Kabus, A.; Brocker, M.; Bott, M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 2012, 5, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Miscevic, D.; Mao, J.Y.; Kefale, T.; Abedi, D.; Moo-Young, M.; Chou, C.P. Strain engineering for high-level 5-aminolevulinic acid production in Escherichia coli. Biotechnol. Bioeng. 2021, 118, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, H.U.; Kim, T.Y.; Lee, S.Y. Systematic engineering of TCA cycle for optimal production of a four-carbon platform chemical 4-hydroxybutyric acid in Escherichia coli. Metab. Eng. 2016, 38, 264–273. [Google Scholar] [CrossRef] [PubMed]
- He, G. Production of 5-Aminolevulinic Acid in the Succinate-Producing Corynebacterium glutamicum by Metabolic Engineering. Master’s Thesis, Tianjin University, Tianjin, China, 28 May 2022. [Google Scholar]
- Rehm, N.; Georgi, T.; Hiery, E.; Degner, U.; Schmiedl, A.; Burkovski, A.; Bott, M. L-glutamine as a nitrogen source for Corynebacterium glutamicum: Derepression of the AmtR regulon and implications for nitrogen sensing. Microbiology 2010, 156, 3180–3193. [Google Scholar] [CrossRef]
- Ge, F.L.; Li, X.K.; Ge, Q.R.; Zhu, D.; Li, W.; Shi, F.H.; Chen, H.J. Modular control of multiple pathways of Corynebacterium glutamicum for 5-aminolevulinic acid production. Amb. Express. 2021, 11, 179. [Google Scholar] [CrossRef]
- Hollenstein, K.; Dawson, R.J.; Locher, K.P. Structure and mechanism of ABC transporter proteins. Curr. Opin. in Struct. Biol. 2007, 17, 412–418. [Google Scholar] [CrossRef]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef]
- Ishige, T.; Krause, M.; Bott, M.; Wendisch, V.F.; Sahm, H. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 2003, 185, 4519–4529. [Google Scholar] [CrossRef]
- Zhang, J.; Kang, Z.; Ding, W.; Chen, J.; Du, G. Integrated optimization of the in vivo heme biosynthesis pathway and the in vitro iron concentration for 5-aminolevulinate production. Appl. Biochem. Biotechnol. 2016, 178, 1252–1262. [Google Scholar] [CrossRef]
- Wuttge, S.; Bommer, M.; Jäger, F.; Martins, B.M.; Jacob, S.; Licht, A.; Scheffel, F.; Dobbek, H.; Schneider, E. Determinants of substrate specificity and biochemical properties of the sn-glycerol-3-phosphate ATP binding cassette transporter (UgpB–AEC2) of Escherichia coli. Mol. Microbiol. 2012, 86, 908–920. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Jiang, M.; Kong, S.; Hong, K.; Zhao, J.; Sun, X.; Cui, Z.; Chen, T.; Wang, Z. Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum. Fermentation 2023, 9, 213. https://doi.org/10.3390/fermentation9030213
Wu J, Jiang M, Kong S, Hong K, Zhao J, Sun X, Cui Z, Chen T, Wang Z. Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum. Fermentation. 2023; 9(3):213. https://doi.org/10.3390/fermentation9030213
Chicago/Turabian StyleWu, Jian, Meiru Jiang, Shutian Kong, Kunqiang Hong, Juntao Zhao, Xi Sun, Zhenzhen Cui, Tao Chen, and Zhiwen Wang. 2023. "Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum" Fermentation 9, no. 3: 213. https://doi.org/10.3390/fermentation9030213
APA StyleWu, J., Jiang, M., Kong, S., Hong, K., Zhao, J., Sun, X., Cui, Z., Chen, T., & Wang, Z. (2023). Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum. Fermentation, 9(3), 213. https://doi.org/10.3390/fermentation9030213





