Abstract
The removal of sulfur by deep hydrodesulfurization is expensive and environmentally unfriendly. Additionally, sulfur is not separated completely from heterocyclic poly-aromatic compounds. In nature, several microorganisms (Rhodococcus erythropolis IGTS8, Gordonia sp., Bacillus sp., Mycobacterium sp., Paenibacillus sp. A11-2 etc.) have been reported to remove sulfur from petroleum fractions. All these microbes remove sulfur from recalcitrant organosulfur compounds via the 4S pathway, showing potential for some organosulfur compounds only. Activity up to 100 µM/g dry cell weights is needed to meet the current demand for desulfurization. The present review describes the desulfurization capability of various microorganisms acting on several kinds of sulfur sources. Genetic engineering approaches on Gordonia sp. and other species have revealed a variety of good substrate ranges of desulfurization, both for aliphatic and aromatic organosulfur compounds. Whole genome sequence analysis and 4S pathway inhibition by a pTeR group inhibitor have also been discussed. Now, emphasis is being placed on how to commercialize the microbes for industrial-level applications by incorporating biodesulfurization into hydrodesulfurization systems. Thus, this review summarizes the potentialities of microbes for desulfurization of petroleum. The information included in this review could be useful for researchers as well as the economical commercialization of bacteria in petroleum industries.
1. Introduction
Sulfur is abundant in fossil fuels in both inorganic and organic forms. Inorganic sulfur is distributed as hydrogen sulfide, elemental sulfur, and pyrites, while organic sulfur is present in the form of hydrocarbons such as disulfides, mercaptans, thiol sulfones, thiophenes, and thioether. Some complex forms of sulfur are also dominantly distributed in petroleum products, including crude oil, petroleum liquids, petroleum distillate fractions, etc. Crude oil is a complex mixture of over 200 different hydrocarbon organic compounds. The quality of crude oil is based on the presence of American Petroleum Institute (API) gravity, specific gravity (SG), and sulfur content (S%). Organic sulfur in crude oils ranges from 5 w% to about 0.1 w% [1,2]. It has been reported that more than 200 sulfur-containing petroleum products are enriched with organic forms [3,4]. They are categorized into three main groups: group (I) contains aliphatic and aromatic thiols and their bisulfides as oxidation products; group (II) covers aliphatic and aromatic thioethers; group (III) includes heterocycles based on the thiophenic ring: thiophene, dibenzothiophene (DBT), benzothiophene (BT), and their alkyl derivatives [5,6]. The physical properties and chemical constituents of petroleum products, including sulfur amounts (0.4–5.0 wt.%), are reported differently in the samples collected from various reservoirs [7]. Moreover, a high percentage of sulfur was observed in crude oil collected from a place of higher density [8,9]. Thioethers, thiols, and thiophenes are predominant forms of organosulfur compounds, mainly reported as condensed thiophenes [10]. High sulfur content was reported in the crude oils of the USA (5.5%), Germany (9.6%), and Utah (13.9%) [6]. Gasoline sulfur is rich in non-thiophenic and thiophenic compounds, while diesel oil sulfur is rich in a high amount of benzothiophene and dibenzothiophenes [11]. The major forms such as thiols, sulfides, disulfides, and thiophenes are observed as organic sulfur, while inorganic moieties are present as pyrites [12]. API gravity and organic sulfur content found in the crude oils produced by different countries are shown in Figure 1 [4].
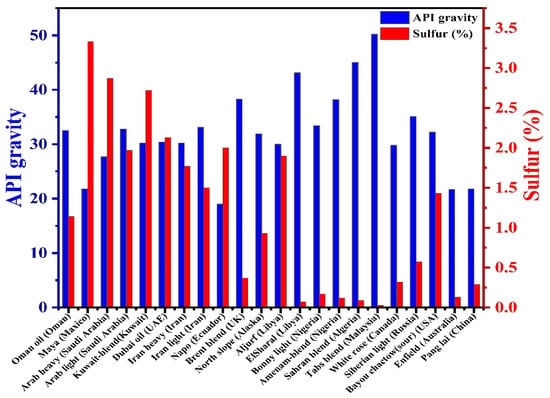
Figure 1.
API gravity and sulfur content of selected crude oils.
As low molecular weight molecules, mercaptans with fewer than eight carbon atoms are commonly found in in crude oil. The refinement of crude oil is performed to create a negligible amount of Mercaptans [13]. The molecular structures of some of these sulfur compounds are represented in Figure 2. Aliphatic sulfides are distributed as cyclic aliphatic sulfides or acyclic aliphatic sulfides and reported as major sulfur-containing contents found in fractions of many petroleum products, including diesel fuels and heating oils. Heavier oil fractions have been characterized by low concentrations of aromatic sulfides, of which thiophenic sulfur is normally the most abundant form. However, it is dependent on the oil’s reservoir history, where the presence of other sulfur compounds in significant amounts has been reported. Thiophenes, an unsaturated five-member heterocyclic ring, have been reported as a significant component of high-sulfur oils [14].
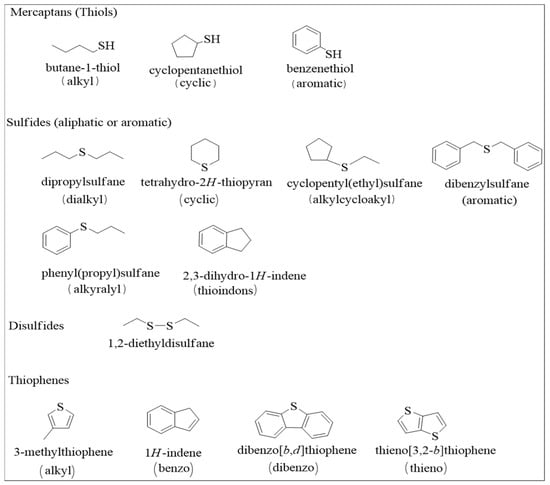
Figure 2.
Different types of sulfur-containing organic compounds identified in crude oil.
2. Problems Caused by Petroleum Sulfur
Petroleum products are a non-renewable source of energy that can be burned to produce a useable amount of energy. Most of the liquid and solid materials, including oil and coal, are contaminated with sulfur-containing compounds. Direct burning of such fuels releases sulfur oxides into the atmosphere [15,16] and results in air toxicity [17]. Sulfur pollution has adverse effects on living beings and environments, causing effects such as respiratory cancer, cardio-respiratory diseases, [18,19] acid rain, and the depletion of the ozone layer [20]. SO2 concentrations in the atmosphere greater than 100 ppm are hazardous to human health and lethal when they reach 400–500 ppm, even when the exposure time is very short. Dust or fog has a noxious effect on plants when it is mixed with SO2, as the plants become damaged in minutes in the presence of 1–2 ppm of SO2 [21].
Moreover, sulfur in oil creates several problems, including corrosion of pipeline and refining equipment and the breakdown of combustion engines. Furthermore, it contaminates many catalysts that are used to refine oil and burn fossil fuels. The burning of diesel and gasoline emits sulfate particulate matter and sulfur dioxide (SO2), resulting in acid rain, which is very harmful for plant and aquatic life, as well as agricultural lands [16,22].
3. Petroleum Refining
Refining of crude oil is a complex chemical process designed to purify petroleum into a number of valuable products with defined properties. Refining begins with the distillation of crude oil and differentiating it into a sequence of streams at definite boiling temperatures. Table 1 indicates some of the distillation fractions and their boiling temperature ranges. Fuels, including diesel, gasoline, and kerosene, are the most valuable products of petroleum [23,24]. Heavier fractions are reprocessed to increase the quantity of these fuels recovered from a single barrel of crude oil. Heavier streams are broken down into smaller molecules to create surplus material in the naphtha range and increase the productivity of the valued product, gasoline. Fluid catalytic cracking (FCC) classically exploits a solid acid zeolite catalyst, which is supported with rare earth metals in a fluidized bed reactor. It has been reported that much of the sulfur content is accumulated as heavy polynuclear aromatics. As a result, cracked naphtha streams frequently contain more sulfur than virgin naphtha [25].

Table 1.
Petroleum distillate fractions and their boiling points.
The quality of other fractionating products and gasoline is improved by two additional processes, such as when n-hexane, e.g., linear paraffins, isomerizes into branched paraffins such as 2,3-dimethylbutane in the reforming process catalyzed by Pt supported on zeolites, sulfated zirconia, and chlorided alumina [26]. In the presence of a catalyst, reactions proceed at different rates, and zeolites offered the lowest reaction rate [27]. Nevertheless, active catalysts are highly susceptible to poisoning by sulfur and water [26]. Removal of sulfur before reforming gasoline streams is thus needed. Alkylation is the procedure applied to expand the quality of gasoline. During this process, 2,2,4-trimethylpentane or isooctane and other branched paraffins are recovered using the reaction of n-butene with isobutane. Liquid acid catalysts are currently used in the alkylation process to avoid excessive cracking during the process. In alkylation, a reactor’s mixture of either hydrofluoric acid or sulfuric acid with butane or isobutene is used to produce alkylate for obtaining high-quality gasoline that is blended into other gasoline streams [28]. The key and last process used in oil refining is hydrodesulfurization (HDS) or hydrotreating (Figure 3). Crude petroleum has a variable sulfur content, depending on where it is recovered as crude petroleum from Norway and the North Sea; Norway typically has the lowest sulfur content (0.1 w%), while the Arabian Peninsula has the highest (3 w%) [4,29]. Table 2 shows the distribution of some aromatic sulfur species and their boiling points found in petroleum. The most common light species denoted as gasoline-range sulfur shown in Table 1 are ethane-, methane-, carbonyl sulfide (COS), t-butanethiol, dimethyl sulfide, and tetrahydrothiophene [30].
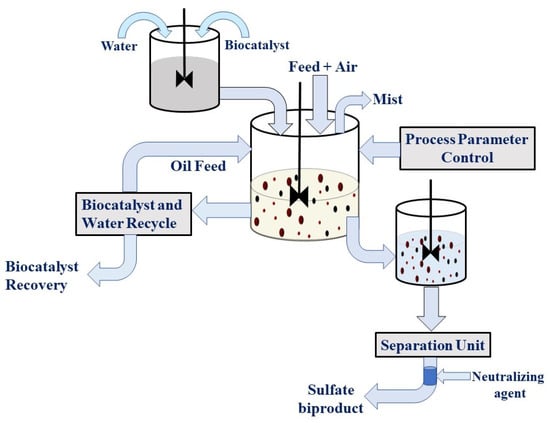
Figure 3.
Typical refining technology hydrodesulfurization plan illustrating desulfurization processing sequences.

Table 2.
Aromatic sulfur species found in petroleum and their boiling point.
4. Desulfurization by Processes Other Than HDS
Released sulfur pollutants are directly related to the amount of sulfur compounds present in petroleum products. It has been shown that pollutant emissions can be reduced by reducing the number of sulfur-containing compounds in petroleum. The biochemical mechanism of desulfurization is mainly dependent on oxidative desulfurization and selective adsorbent processes. Moreover, the microbial metabolisms of sulfur-containing compounds also play a significant role in the desulfurization process [31]. Oxidative desulfurization (ODS) is a key process used to remove sulfur present in petroleum products. This process is routinely used by petrochemical industries to remove heavy sulfur as polynuclear aromatics. The oxidation of sulfur is carried out by the addition of one or two moles of oxygen without breaking the C–C bond, producing sulfone and sulfoxide [32,33]. A catalyst, molybdenum/modified medicinal stone (Mo/MMS), has been reported to remove dibenzothiophene (DBT) up to 97.5% within 60 min [34], while the catalyst MoO3/4 has been reported to remove DBT from 500 to 5.0 ppm in octane under optimal conditions [35]. The oxidized compounds from this process can then be successfully separated via adsorption or extraction from the petroleum stream [36]. A mixture of solvents N,N-dimethylformamide (DMF), N,N-dimethylacetamide (DMAC), and tetramethylsulfone (TMS) has been analyzed as a powerful agent used to separate DBT and DBT derivatives from gasoline [37]. The adsorbents used in the desulfurization process have reported low adsorption capacities, such as 0.123 moles sulfur per moles of metal for a vague transition metal on silica adsorbent [38]. The adsorbent capacities of economical clay materials are investigated at 1–4 mg sulfur compound per gram clay [39]. Thus, enormous amounts of adsorbent are required for desulfurization. In pressure swing adsorption, large adsorbent beds and multiple beds have been used for desulfurization. Among these adsorbent pressure swings, adsorption is reported to be less effective compared to large adsorbent beds and is preferred to reduce the number of turnovers; thus, multiple beds are required to maintain a refinery on-stream run. Continuous use of calcinations has also been reported to damage the surface area because of sintering, minimizing the amount of sulfur available. Efforts have been made in this field to make the process more cost effective [40] and to use materials with a larger surface area [41,42].
Currently, biotechnology and microbiology have been effectively used for desulfurization. Several bacteria have the capacity to metabolize organosulfur compounds in any one of three possible ways: reductive C–S, oxidative C–C bond cleavage, or oxidative C–S bond cleavage. Pseudomonas and Desulfovibrio desulfuricans can reduce dibenzothiophene and produce the sulfur as H2S, along with denitrification [39]. Rhodococcus erythropolis strain IGTS8 has a 4S pathway, which catalyzes the oxidative C–S cleavage-based oxidation of organosulfur to sulfite or sulphate [40]. Sulfur dioxide in flue gas can also be removed chemically by reacting with H2O to form sulfite, which is then transformed into sulfate [43].
Although microbial processes are eco-friendly and economical, they have many limitations that prevent them being commercialized today [44]. The conversion rate of sulfur compounds into their metabolites is generally slow compared to chemical reactions. The sulfur transformation capacity of bacteria has also been established to decrease the sulfur content of diesel fuel from 535 ppm to 75 ppm in 24 h. The rate-limiting step and mass transfer from the oil/water interface control the metabolizing process of sulfur [45,46]. Furthermore, for biotransformation to be catalyzed in various physio-chemical conditions, a microbial system requires approximately 2.5 g of biomass per g of sulfur and must be alive. Olma et al. stated that the conversion rate in the process of desulfurization depends on dissolved oxygen, concentration, temperature, and pH [47]. Moreover, the immobilized cell catalyzes desulfurization at a slower rate, and separating cells from the oil can also be complicated [48]. Thermophilic bacteria are still in focus for biodesulfurization in comparison to mesophilic bacteria because desulfurization takes place at high temperatures in the real desulfurization industry [49].
Combining concepts from different technologies has been recommended for desulfurization for short-term desulfurization by focusing on the benefits of each technology used. Most commercial desulfurization processes, such as ExxonMobil’s SCAN refining process [50] and UniPure’s ASR-29 [51], contain combinations of several desulfurization technologies that exist in refining. These combinations eliminate sulfur at small levels by focusing on the strengths of each technology used. Another example is using amine scrubbing and mild HDS to remove H2S. An oxidative desulfurization (ODS) process comprises two extractions and several adsorption steps. Despite this, HDS remains the main sulfur removal method in petroleum refining. The high use of catalytic hydrogenation for sulfur removal suggests that improved techniques for desulfurization may be found in typical hydrogenation reactors.
4.1. Hydrodesulfurization Reaction
HDS diminishes sulfur-containing compounds in a stream using a catalyst and hydrogen gas. There is no flexibility to perform HDS after FCC or before, which is facilitated by refinery design. HDS must be conducted before reforming because of the poisoning effect of sulfur on the Pt catalyst. In all HDS reactors, sulfur compounds are reduced to H2S, which is recovered into the flue gas using amine scrubbing. HDS is the primary and most frequent desulfurization method used today, while low-molecular weight thiols are removed via caustic washing. The majority HDS operations also remove some metal impurities and, in particular, nitrogen compounds [52].
HDS reactors operated at a pressure of 35–270 bar and a temperature 300–450 °C [53]. Therefore, hydrogenation of olefins under these conditions results in a loss of hydrogen consumption and a loss of octane rating. In mild HDS, the produced H2S reacts with alkene to form linear or branched thiols with 5–12 carbons, which are then recombinant mercaptans. The reaction follows a similar pattern to the addition of water to produce alcohol from alkene. The sulfur can be recovered from recombinant mercaptans, limiting the effectiveness of the HDS process. The balance between fuel quality degradation and degree of sulfur removal provides a push for increased research into HDS catalysts and process designs. The catalysts used in the HDS process are nickel or cobalt promoted molybdenum sulfide. This catalyst was originally characterized by Henrik Topsoe and his co-workers as cobalt promoted molybdenum catalysts, and research is still in progress regarding the development of nickel promoted molybdenum sulfide catalysts for the HDS process [54]. Refinement of products through desulfurization also depends on a number of parameters, such as excess desulfurization equipment, the nature of refinery organization, the quality of used crude oil, and the quality of the products being produced. Desulfurization of organically bound sulfur is carried out to a certain limit through the reductive HDS process. In the conventional process of desulfurization, hydrotreatment is conducted with the simultaneous unidirectional flow of hydrocarbons over a catalyst bed, and hydrogen gas is used to remove sulfur. Although different kinds of hydro-reactor designs appear in practice, they all are based on the same working principle (Figure 3).
The hydrodesulfurization process condition has determined the degree of desulfurization; for example, the inlet mixture is mixed with a co-current of hydrogen and maintained at a pressure of 5–10 MPa and a temperature of 260–430 °C with a supply of cobalt or molybdenum oxides. Catalysts activate hydrogen to react with nitrogen and sulfur to produce ammonia and hydrogen sulfide. After separating the reaction products and cooling them, the oil–feed–gas blend is separated in a stripper column. The H2S produced could be toxic to the catalyst, complicating the desulfurization process [55]. Thus, the effectiveness of HDS management differs because of the broad variety of chemical hydrocarbons. Organic sulfur that appears in the form of thioethers, mercaptans, and disulfides can be simply desulfurized by HDS because of its uncertainty, but this method is not appropriate for polyaromatic sulfur heterocycles (PASHs) present in heavier fractions, a key class of HDS-refractory organic sulfur-rich molecules. Up to 70% of sulfur in fuels is found as refractory molecules such as substituted DBTs and dibenzothiophene (DBT) [56]. Thus, HDS helps eliminate the obstinate molecules at extreme pressures and temperatures that would result in damaging the valuable hydrocarbons of the petroleum product being processed [56]. Because the HDS technique is quite expensive, the development of novel catalysts, innovations, and HDS process configurations is required to achieve an optimal level of desulfurization. Thus, researchers are attempting to move towards biological methods that have several advantages.
4.2. Biodesulfurization
Microbial desulfurization of crude oil came to light in 1935 [57]. Since then, research has been accelerated to determine the significance of microbial biodesulfurization (BDS) processes in the petroleum industry. In the early 1950s, a number of U.S. patents were issued for the application of microbes to carry out the desulfurization of petroleum products to reduce the sulfur contents. Nevertheless, early efforts were ineffective for desulfurization due to its inability to regulate bacteria [16]. Between the 1970s and 1980s, the United States Department of Energy (DOE), in collaboration with other related bodies, attempted to investigate the scope of technology development for desulfurization. Throughout this time, research has been accelerated and scaled up in order to establish desulfurization. Initially, it was reported that bacteria that metabolize hydrocarbons were not of interest [58], but later, bacteria that carried out desulfurization without degrading hydrocarbons were isolated and characterized [59,60]. Many processes have reported that microbes are very specific to their process conditions. Various benefits of BDS over HDS have been described with biological desulfurization, but one of the most interesting was biotransformation at low temperature and pressure. Moreover, it was reported as being rapid, cost effective, easy to handle, and offering a wide range of petroleum products with better safety. This discovery drew the attention of many in the petroleum industry, and they have attempted to adapt to this method for economic purposes. A Texas refinery in the Woodlands reported surprising capital cost savings of 50% and operating cost reductions of 10–20% [61]. The BDS process can also be recorded as eco-friendly. There was a nearly 80% reduction in emissions of greenhouse gases compared to HDS, and this could be because of the operation of BDS at low temperature and pressure. Figure 4 depicts a schematic representation of the biocatalytic desulfurization process by Fungi and Bacteria for Dibenzyl sulfide, a non-thiophenic organosulfur compound in crude oil.
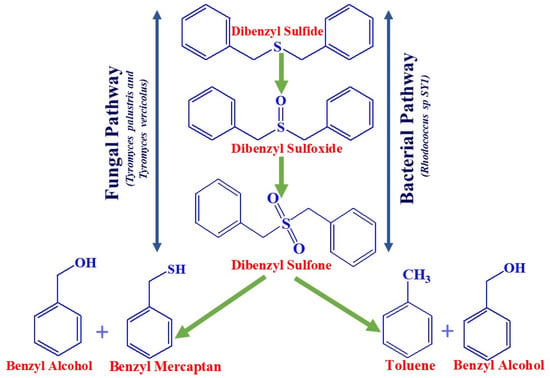
Figure 4.
Pathways for the desulfurization of DBS.
A model has been proposed for the industrial application of BDS. A mixture of petroleum products to be desulfurized, a slurry of biocatalyst, and oxygen are stirred in reactors. Desulfurized petroleum products and biocatalysts are separated as aqueous outlets. Sulfur as sulfate, present in the aqueous phase, can be removed as salts of ammonium sulfate and sodium sulfate. Biocatalysts from water are recovered, and water is recycled back to the reactor. In the last decade, many researchers have explored and accelerated research in the field of biodesulfurization, and much more progress has been made in the establishment of the BDS mechanism [59]. However, significant desulfurization from organic compounds in petroleum products is needed and needs to be explored more. The BDS process has been characterized by microbial stability, selectivity, and activity. Thus, a microbial mixture is recommended for efficient BDS [45,62]. Moreover, due to the evaluation of modern branches of genetic engineering, protein engineering, and metabolic engineering, the efficiency of desulfurization could be increased using multiple copies of the respective gene, inducing gene expression, and increasing gene efficiencies to carry out significant desulfurization [63,64,65]. Kobayashi et al. in 2001 and Folsom et al. in 1999 have described the use of multiple copies of the dsz-genes of Rhodococcus erythropolis for metabolism of sulfur compounds, which increased the activity of BDS by 25% compared to its wild type [66,67]. Recently, surface displays of the dszC gene have been shown in recombinant E. coli that have increased the desulfurization activity threefold in comparison to the control [68]. The genome of Baccilus sp., a thermophilic bacterium, has also been elucidated, which could be utilized for better programming of biosynthetic engineering [69].
5. Microbial Reaction Pathways
The microbial factory is thought to be catalyzing many remediation reactions. In this system, bacteria are found to be the most dominant compared to fungi, algae, yeast, protozoa, and mold. Many bacteria have been found to produce large amounts of enzymes that aid in the metabolization of undesirable molecules such as metals, and their metabolic products have been described as nanoparticles [70,71]. Fungi, algae, yeast, protozoa, and mold were reported as being inefficient to grow due to size limitations and their tolerance of harsh conditions such as high concentrations of NaCl at petroleum sites. However, many bacteria have been reported to grow at high salt concentrations and have been able to produce biosurfactants and polymers necessary for the survival of petroleum reservoirs [72,73].
Dibenzothiophene (DBT) is found as a main component of residual post-HDS sulfur and has been applied as a model compound of BDS. Thus, scientists have accelerated their research, focusing mainly on the mechanism of biotransformation of DBT. Alkyl-derivative of DBT is thought to be more refractory to HDS management (Scheme 1). Most of the crude oils are reported to be rich in DBT, which is responsible for the majority of sulfur compounds [73]. As a result, DBT is well thought out as a model refractory sulfur composed of many heterocyclic molecules in the development of new sulfur removal methods or processes [74]. Thus, microbial desulfurization of model compounds can be thought of as the parallel degradation of other such molecules. Therefore, as a preliminary step, it is concentrated to remove the sulfur from DBT, which could be a major part of the thiophenic sulfur found in most fuels.
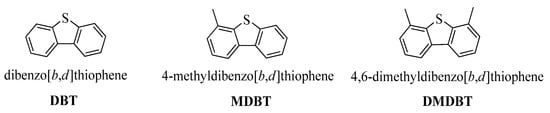
Scheme 1.
Structural formulas of refractory dibenzothiophene (DBT), methyl dibenzothiophene (MDBT), and dimethyl dibenzothiophene (DMDBT).
Researchers have described two pathways for the removal of specific sulfur from petroleum products. In their study, McFarland et al. describe that the Kodama pathway (Scheme 2) is the primary pathway for biodesulfurization of DBT [75]. The major steps in this pathway are ring cleavage and oxidation of one of the cyclic rings of DBT. However, the formation of sulfoxide or sulfone has also been reported in some cases [76]. Moreover, formyl benzothiophene is a dead-end product, and no specific sulfur was released at the end of the reaction. Moreover, it is stated that residue of benzothiophene’s dead-end product has not been favored to release sulfur [75].
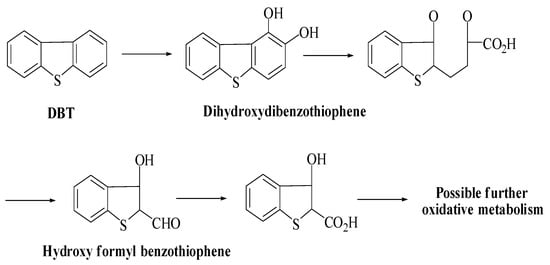
Scheme 2.
Kodama pathway for the degradation of DBT [58].
The sulfur moiety is held by an oxidative and ring-cleaved product, hydroxyl formyl benzothiophene, which is thought to be an inaccurate biodesulfurization mechanism. Thus, in 1990, Kilbane postulated another sulfur-specific pathway, which is popularly known as the 4S pathway [77]. In this pathway, microbe-mediated DBT is facilitated by the selective oxidation of the sulfur moiety of DBT without the C–C bonds breaking, hence maintaining fuel value. Therefore, it has been discovered that microbes mediate sulfur removal from petroleum products [78]. This pathway progresses through sequential and selective oxidation of the sulfur moiety followed by carbon–sulfur bond breaking to form a product, 2-hydroxy biphenyl (HBP), and the c-c bond remains uncleaved [79]. This pathway is described by four specific sequential phases catalyzed by specific enzymes. Therefore, this is also known as 4S pathway (Scheme 3). The sulfur fraction of the fuel is specifically targeted by an enzyme, and the hydrocarbon portion remains unchanged in the 4S pathway. A thermophilic bacterium, Geobacillus thermoglucosidasius W-2, has shown alkanesulfonate monooxygenase systems SsuD1/SsuE1 and SsuD2/SsuE2 for desulfurization of heavy oils with thermal and pH stability, and a higher substrate range. A new pathway system other than the 4S pathway is reported to be found in this thermophilic Geobacillus thermoglucosidasius W-2 bacterium and has potential for industrial application [80].
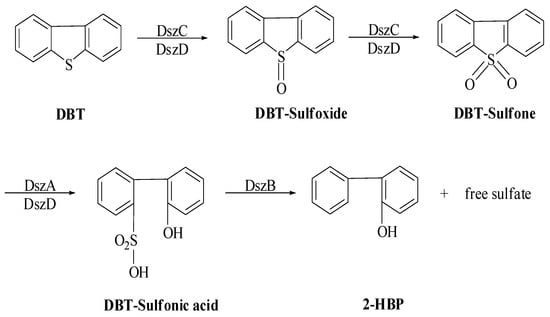
Scheme 3.
Proposed (4S) pathway for DBT desulfurization by R. erythropolis IGTS8 [76].
The specific enzymes DszA, DszB, DszC, and DszD were involved in the sulfur removal of DBT. These reactions can be described mainly in three phases: (i) oxidation of the sulfur moiety and activation of the thiophene ring for cleavage; (ii) aromatic sulfinate production by ring cleavage; and (iii) release of the sulfinate group. The products, HBP and sulfate, were discharged in the cytoplasm and blended. R. erythropolis was thus unable to further metabolize HBT in the presence of DBT, and it accumulated in the medium [75,81].
Van Hamme et al. described that the sulfur removal process in fungus, Dsz enzymes catalyzes and produces 2-hydroxybiphenyl (2-HBP) and sulfate [72]. Desulfurization is carried out by two monooxygenases, which produce the end products. S-oxidation of DBT occurs in two steps, catalyzed by monooxygenase (DszC). The first product, dibenzothiophene-5-oxide (DBTO), formed in step one, is then converted to dibenzothiophene 5, 5-dioxide (DBTO2), which is converted to 2-(2′-hydroxyphenyl) benzene sulfinate (HBPS) by the second monooxygenase. The final step is operated by a sulfinase (DszB) and produces sulfate and 2-HBP as the final products. DszD, which is FMN reductase, helps DszC and DszA in the 4S pathway in oxidation. Brevibacterium sp., on the other hand, uses a different mechanism (Scheme 4) to produce stoichiometric amounts of benzoate and sulfite from DBT. In the ring opening of DBT, DBT sulfones (DBTS) are produced, then aromatic dioxygenase catalyzes the opening of the thiophenic ring and yields 2,3-dihydroxybiphenyl 2′-sulfinate. In the next step, enzyme dioxygenase catalyzes the opening of 2,3-dihydroxybenzene nucleus. Benzoate is converted to CO2 and H2O, with dissolved organic carbon accounting for only 9% of the carbon in DBT [82]. DBT is metabolized by microbes to meet their own nutritional needs for carbon or sulfur. The hydrocarbon is finally converted to CO2 and H2O, which necessarily aggravates the loss of the chemical energy of the crude oils. Although the DBT degradation pathway is of less importance, bacteria using this biotransformation mechanism are potentially beneficial in the development of heterogenous microbial inoculum for the bioremediation process of polyaromatic hydrocarbons. In 2014 and 2015, Ahmed et al. described similar pathway of 4S desulfurization for dibenzyl sulfide and thianthrene degradation found in petroleum products with isolated Gordonia sp. IITR100. This strain desulfurizes dibenzyl sulfide to produce benzoic acid as a byproduct. This bacterium (IITR100) has also shown desulfurization potential for benzonaphthothiophene, as shown by Chauhan et al., 2015. Gordonia sp. IITR100 has a wide substrate range for aliphatic and aromatic organosulfur compounds [83].
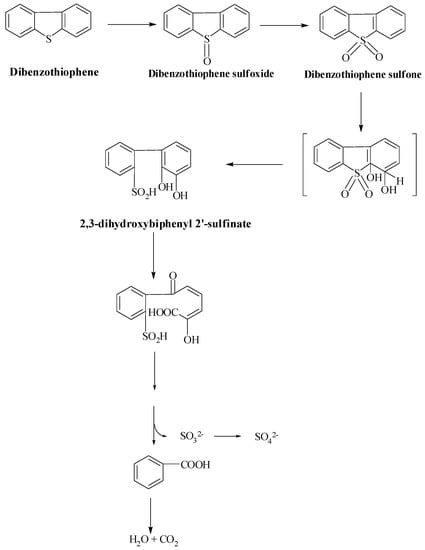
Scheme 4.
Van Afferden Pathway: oxidation of dibenzothiophene-to-dibenzothiophene sulfone [82].
6. 4S Pathway Enzyme Genetics
The primary biocatalyst of the 4S pathway that uses DBT is a monooxygenase (DszC), which completes the oxidation of DBT to dibenzothiophene sulfone (DBTS) in two phases. The product 2-(2′-hydroxyphenyl) benzenesulfonate is produced by the catalytic action of the second enzyme monooxygenase (DszA) on DBTS; lastly, 2-hydroxybiphenyl and sulfite are produced by a lyase (DszB). Moreover, reduced flavin is needed to maintain monooxygenase activity, which is catalyzed by FMN-reductase (DszD) (Scheme 3). More emphasis is now being placed on pathway engineering and metabolic upgradation in order to increase overall activity to meet the demand for industrial-level desulfurization of petroleum products, which is why details of all enzymes are being revealed. The other enzyme is DBT-monooxygenase (DszC), which metabolizes DBT to DBT sulfone using FMNH2 as a co-substrate [84].
This enzyme has been reported to be produced by R. erythropolis D-1 and Paenibacillus sp. strain A11-2 [85]. DBT-sulfone monooxygenase (DszA) has also been reported as an oxidizing enzyme of DBT sulfone to 2-(2′-hydroxyphenyl)benzene sulfinate (HPBS) with the help of FMNH2 as a co-substrate [84]. The reaction rate of monooxygenase (DszA) is 5–10-fold higher than that of DszC [86]. Bacillus subtilis WU-S2B and Paenibacillus sp. strain A11-2 enzymes with higher desulfurizing potential than the DszC enzyme have been reported [85,87]. The 2′-hydroxybiphenyl-2-sulfinate desulfinase enzyme metabolizes 2-(2′-hydroxyphenyl) benzene sulfinate (HPBS) to 2-HBP, and NADH-FMN oxidoreductase has also been reported [88,89].
Various desulfurizing microbes have the dszABC operon, which produces DszA, DszB, and DszC enzymes, respectively. DszD, a product of dszD, helps DszA, DszB, and DszC proteins for specific sulfur removal. The dszABC genes comprise a single operon carried on a transmissible plasmid, with the dszD gene unlinked to the others. There are also cases of chromosomally integrated dsz operons. Certain thermophiles have analogous operons (tds), which encode thermophilic versions of the dsz-encoded proteins. The activities of TdsA, TdsB, and TdsC from Paenibacillus sp. A11-2 differ in comparison to DszA, DszB, and Dsz C from Rhodococcus erythropolis IGTS8, but they have sequence homology. Overlap between dszA and dszB follows the same sequence (ATGA) in thermophilic bacteria, Paenibacillus sp. A11-2 and Rhodococcus erythropolis IGTS8. These tds and related operons are found in a 120 kb plasmid [90]. Other reported 4S pathway-based microbes have almost same type of genetic systems for Biodesulfurization because of the dsz operon carrying plasmid’s broad host range transmissibility. Identical dsz genes are also reported in different thermophilic species as in Bacillus subtilis WU-S2B and Mycobacterium phlei WU-F1 [91,92]. Enzymes from thermophilic bacteria, Paenibacillus sp. A11-2 and Bacillus subtilis WU-S2B can show better activity even at 50 °C, which gives us good hope for industrially suitable applications of biodesulfurization. Crystal structures for TdsC have been elucidated for a better substrate range of organosulfur compounds [93]. A better and different substrate range for organosulfur in microbes is important because not all biodesulfurization microbes have shown a broad substrate range for different organosulfur compounds. So, a better substrate range is desirable for commercial-level biodesulfurization [94]. MdsA, MdsB, and MdsC proteins which are gene products of mdsA, mdsB, and mdsC, respectively, from Mycobacterium sp. G3 are cloned in E. coli JM109, and have shown good potential for alkylated Dibenzothiophenes [95]. Expression of the dsz operon is negatively controlled by inorganic sulfate, methionine, and cysteine, meaning if any of these sulfur containing compounds are present, expression of the 4S pathway will not happen or may be suppressed. Knockouts of the reverse transsulfuration pathway enzyme genes cbs and metB in Rhodococcus sp. strain IGTS8 have eliminated the inorganic sulfur repression [96]. Repression of dsz operon has been removed by recombinant operon [97]. The presence of 2-HBP, which is the end product of the 4S pathway, is reported to decrease the activities of 4S pathway enzymes DszA, DszB, and DszC [98]. Various mechanistic approaches such as metabolic engineering and repression removal in the dsz operon are being employed to understand even more of the molecular aspects of the 4S pathway and feedback inhibition by 2-HBP to achieve commercial-level biodesulfurization [99].
7. Biodesulfurization Application
Biodesulfurization is the process of desulfurizing petroleum products in refineries using microorganisms. This process has not yet been commercialized [100]. However, some factors, including continuous operation, high throughput, mass transfer from liquid–liquid or gas–liquid transport, ability to recover biocatalyst, and emulsion breaking, are significant. Continuous operation and high throughput are important considerations when selecting a reactor for aqueous–organic contact [7]. The intact cells have been used in the biodesulfurization of DBT, and they are not required for Dsz enzyme recovery [101]. The immobilized whole cell can be used to eliminate sulfur from DBT for the cell’s lifetime, which is 600 h longer than it is when using a biocatalyst [102,103]. Many supports, including Sepiolite (Sep), Alumina (Al) and Silica (Si) have been used to immobilize Rodococcus rhodochrous IGTS8 for desulfurizing petroleum products [102,104]. Dinamarca et al. reported that the synergistic action of surfactant with the catalytic system results in enhanced desulfurization of gaseous oil and DBT [102].
Regarding industrial-scale use, many types of reactors have been tested, but usually impeller-based stirred bioreactors are reported to be effective, since they provide better mixing and maintain homogeneity throughout the reactors. They also have a high surface area due to the formation of micrometer-sized droplets and the good connection between the organic and aqueous phases. More energy consumption was reported to achieve smaller droplets, with a five-fold increase in reactor consumption required to achieve droplets of about 5 µm [105]. A bacterial system catalyzes the biodesulfurization process by adding oil to the aqueous phase, which results in an oil-water-bacteria mixture, and batch mode was adapted to obtain oil [106]. Energy BioSystems Corporation (ECB) has developed new technologies for treating diesel, as well as a value-added surfactant byproduct that makes the process cost effective [59,107].
8. Oil Biodesulfurization
Biodesulfurization of various petroleum products by some bacterial strains at optimized conditions is shown in Table 3. The desulfurization rate of diesel oil is much smaller in a hydrophilic system that has only slight similarities to the conditions of a biocatalyst than those obtained for pure DBT [79]. Mid distillates have been reported to remove nearly 30 and 70% of sulfur through biodesulfurization [108], 25–60% for crude oils [75], 20–60% for light gas oils [109,110], and 24–78% for hydro-treated diesel [79,111], respectively.
Grossman et al. discovered that Rhodococcus sp. strain ECRD-1 of middle distillate oil removed 92% of the sulfur (669 ppm to 56 ppm) from DBTs [108]. The time-dependent progressive reduction of sulfur, which occurred after 0, 1, 3, and 6 h for concentrations of 1850, 1620, 1314, and 949 ppm, respectively, has been observed through Rhodococcus erythropolis I-19 involving hydro-desulfurized middle distillate oil. The reduction in total sulfur at the time intervals after 1 h, between 1 and 3 h, and at 3 h was 230 ppm, 300 ppm, and 365 ppm, respectively. Between 3 and 6 h, desulfurization changed to the higher-boiling-range sulfur compounds, resulting in an additional 365 ppm drops in total sulfur. Thus, a sulfur removal range of 1850 to 615 ppm was described, with residual sulfur estimated to be 75% thiophenes and 2% sulfoxides, 11% sulfides, and 12% sulfones [67].
Many petroleum compounds have also been reported to be desulfurized by thermophilic bacteria. Light gas oil is reported to contain cover trace amounts of sulfur and a trace amount of heterocyclic compounds, the majority of which are alkylated derivatives of DBT [49]. Paenibacillus sp. strains A11-1 and A11-2 were reported to grow at higher temperatures, catalyze the biotransformation of sulfur present in light gas oil at 800 ppm at high temperatures, and efficiently reduce the sulfur [112]. Recently, interactive cultures of two Paenibacillus species have shown moderately thermophilic biodesulfurization [113]. Paenibacillus validus strain PD2 has shown good potential in aqueous and biphasic (model oil) systems, which is a good indicator of its applicability at the industrial level [114].
However, R. rhodochrous is reported as more significant (high desulfurization) compared to Paenibacillus sp. [115,116]. As such, thermophiles are not required to cool down oil fractions after the HDS, which offers this process at a compatible cost [117]. New approaches to further enhance the desulfurization potential of thermophilic isolates can be explored, e.g., the use of nanomaterials with immobilized thermophilic strains; the activity–stability tradeoff can be avoided by genetic and chemical modifications [118,119,120,121]. Moreover, they also reduced the chances of contamination by other bacteria, which may adversely affect the BDS process [122]. Although the obtained sulfur removal activity is significant, it is not at the desired level [108,122]. So, more such smart efforts and approaches are needed.

Table 3.
Biodesulfurization of petroleum products by bacterial strains at optimized conditions.
Table 3.
Biodesulfurization of petroleum products by bacterial strains at optimized conditions.
| S. No. | Desulfurizing Bacteria | Model Petroleum/Oil Fraction | Sulfur Rmoval (%) | References |
|---|---|---|---|---|
| 1. | Gordonia alkanivorans RIPI90A | Hexadecane | 90 | [123] |
| 2. | Mycobacterium phlei WU-0103 (growing cells) | 12-fold diluted straight run light gas oil (1000 ppm) | 52 | [110] |
| 3. | Pseudomonas stutzeri UP-1 | n-Hexadecane | 74 | [124] |
| 4. | Mycobacterium sp. X7B | Diesel oil | 86 | [46] |
| 5. | Gordonia sp. SYKS1 (resting cells) | Diesel fuel (250 ppm) | 76 | [125] |
| 6. | Gordonia alkanivorans 1B | n-heptane | 63 | [126] |
| 7. | Pseudomonas delafieldii R-8 (resting cells) | HDS-treated diesel oil (591 ppm) | 90.5 | [127] |
| 8. | Pseudomonas delafieldii R-8 (growing cells) | Diesel fuel | 47 | [128] |
| 9. | Mycobacterium goodie X7B | Liaoning crude oil | 59 | [129] |
| 10. | Gordonia sp. P32C1 (resting cells) | Light diesel fuel | 48.5 | [130] |
| 11. | Rhodococcus sp. and Athrobacter sulfurcus | Diesel oil | 50 | [131] |
| 12. | Gordonia sp. CYKS1 | Middle distillate unit feed | 70 | [79] |
| 13. | Caldariomyces fumago | Diesel fuel | 99 | [132] |
| 14. | Rhodococcus sp. ECRD-1 (growing cells) | Middle-distillate (diesel range) | 8.1 | [108] |
| 15. | Rhodococcus erythropolis XP | Diesel oil | 94.5 | [133] |
| 16. | Rhodococcus sp. ECRD-1 (growing cells) | Medium fraction of light gasoil | 92 | [134] |
| 17. | Sphingomonas subarctica T7b | Light gas oil | 94 | [135] |
| 18. | Bacillus subtilis WU-S2B | n-tridecane | 50 | [122] |
| 19. | Gordonia sp. CYKS1 (resting cells) | Light gasoil | 50 | [79] |
| 20. | Mycobacterium sp. ZD-19 | Hexadecane | 100 | [136] |
| 21. | Rhodococcus erythropolis ATCC 53968 | Decane | 90 | [137] |
| 22. | Rhodococcus sp. SA11 | Hexane soluble fraction (heavy crude oil) | 18 | [138] |
| 23. | Rhodococcus erythropolis I-19 (resting cells) | Medium distillate (1850 ppm) | 67 | [67] |
| 24. | Gordonia sp. SYKS1 (resting cells) | Light gasoil (3000 ppm) Middle distillate unit feed (1500 ppm) | 35 60 | [139] |
9. Bacteria Desulfurizing Sulfidic Compounds
Heavy crude oils are distinguished by their high viscosity, high molecular weight, and the presence of asphaltene. Asphaltenes are polycyclic aromatic compounds that are linked together by aliphatic chains of varying lengths to form aliphatic bridges. These bridges are distinguished by the inclusion of esters, sulfides, and ethers as connecting components [140,141]. It has been demonstrated that using a low-molecular weight asphaltene fraction reduces crude oil viscosity. Viscosity reduction is a difficult task because it is associated with various problems such as thermal visbreaking, creation of unstable oil, and the avoidance of hydrogen sulfide liberation, which might be minimized by employing a biocatalyst. The desulfurization of aromatic compounds such as dibenzothiophene and alkyl-substituted dibenzothiophenes is also a major challenge for petroleum products. Many bacteria can catalyze the 4S route desulfurization of dibenzothiophene without changing the carbon skeleton to 2-hydroxybiphenyl [79,142]. Although the molecular weight remains unchanged because of this action, it cannot contribute significantly to the change in viscosity. Furthermore, a biocatalyst would disrupt the carbon–sulfur bond but not the dibenzothiophene bond, so in addition to the biodesulfurization of petroleum products, viscosity reduction is a key concern that can be achieved by breaking carbon–sulfur linkages to depolymerize asphaltene molecules. The carbon–sulfur breakdown of high-molecular-weight aliphatic sulfides has received less attention. Low-molecular-weight molecules, such as alkyl sulfides or their derivatives, such as methyl, ethyl, and butyl sulfides [143,144], 2-chloroethyl ethyl sulfide [145], and thiodiglycol [146], are cut at the sulfur atom. Jenisch-Anton et al. (2000) reported microbial bioremediation of phytanyl octadecyl sulfide’s n-alkyl chain. Not only bacteria catalyze desulfurization; some fungi have also been discovered to catalyze sulfur oxidation of dibenzosulfide but are unable to rupture the carbon–sulfur bond [147]. R. erythropolis IGTS8 uses di-n-octyl sulfide, which is not a common substrate for the wild-type organism [65]. To fulfill the sulfur requirement, Rhodococcus sp. JVH1 catalyzes the fluorinated chemical bis-(3-pentafluorophenylpropyl) sulfide. Van Hamme et al. (2004) used the fluorinated chemical bis-(3-pentafluorophenylpropyl) sulfide to identify Rhodococcus sp. JVH1. While JVH1 does not use dibenzothiophene as a sulfur source, bacteria use it as a sulfur source via a method comparable to the 4S route [148]. The desulfurization of sulfur, which is found in alkyl chains such as thiophenes and DBTs, has received less attention. The bridge structure is said to be necessary for high-molecular-weight asphaltene [149]. Furthermore, efficiently metabolizing thiophenes and DBTs results in a significant reduction in crude oil viscosity. 1-Chloroethyl sulfide is metabolized by IGTS8 [145], dibenzyl sulfide (DBS) by Gordonia CYKS1 [79], and fungi mediated DBS metabolism to benzyl alcohol and benzyl mercaptans by T. versicolor FO 30340 and Tyromyces palustris IFO 303 [150]. Gordonia sp. IITR100 metabolizes dibenzyl sulfide to benzoic acid, which is the preferred sulfur source over DBT [151]. This bacterium has also demonstrated an optimum desulfurization rate from crude oil and diesel oil of up to 76% and 98%, respectively [152].
10. Bacteria Desulfurizing Non-Thiophenic Organosulfur Compounds
Thianthrene (TA) and 1,4-dithiane are non-thiophenic sulfur compounds, and desulfurization of these compounds is not well explored [153,154]. Some bacterial strains, such as Pseudomonas sp. K1OA, and Rhodococcus sp. such as Rhodococcus erythropolis EPWF and K1OA, have been shown to grow under restrictive conditions on such sulfur chemicals, although their metabolism is limited. DBT reduced indicates sulfur has been removed by the bacterium Rhodococcus sp. IGTS8 was reported to be up to 90% [153]. Similarly, when TA is metabolized by the bacterium Rhodococcus sp. IGTS8 for sulfur as a nutrient source, thianthrene inhibits cell growth. Furthermore, no information on the generated metabolite was provided under these conditions [155]. Schreiner et al. reported in another study that Phanerochaete chrysosporium generates ligninase, which catalyzes the conversion of TA to TA-monosulfoxide [156]. Gordonia sp. IITR100 has the ability to desulfurize thianthrene into o-hydroxyphenyl phenyl sulfone [151].
11. Improvements Made to the Biodesulfurization Process
11.1. Search for Novel Strains
Numerous microorganisms that can break down and remove sulfur from organosulfur compounds have been isolated, studied, and developed for better desulfurization [157]. However, the selectivity and reaction rate catalyzed by the microbial enzymatic system determine the industrial use of the isolated strain [158]. Up to this point, the majority of isolated and identified bacteria catalyzed the biodesulfurization process at a slower rate, which needs to be enhanced by up to 500 times to make the procedure or technology effective [59]. Bacteria isolated from an oil habitat more effectively break down sulfur compounds, determining the quantity, yield, and quality of petroleum [159]. Numerous efficient isolates for better desulfurization have been found, and they are now garnering attention for prospective use in desulfurization reactions. DBT has been described as a traditional recalcitrant sulfur-containing molecule for the petroleum sector [59]. Isbister and Koblynski in 1985 isolated and discovered a Pseudomonas species that catalyzes the desulfurization of DBT, but sadly, before the reaction pathway could be fully defined, this Pseudomonas species was lost [160]. Kilbane isolated the Rhodococcus erythropolis IGTS8 bacterium through an ongoing study over a protracted period of time [76]. Since then, numerous bacterial strains have been found to catalyze the 4S pathway’s breakdown of DBT. Researchers are always looking for new microorganisms that are even more promising than the ones already discovered. This area has always been a thrust area for biodesulfurization in nature. There may be yet undiscovered microbes that can meet the demand for a 500× increase in activity.
11.2. Bacteria Characterizing for the Desulfurization of a Model Thiophene Organosulfur Compounds (Dibenzothiophene and Benzothiophene)
Chemical desulfurization of thiophene, benzothiophene (BT), benzonaphthothiophene, and alkyl derivatives containing heterocyclic organic molecules is expected to be complicated, but bacteria efficiently catalyze sulfur-containing compounds. Desulfurization of such compounds by microbial factories may thus be an alternative option [161].
Many microorganisms have been identified that are capable of desulfurizing petroleum-based products. The role of Rhodococcus sp. IGTS8 [162], N1-36 [163], and Rhodococcus erythropolis in the generation of sulfite ions and 2-hydroxybiphenyl has been investigated. Furthermore, microorganisms have been observed to be vulnerable to benzothiophene. Kropp et al. discovered that Pseudomonas sp. BT1 can convert 3-methyl benzothiophene to sulfoxide and sulfone, as well as benzothiophene to benzothiophene-2, 2-dione [164]. The UV4 enzyme in Pseudomonas putida catalyzes the conversion of benzothiophene to three dihydrodiols: cis- and trans-2,3-dihydro benzothiophene, and cis-4,5-dihydro benzothiophene. Boyd et al. used Gordonia sp. 213E to desulfurize benzothiophene [89]. Microbial growth for sulfur requirements on mineral salt medium containing dibenzothiophene could provide hope for the isolation of benzothiophene-metabolizing microorganisms. Gordonia sp. 213E (NCIMB 40816) was isolated and shown to be unable to grow in salt medium containing dibenzothiophene as the only sulfur source [165], whereas thermophilic Paenibacillus sp. A11-2 can desulfurize dibenzothiophene [117]. The selective desulfurization of DBT by G. alkanivorans strain 1B resulted in the formation of 2-hydroxybiphenyl. G. alkanivorans 1B used an equal ratio of DBT and BT as a sulfur source in a sequential manner. However, BT has been described as the predominant source of sulfur, and DBT is used if very little BT is present [3]. However, some bacteria, such as Rhodococcus sp. KT462, have been observed to use both DBT and BT as major sulfur sources. The BT desulfurization route is shared by Rhodococcus sp. KT462, Sinorhizobium sp. KT55, and Paenibacillus sp. A11-2. Furthermore, the strain KT462 was discovered to produce the same product, 2-hydroxybiphenyl, as R. erythropolis IGTS8 and R. erythropolis KA2-5-1. R. erythropolis KA2-5-1 has been found to degrade alkyl compounds of BT and DBT [166]. Some Gram-negative isolates, including Sinorhizobium sp. KT55, and Gram-positive strains such as Gordonia rubropertinctus T08, utilize the benzothiophene desulfurization pathway for benzothiophene to complete sulfur requirements [167]. Some bacteria, such as Rhodococcus sp. SA31 resting and growing cells, have also been reported to catalyze the desulfurization of alkyl derivatives BT and DBT containing petroleum hydrocarbon [138]. Gordonia rubropertinctus T08 catalyzes the desulfurization of benzothiophene to produce benzothiophene sulfone, benzothiophene sulfoxide, benzo[e]-[1,2] oxathiin S-oxide (BT-sultine), 2-coumaranone, but not 2-(2′-hydroxyphenyl) ethan-1-al, o-hydroxystyrene, and benzo [1,2] oxathiin S-dioxide (BT-sultone) [168].
Naphtho [2,1-b] thiophene (NTH) or the structural isomer of dibenzothiophene or NTH derivatives are also components of diesel oil and have undergone desulfurization and hydrodesulfurization processes. The growth of some bacteria has been observed with or without BT or DBT on NTH, including a bacterium such as Rhodococcus sp. WU-K2R metabolizes NTH as a primary source to accomplish the nutritional requirements of sulfur. It grew on benzothiophene (BT), NTH sulfone, 5-methyl-BT, or 3-methyl-BT but did not utilize NTH or BT as the primary source of carbon. The identified metabolites of this strain were NTH sulfone, naphtha [2, 1-b] furan, and 2′-hydroxynaphthylethene. Similarly, benzo[c][1,2] oxathiin S-oxide, BT sulfone, benzo[c][1,2] oxathiin S, S-dioxide, o-hydroxystyrene, benzofuran, and 2-(2′-hydroxyphenyl) ethan-1-al were observed BT metabolites. Metabolomics analysis of the WU-K2R strain concluded that the organism catalyzes sulfur-specific degradation pathways with the sulfur-selective cleavage of the carbon–sulfur bond [169]. Four Rhodococcus spp. displayed the ability to use 4,4′-dithiodibutyric acid (DTDB) as a solitary carbon source for growth. Nox is responsible for the initial cleavage of DTDB into two molecules of 4-mercaptobutyric acid [170].
11.3. Improvement of Biodesulfurization by Genetic Engineering
Techniques for genetic engineering have been applied to many microbial systems, including Mycobacterium goodie X7B [171], Gordonia sp. IITR100 [172], Rhodococcus sp. IGTS8 [173], Rhodococcus erythropolis MC1109, Rhodococcus erythropolis KA2-5-1, Rhodococcus erythropolis DS-3, Bacillus subtilis, and many more. Mycobacterium sp. G3 in the phsp60 promoter lead to an increase in activity of 1.2 times [174]. Rhodococcus qingshengii were transformed with the kstD promoter, and the resulting strains CW25[pRESX-dszABC] and CW25[pRESX-dszAS1BC]) were exposed to adaptive selection. It was found that the resultant strains were stable and 13 times more active than the natural strains [175]. The recombinant strains have been tested for the desulfurization of DBT and BT, which are used as a primary source of sulfur by the microbes [168]. It has been observed that the acceptance of sulfur compounds from petroleum product become hard to uptake for metabolism. In this regard, improvements have been made through gene transfer technology, in which dsz genes from Rhodococcus erythropolis KA2-5-1 have been transferred into Rhodococcus erythropolis MC1109, which doubled the uptake rate of sulfur compared to the previous strain [176]. In the membrane, topoisomerase mutants resulted in an improvement in the uptake of petroleum compounds for enhanced desulfurization [176]. Nearly nine times improved activity has been reported with the design of a plasmid with the dszB gene, in which 16 nucleotides at the 5′untranslated region were removed [177]. When the phsp60 promoter was used, there was a 1.2-fold increase in desulfurization activity with the Mycobacterium sp. G3 gene [174]. It has also been observed that not only gene integration, but enhanced expression is utilized for better desulfurization. Altering desulfurization gene orders from dszABC to dszBCA is also responsible for enhanced desulfurization, and it is 12 times higher than that of the native form [178]. Pseudomonas putida DS23 was grown in organic solvent-tolerant conditions with expression vectors, and it degraded 56% of 0.5 mM DBT in 12 h in a biphasic reaction containing 33.3% (v/v) n-hexane, whereas induction by isopropyl -D-1-thiogalactopyranoside degraded only 26% [179]. A model bacterial “chassis”, Pseudomonas putida KT2440, has been tested with synthetic Dsz cassettes for metabolic studies of the desulfurization process. The complete dszB1A1C1-D1 cassette behaved as an attractive alternative for the conversion of DBT into 2HBP [99]. Recombinant K. oxytoca ISA4, which has 3.8 kb of dszABC genes, has the maximum desulfurization ability (48%) compared to R. erythropolis IGTS8 (42%), and Pseudomonas aeruginosa pTSOX4 (46%) [180]. An engineered strain of Rhodococcus opacus with sulpeptide genes, combined with DBT-induced selective pressure for rapid growth, produced 20-fold more activity than the original dszABC-only strain [181]. Several in silico experiments and flux balance analyses have been proposed to use minimal media, determine gene and reaction essentiality, and compare the efficacy of carbon, nitrogen, and sulfur sources on the desulfurization activity of wild and mutant strains of Rhodococcus sp. and E. coli [182]. While the genome sequence drafts of the thermophilic bacteria Aeribacillus pallidus W-12 [80] and Gordonia sp. IITR100 [152] have been announced, it may be helpful to design a new synthetic pathway for further improving the catalytic activity of desulfurization. Metabolic engineering can also be achieved by transferring the characteristics of one host to another in biodesulfurization [183,184]. To achieve commercial biodesulfurization, it is critical to apply all of the information and techniques obtained at the metabolic and genetic levels to improve process level engineering, such as biocatalyst separation, increasing mass transfer rates, scaling up biphasic systems, kinetic modeling, and so on, in addition to the development of bioreactors [185]. A bioreactor with immobilized bacteria can be designed and used to commercialize the BDS into HDS in the petroleum refinery [186]. Overcoming product inhibition caused by 2-hydroxybiphenyl (2-HBP) is a very crucial and important concern to make it feasible for industrial application and to create more robust biocatalysts [187,188]. Therefore, Murarka et al. explored the molecular biology of 4S pathways. Murarka et al. discovered that TetR family proteins bind to the promoter region of dsz operons [96,189,190]. To activate the dszABC operon for biodesulfurization, TetR must bend [191]. This finding might be helpful when carrying out bioengineering on the 4S pathway, as the whole genome sequences of Gordonia sp. IITR100 have been revealed.
12. Conclusions
Sulfur contents (organosulfur compounds) vary in concentrations in various crude oil wells in different countries because they define the quality of crude oil; more sulfur content means poor-quality crude oil, and less sulfur or no sulfur means better-quality crude oil. These sulfur compounds, in the form of organosulfur compounds, are problematic in crude oil, gasoline, and diesel. These sulfur compounds are removed by hydrodesulfurization (HDS) during the refining of crude oils. Some of the recalcitrant organosulfur compounds (DBT, DMDBT, BT, etc.) still remain there even after treatment. Interestingly, some good microbes have been described in nature in soil; in this review, we discussed them. These sulfur compounds can desulfurize via several known pathways found in microbes, such as the Kodama pathway, the reductive pathway, and the 4S pathway. The 4S pathway is a desirable pathway for microbial desulfurization because, in this pathway, compounds’ fuel value remains conserved. However, reported activity for the different microbes needs to increase even more (up to 100 times than its current activity level). Various approaches have been implemented to increase the activity of the 4S pathway. To improve the desulfurizing activity of reported microbes, numerous strategies and approaches have been devised and are being developed. Among them are the biocatalytic activity of microbes at the oil/water interface, the mass transfer rate, and the stability of the biological system, which are different parameters that are very useful in increasing the activity of the process. The search for new bacterial strains that can act on a wide range of substrates at high temperatures is being explored for the commercial bioprocessing of microbially mediated desulfurization for the development of sustainable biotechnological process technologies. Through the use of tools and techniques of genetic engineering, synthetic biology is helping to solve issues in the 4S pathway by enhancing the activity of 4S pathway-related enzymes. Feedback inhibition due to an end-product of the 4S pathway, 2-HBP, is explored. Some proteins of the TetR family have been identified as playing a role in decreasing the activity of Dsz enzymes. Therefore, more accelerated research must be conducted in the field of biodesulfurization, taking into account Dsz enzymes and various novel microbes that have a broad substrate range for sulfur compounds in petroleum products. A lab-scale biotrickling filter is being developed to optimize the use of H2S in biodesulfurization. To achieve commercial-scale biodesulfurization, all information, tools, and techniques obtained at the metabolic and genetic levels must be applied to improve process-level engineering, such as biocatalyst separation, increasing mass transfer rates, scaling up biphasic systems, kinetic modeling, use of nanoparticles, and so on, in addition to the development of large-scale bioreactors. As a result, it is concluded that these findings, along with the multidimensional approaches reported and suggested, could be useful for the development of cost-effective petroleum product desulfurization using microbes as factories. Furthermore, the reported bacterial strain may be useful in the remediation of petroleum-contaminated sites. All these explorations and industrial scale-based discoveries for biodesulfurization may open a new door to making sulfur desulfurization an economical and environmentally friendly.
Author Contributions
A.A.: original draft, writing and review, M.A.Z.: review and editing, V.A.: review and editing, S.A.-T.: review and editing revised draft, M.S.A.: review and editing, M.J.K.: original draft, writing and editing draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this article.
Acknowledgments
This work was sponsored by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, Saudi Arabia, under grant no. (KEP-34-130-41). The authors, therefore, acknowledge and thank DSR for its financial and technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Soleimani, M.; Bassi, A.; Margaritis, A. Biodesulfurization of refractory organic sulfur compounds in fossil fuels. Biotechnol. Adv. 2007, 25, 570–596. [Google Scholar] [CrossRef] [PubMed]
- Dickson, U.J.; Udoessien, E.I. Physicochemical Studies of Nigeria’s Crude Oil Blends. Pet. Coal 2012, 54, 243–251. [Google Scholar]
- Alves, L.; Salgueiro, R.; Rodrigues, C.; Mesquita, E.; Matos, J.; Gírio, F.M. Desulfurization of dibenzothiophene, benzothiophene, and other thiophene analogs by a newly isolated bacterium, Gordonia alkanivorans strain 1B. Appl. Biochem. Biotechnol. 2005, 120, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.; Ni, W.; Ma, L.; Liu, P.; Li, Z. The Use of Energy in Malaysia: Tracing Energy Flows from Primary Source to End Use. Energies 2015, 8, 2828–2866. [Google Scholar] [CrossRef]
- French, K.L.; Birdwell, J.E.; Lewan, M.D. Trends in thermal maturity indicators for the organic sulfur-rich Eagle Ford Shale. Mar. Pet. Geol. 2020, 118, 104459. [Google Scholar] [CrossRef]
- Kropp, K.G.; Fedorak, P.M. A review of the occurrence, toxicity, and biodegradation of condensed thiophenes found in petroleum. Can. J. Microbiol. 1998, 44, 605–622. [Google Scholar] [CrossRef]
- Boniek, D.; Figueiredo, D.; dos Santos, A.F.B.; de Resende Stoianoff, M.A. Biodesulfurization: A mini review about the immediate search for the future technology. Clean Technol. Environ. Policy 2015, 17, 29–37. [Google Scholar] [CrossRef]
- Kropp, K.G.; Andersson, J.T.; Fedorak, P.M. Bacterial transformations of 1,2,3,4-tetrahydrodibenzothiophene and dibenzothiophene. Appl. Env. Microbiol. 1997, 63, 3032–3042. [Google Scholar] [CrossRef]
- Sousa, S.F.; Sousa, J.F.M.; Barbosa, A.C.C.; Ferreira, C.E.; Neves, R.P.P.; Ribeiro, A.J.M.; Fernandes, P.A.; Ramos, M.J. Improving the Biodesulfurization of Crude Oil and Derivatives: A QM/MM Investigation of the Catalytic Mechanism of NADH-FMN Oxidoreductase (DszD). J. Phys. Chem. A 2016, 120, 5300–5306. [Google Scholar] [CrossRef]
- Zherebker, A.; Kostyukevich, Y.; Volkov, D.S.; Chumakov, R.G.; Friederici, L.; Rüger, C.P.; Kononikhin, A.; Kharybin, O.; Korochantsev, A.; Zimmermann, R. Speciation of organosulfur compounds in carbonaceous chondrites. Sci. Rep. 2021, 11, 7410. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, Y.; Wu, J.; Wang, Y.; Li, J.; Shi, Q.; Xu, C.; Hsu, C.S. Comprehensive Composition, Structure, and Size Characterization for Thiophene Compounds in Petroleum Using Ultrahigh-Resolution Mass Spectrometry and Trapped Ion Mobility Spectrometry. Anal. Chem. 2021, 93, 5089–5097. [Google Scholar] [CrossRef]
- Meshoulam, A.; Said-Ahmad, W.; Turich, C.; Luu, N.; Jacksier, T.; Shurki, A.; Amrani, A. Experimental and theoretical study on the formation of volatile sulfur compounds under gas reservoir conditions. Org. Geochem. 2021, 152, 104175. [Google Scholar] [CrossRef]
- Ndagijimana, P.; Liu, X.; Li, Z.; Xing, Z.; Pan, B.; Yu, G.; Wang, Y. Adsorption performance and mechanisms of mercaptans removal from gasoline oil using core-shell AC-based adsorbents. Environ. Sci. Pollut. Res. 2021, 28, 67120–67136. [Google Scholar] [CrossRef]
- Payzant, J.; Montgomery, D.; Strausz, O. Sulfides in petroleum. Org. Geochem. 1986, 9, 357–369. [Google Scholar] [CrossRef]
- Monticello, D.J. Biodesulfurization and the upgrading of petroleum distillates. Curr. Opin. Biotechnol. 2000, 11, 540–546. [Google Scholar] [CrossRef]
- Monticello, D.J. Riding the fossil fuel biodesulfurization wave. Chemtech 1998, 28, 38–45. [Google Scholar]
- Manisalidis, I.; Stavropoulou, E.; Stavropoulos, A.; Bezirtzoglou, E. Environmental and health impacts of air pollution: A review. Front. Public Health 2020, 8, 14. [Google Scholar] [CrossRef]
- Almetwally, A.A.; Bin-Jumah, M.; Allam, A.A. Ambient air pollution and its influence on human health and welfare: An overview. Environ. Sci. Pollut. Res. 2020, 27, 24815–24830. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Santos, J.M.; Mill, J.G.; Reis Júnior, N.C.; Andreão, W.L.; de Albuquerque, T.T.A.; Stuetz, R.M. Mortality risks due to long-term ambient sulphur dioxide exposure: Large variability of relative risk in the literature. Environ. Sci. Pollut. Res. 2020, 27, 35908–35917. [Google Scholar] [CrossRef]
- Barnes, P.W.; Robson, T.M.; Neale, P.J.; Williamson, C.E.; Zepp, R.G.; Madronich, S.; Wilson, S.R.; Andrady, A.L.; Heikkilä, A.M.; Bernhard, G.H.; et al. Environmental effects of stratospheric ozone depletion, UV radiation, and interactions with climate change: UNEP Environmental Effects Assessment Panel, Update 2021. Photochem. Photobiol. Sci. 2022, 21, 275–301. [Google Scholar] [CrossRef]
- Schmidt, M.A.X.; Siebert, W. 23—SULPHUR. In The Chemistry of Sulphur, Selenium, Tellurium and Polonium; Schmidt, M., Siebert, W., Bagnall, K.W., Eds.; Pergamon: Oxford, UK, 1973; pp. 795–933. [Google Scholar]
- Liu, W.; Liu, X.; Gu, Y.; Liu, Y.; Yu, Z.; Lyu, Y.; Tian, Y. A new composite consisting of Y zeolite and ZrO2 for fluid catalytic cracking reaction. Compos. Part B Eng. 2020, 200, 108317. [Google Scholar] [CrossRef]
- Shah, N.K.; Li, Z.; Ierapetritou, M.G. Petroleum Refining Operations: Key Issues, Advances, and Opportunities. Ind. Eng. Chem. Res. 2011, 50, 1161–1170. [Google Scholar] [CrossRef]
- Lv, D.; Lu, S.; Tan, X.; Shao, M.; Xie, S.; Wang, L. Source profiles, emission factors and associated contributions to secondary pollution of volatile organic compounds (VOCs) emitted from a local petroleum refinery in Shandong. Environ. Pollut. 2021, 274, 116589. [Google Scholar] [CrossRef]
- Usman, A.; Aitani, A.; Al-Khattaf, S. Catalytic Cracking of Light Crude Oil: Effect of Feed Mixing with Liquid Hydrocarbon Fractions. Energy Fuels 2017, 31, 12677–12684. [Google Scholar] [CrossRef]
- Jones, D.S.; Pujadó, P.P. Handbook of Petroleum Processing; Springer Nature: Dordrecht, The Netherlands, 2006; Volume 1353. [Google Scholar] [CrossRef]
- Stratiev, D.; Shishkova, I.; Ivanov, M.; Dinkov, R.; Georgiev, B.; Argirov, G.; Atanassova, V.; Vassilev, P.; Atanassov, K.; Yordanov, D.; et al. Role of Catalyst in Optimizing Fluid Catalytic Cracking Performance During Cracking of H-Oil-Derived Gas Oils. ACS Omega 2021, 6, 7626–7637. [Google Scholar] [CrossRef]
- Galadima, A.; Muraza, O. Role of zeolite catalysts for benzene removal from gasoline via alkylation: A review. Microporous Mesoporous Mater. 2015, 213, 169–180. [Google Scholar] [CrossRef]
- Alfke, G.; Irion, W.W.; Neuwirth, O.S. Oil Refining. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH: Weinheim, Germany, 2007; Volume 25, p. 261. [Google Scholar]
- Saha, B.; Vedachalam, S.; Dalai, A.K. Review on recent advances in adsorptive desulfurization. Fuel Process. Technol. 2021, 214, 106685. [Google Scholar] [CrossRef]
- Bordoloi, N.K.; Bhagowati, P.; Chaudhuri, M.K.; Mukherjee, A.K. Proteomics and metabolomics analyses to elucidate the desulfurization pathway of Chelatococcus sp. PLoS ONE 2016, 11, e0153547. [Google Scholar] [CrossRef]
- Kaiser, D.; Klose, I.; Oost, R.; Neuhaus, J.; Maulide, N. Bond-Forming and -Breaking Reactions at Sulfur(IV): Sulfoxides, Sulfonium Salts, Sulfur Ylides, and Sulfinate Salts. Chem. Rev. 2019, 119, 8701–8780. [Google Scholar] [CrossRef]
- Stanislaus, A.; Marafi, A.; Rana, M.S. Recent advances in the science and technology of ultra low sulfur diesel (ULSD) production. Catal. Today 2010, 153, 1–68. [Google Scholar] [CrossRef]
- Qiu, L.; Cheng, Y.; Yang, C.; Zeng, G.; Long, Z.; Wei, S.; Zhao, K.; Luo, L. Oxidative desulfurization of dibenzothiophene using a catalyst of molybdenum supported on modified medicinal stone. RSC Adv. 2016, 6, 17036–17045. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, K.; Cheng, Y.; Zeng, G.; Zhang, M.; Shao, J.; Lu, L. Catalytic oxidative desulfurization of BT and DBT from n-octane using cyclohexanone peroxide and catalyst of molybdenum supported on 4A molecular sieve. Sep. Purif. Technol. 2016, 163, 153–161. [Google Scholar] [CrossRef]
- Liu, W.; Liao, Y.; Pan, Y.; Jiang, B.; Zeng, Q.; Shi, Q.; Hsu, C.S. Use of ESI FT–ICR MS to investigate molecular transformation in simulated aerobic biodegradation of a sulfur-rich crude oil. Org. Geochem. 2018, 123, 17–26. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, Y.; Liu, H.; Yang, C.; Qiu, L.; Zeng, G.; He, H. Extractive desulfurization of dibenzothiophene by a mixed extractant of N,N-dimethylacetamide, N,N-dimethylformamide and tetramethylene sulfone: Optimization by Box–Behnken design. RSC Adv. 2015, 5, 66013–66023. [Google Scholar] [CrossRef]
- Fukunaga, T.; Katsuno, H.; Matsumoto, H.; Takahashi, O.; Akai, Y. Development of kerosene fuel processing system for PEFC. Catal. Today 2003, 84, 197–200. [Google Scholar] [CrossRef]
- Chen, J.; Li, B.; Zheng, J.; Chen, J. Control of H2S generation in simultaneous removal of NO and SO2 by rotating drum biofilter coupled with FeII (EDTA). Environ. Technol. 2019, 40, 1576–1584. [Google Scholar] [CrossRef]
- Thompson, D.; Cognat, V.; Goodfellow, M.; Koechler, S.; Heintz, D.; Carapito, C.; Van Dorsselaer, A.; Mahmoud, H.; Sangal, V.; Ismail, W. Phylogenomic classification and biosynthetic potential of the fossil fuel-biodesulfurizing Rhodococcus strain IGTS8. Front. Microbiol. 2020, 11, 1417. [Google Scholar] [CrossRef]
- Jeevanandam, P.; Klabunde, K.; Tetzler, S. Adsorption of thiophenes out of hydrocarbons using metal impregnated nanocrystalline aluminum oxide. Microporous Mesoporous Mater. 2005, 79, 101–110. [Google Scholar] [CrossRef]
- Peh, S.; Mu, T.; Zhong, W.; Yang, M.; Chen, Z.; Yang, G.; Zhao, X.; Sharshar, M.M.; Samak, N.A.; Xing, J. Enhanced Biodesulfurization with a Microbubble Strategy in an Airlift Bioreactor with Haloalkaliphilic Bacterium Thioalkalivibrio versutus D306. ACS Omega 2022, 7, 15518–15528. [Google Scholar] [CrossRef]
- Philip, L.; Deshusses, M.A. Sulfur Dioxide Treatment from Flue Gases Using a Biotrickling Filter−Bioreactor System. Environ. Sci. Technol. 2003, 37, 1978–1982. [Google Scholar] [CrossRef]
- Gupta, N.; Roychoudhury, P.; Deb, J. Biotechnology of desulfurization of diesel: Prospects and challenges. Appl. Microbiol. Biotechnol. 2005, 66, 356–366. [Google Scholar] [CrossRef]
- Marcelis, C.; Van Leeuwen, M.; Polderman, H.; Janssen, A.; Lettinga, G. Model description of dibenzothiophene mass transfer in oil/water dispersions with respect to biodesulfurization. Biochem. Eng. J. 2003, 16, 253–264. [Google Scholar] [CrossRef]
- Li, F.L.; Xu, P.; Ma, C.Q.; Luo, L.L.; Wang, X.S. Deep desulfurization of hydrodesulfurization-treated diesel oil by a facultative thermophilic bacterium Mycobacterium sp. X7B. FEMS Microbiol. Lett. 2003, 223, 301–307. [Google Scholar] [CrossRef]
- Del Olmo, C.H.; Santos, V.E.; Alcon, A.; Garcia-Ochoa, F. Production of a Rhodococcus erythropolis IGTS8 biocatalyst for DBT biodesulfurization: Influence of operational conditions. Biochem. Eng. J. 2005, 22, 229–237. [Google Scholar] [CrossRef]
- Konishi, M.; Kishimoto, M.; Omasa, T.; Katakura, Y.; Shioya, S.; Ohtake, H. Effect of sulfur sources on specific desulfurization activity of Rhodococcus erythropolis KA2-5-1 in exponential fed-batch culture. J. Biosci. Bioeng. 2005, 99, 259–263. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, C.; Liu, Q.; Zang, M.; Liu, C.; Zhang, Y. Thermophilic biodesulfurization and its application in oil desulfurization. Appl. Microbiol. Biotechnol. 2018, 102, 9089–9103. [Google Scholar] [CrossRef]
- Ghosh, P.; Andrews, A.T.; Quann, R.J.; Halbert, T.R. Detailed kinetic model for the hydro-desulfurization of FCC naphtha. Energy Fuels 2009, 23, 5743–5759. [Google Scholar] [CrossRef]
- Boshagh, F.; Rahmani, M.; Zhu, W. Recent Advances and Challenges in Developing Technological Methods Assisting Oxidative Desulfurization of Liquid Fuels: A Review. Energy Fuels 2022, 36, 12961–12985. [Google Scholar] [CrossRef]
- Rodríguez, M.A.; Elizalde, I.; Ancheyta, J. Modeling the performance of a bench-scale reactor sustaining HDS and HDM of heavy crude oil at moderate conditions. Fuel 2012, 100, 152–162. [Google Scholar] [CrossRef]
- Sinfelt, J.H. Chemistry of catalytic processes, by Bruce C. Gates, James R. Katzer, and G. C. A. Schuit. Mcgraw-Hill, 1979, 464 pp. $28.50. AIChE J. 1979, 25, 734. [Google Scholar] [CrossRef]
- Clausen, B.S.; Topsoe, H.; Candia, R.; Villadsen, J.; Lengeler, B.; Als-Nielsen, J.; Christensen, F. Extended x-ray absorption fine structure study of the cobalt-molybdenum hydrodesulfurization catalysts. J. Phys. Chem. 1981, 85, 3868–3872. [Google Scholar] [CrossRef]
- Speight, J.G. Heavy and Extra-Heavy Oil Upgrading Technologies; Gulf Professional Publishing: Oxford, UK, 2013. [Google Scholar]
- Safa, M.A.; Bouresli, R.; Al-Majren, R.; Al-Shamary, T.; Ma, X. Oxidative desulfurization kinetics of refractory sulfur compounds in hydrotreated middle distillates. Fuel 2019, 239, 24–31. [Google Scholar] [CrossRef]
- Elmore, B. Biodesulfurization Techniques: Application of Selected Microorganisms for Organic Sulfur Removal from Coals; Final Report; Louisiana Polytechnic Institute: Ruston, LA, USA, 1993. [Google Scholar]
- Kodama, K.; Umehara, K.; Shimizu, K.; Nakatani, S.; Minoda, Y.; Yamada, K. Identification of Microbial Products from Dibenzothiophene and Its Proposed Oxidation Pathway. Agric. Biol. Chem. 1973, 37, 45–50. [Google Scholar] [CrossRef]
- Kilbane II, J.J. Microbial biocatalyst developments to upgrade fossil fuels. Curr. Opin. Biotechnol. 2006, 17, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Liu, L.; Shi, J.; Yan, W.; Chen, X.; Li, X. Biodesulfurization of thiosulfate by a Pseudomonas strain PSP1 and the investigation of underlying metabolic mechanisms. Environ. Sci. Pollut. Res. 2022, 29, 33764–33773. [Google Scholar] [CrossRef]
- Ahmad, A.; Khan, M.J.; Gupta, S.P.; Khan, M.M.; Kashif, M.; Khan, M.A.; Bee, A. Diverse Groups of Isolated Bacteria for Biodesulfurization of Petroleum Products in Petroleum Refinery Industry Curr. Res. Microbiol. 2017, 2, 1–25. [Google Scholar]
- Gupta, S.; Plugge, C.M.; Klok, J.; Muyzer, G. Comparative analysis of microbial communities from different full-scale haloalkaline biodesulfurization systems. Appl. Microbiol. Biotechnol. 2022, 106, 1759–1776. [Google Scholar] [CrossRef]
- Nuhu, A.A. Bio-catalytic desulfurization of fossil fuels: A mini review. Rev. Environ. Sci. Bio/Technol. 2013, 12, 9–23. [Google Scholar] [CrossRef]
- Fallahzadeh, R.; Bambai, B.; Esfahani, K.; Sepahi, A.A. Simulation-based protein engineering of R. erythropolis FMN oxidoreductase (DszD). Heliyon 2019, 5, e02193. [Google Scholar] [CrossRef]
- Arensdorf, J.J.; Loomis, A.K.; DiGrazia, P.M.; Monticello, D.J.; Pienkos, P.T. Chemostat approach for the directed evolution of biodesulfurization gain-of-function mutants. Appl. Environ. Microbiol. 2002, 68, 691–698. [Google Scholar] [CrossRef]
- Kobayashi, M.; Horiuchi, K.; Yoshikawa, O.; Hirasawa, K.; Ishii, Y.; Fujino, K.; Sugiyama, H.; Maruhashi, K. Kinetic analysis of microbial desulfurization of model and light gas oils containing multiple alkyl dibenzothiophenes. Biosci. Biotechnol. Biochem. 2001, 65, 298–304. [Google Scholar] [CrossRef]
- Folsom, B.; Schieche, D.; DiGrazia, P.; Werner, J.; Palmer, S. Microbial desulfurization of alkylated dibenzothiophenes from a hydrodesulfurized middle distillate by Rhodococcus erythropolis I-19. Appl. Environ. Microbiol. 1999, 65, 4967–4972. [Google Scholar] [CrossRef]
- Rangra, S.; Kabra, M.; Gupta, V.; Srivastava, P. Improved conversion of Dibenzothiophene into sulfone by surface display of Dibenzothiophene monooxygenase (DszC) in recombinant Escherichia coli. J. Biotechnol. 2018, 287, 59–67. [Google Scholar] [CrossRef]
- Kashif, M.; Lu, Z.; Sang, Y.; Yan, B.; Shah, S.J.; Khan, S.; Hussain, M.A.; Tang, H.; Jiang, C. Whole-Genome and Transcriptome Sequencing-Based Characterization of Bacillus cereus NR1 from Subtropical Marine Mangrove and Its Potential Role in Sulfur Metabolism. Front. Microbiol. 2022, 13, 120187. [Google Scholar] [CrossRef]
- Kalita, M.; Chutia, M.; Jha, D.K.; Subrahmanyam, G. Mechanistic Understanding of Gordonia sp. in Biodesulfurization of Organosulfur Compounds. Curr. Microbiol. 2022, 79, 82. [Google Scholar] [CrossRef]
- Khan, J.; Ali, M.I.; Jamal, A.; Ahmad, M.; Achakzai, J.K.; Zafar, M. Response of mixed bacterial culture towards dibenzothiophene desulfurization under the influence of surfactants and microscopically (SEM and TEM) characterized magnetic Fe3O4 nanoparticles. Microsc. Res. Technol. 2022, 85, 3838–3849. [Google Scholar] [CrossRef]
- Van Hamme, J.D.; Wong, E.T.; Dettman, H.; Gray, M.R.; Pickard, M.A. Dibenzyl sulfide metabolism by white rot fungi. Appl. Environ. Microbiol. 2003, 69, 1320–1324. [Google Scholar] [CrossRef]
- Zhuo, Y.; Yang, P.; Zhou, M.; Peng, D.; Han, Y. Low H2S content biogas biodesulfurization from high solid sludge anaerobic digestion using limited external aeration biotrickling filter: Effect of gas-liquid pattern on oxygen utilization performance. J. Environ. Manag. 2022, 314, 115084. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.-D.; Chen, H.; Chen, J.-M.; Shi, Y. Biodesulfurization of dibenzothiophene by growing cells of Gordonia sp. in batch cultures. Biotechnol. Lett. 2006, 28, 1175–1179. [Google Scholar] [CrossRef]
- McFarland, B.L.; Boron, D.J.; Deever, W.; Meyer, J.; Johnson, A.R.; Atlas, R.M. Biocatalytic sulfur removal from fuels: Applicability for producing low sulfur gasoline. Crit. Rev. Microbiol. 1998, 24, 99–147. [Google Scholar] [CrossRef]
- Kilbane II, J.J. Sulfur-specific microbial metabolism of organic compounds. Resour. Conserv. Recycl. 1990, 3, 69–79. [Google Scholar] [CrossRef]
- Kilbane, J. Toward sulfur-free fuels. Chemtech 1990, 20, 747–751. [Google Scholar]
- Bressler, D.C.; Norman, J.A.; Fedorak, P.M. Ring cleavage of sulfur heterocycles: How does it happen? Biodegradation 1997, 8, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.-K.; Chang, J.H.; Chang, Y.K.; Chang, H.N. Desulfurization of dibenzothiophene and diesel oils by a newly isolated Gordona strain, CYKS1. Appl. Environ. Microbiol. 1998, 64, 2327–2331. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Huang, D.; Shi, Y.; Zhang, B.; Sun, L.; Li, M.; Deng, X.; Wang, W. Comparative transcriptomic analysis revealed the key pathways responsible for organic sulfur removal by thermophilic bacterium Geobacillus thermoglucosidasius W-2. Sci. Total Environ. 2019, 676, 639–650. [Google Scholar] [CrossRef]
- Gray, K.A.; Mrachko, G.T.; Squires, C.H. Biodesulfurization of fossil fuels. Curr. Opin. Microbiol. 2003, 6, 229–235. [Google Scholar] [CrossRef]
- Van Afferden, M.; Schacht, S.; Klein, J.; Trüper, H.G. Degradation of dibenzothiophene by Brevibacterium sp. DO. Arch. Microbiol. 1990, 153, 324–328. [Google Scholar] [CrossRef]
- Ahmad, A.; Chauhan, A.K.; Kushwaha, H.N.; Javed, S.; Kumar, A. Preferential desulfurization of dibenzyl sulfide by an isolated Gordonia sp. IITR100. 3 Biotech 2015, 5, 237–243. [Google Scholar] [CrossRef]
- Xi, L.; Squires, C.H.; Monticello, D.J.; Childs, J.D. A Flavin Reductase Stimulates DszA and DszC Proteins of Rhodococcus erythropolis IGTS8 In Vitro. Biochem. Biophys. Res. Commun. 1997, 230, 73–75. [Google Scholar] [CrossRef]
- Konishi, J.; Okada, H.; Hirasawa, K.; Ishii, Y.; Maruhashi, K. Comparison of the substrate specificity of the two bacterial desulfurization systems. Biotechnol. Lett. 2002, 24, 1863–1867. [Google Scholar] [CrossRef]
- Gray, K.A.; Pogrebinsky, O.S.; Mrachko, G.T.; Xi, L.; Monticello, D.J.; Squires, C.H. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat. Biotechnol. 1996, 14, 1705–1709. [Google Scholar] [CrossRef]
- Ohshiro, T.; Ishii, Y.; Matsubara, T.; Ueda, K.; Izumi, Y.; Kino, K.; Kirimura, K. Dibenzothiophene desulfurizing enzymes from moderately thermophilic bacterium Bacillus subtilis WU-S2B: Purification, characterization and overexpression. J. Biosci. Bioeng. 2005, 100, 266–273. [Google Scholar] [CrossRef]
- Watkins, L.M.; Rodriguez, R.; Schneider, D.; Broderick, R.; Cruz, M.; Chambers, R.; Ruckman, E.; Cody, M.; Mrachko, G.T. Purification and characterization of the aromatic desulfinase, 2-(2′-hydroxyphenyl)benzenesulfinate desulfinase. Arch. Biochem. Biophys. 2003, 415, 14–23. [Google Scholar] [CrossRef]
- Boyd, D.R.; Sharma, N.D.; Stevenson, P.J.; Hoering, P.; Allen, C.C.R.; Dansette, P.M. Monooxygenase- and Dioxygenase-Catalyzed Oxidative Dearomatization of Thiophenes by Sulfoxidation, cis-Dihydroxylation and Epoxidation. Int. J. Mol. Sci. 2022, 23, 909. [Google Scholar] [CrossRef]
- Ishii, Y.; Konishi, J.; Okada, H.; Hirasawa, K.; Onaka, T.; Suzuki, M. Operon Structure and Functional Analysis of the Genes Encoding Thermophilic Desulfurizing Enzymes of Paenibacillus sp. A11-2. Biochem. Biophys. Res. Commun. 2000, 270, 81–88. [Google Scholar] [CrossRef]
- Kirimura, K.; Harada, K.; Iwasawa, H.; Tanaka, T.; Iwasaki, Y.; Furuya, T.; Ishii, Y.; Kino, K. Identification and functional analysis of the genes encoding dibenzothiophene-desulfurizing enzymes from thermophilic bacteria. Appl. Microbiol. Biotechnol. 2004, 65, 703–713. [Google Scholar] [CrossRef]
- Furuya, T.; Takahashi, S.; Iwasaki, Y.; Ishii, Y.; Kino, K.; Kirimura, K. Gene cloning and characterization of Mycobacterium phlei flavin reductase involved in dibenzothiophene desulfurization. J. Biosci. Bioeng. 2005, 99, 577–585. [Google Scholar] [CrossRef]
- Hino, T.; Hamamoto, H.; Suzuki, H.; Yagi, H.; Ohshiro, T.; Nagano, S. Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity. J. Biol. Chem. 2017, 292, 15804–15813. [Google Scholar] [CrossRef]
- Konishi, J.; Ishii, Y.; Onaka, T.; Maruhashi, K. Purification and characterization of the monooxygenase catalyzing sulfur-atom specific oxidation of dibenzothiophene and benzothiophene from the thermophilic bacterium Paenibacillus sp. strain A11–2. Appl. Microbiol. Biotechnol. 2002, 60, 128–133. [Google Scholar]
- Nomura, N.; Takada, M.; Okada, H.; Shinohara, Y.; Nakajima-Kambe, T.; Nakahara, T.; Uchiyama, H. Identification and functional analysis of genes required for desulfurization of alkyl dibenzothiophenes of Mycobacterium sp. G3. J. Biosci. Bioeng. 2005, 100, 398–402. [Google Scholar] [CrossRef]
- Martzoukou, O.; Glekas, P.D.; Avgeris, M.; Mamma, D.; Scorilas, A.; Kekos, D.; Amillis, S.; Hatzinikolaou, D.G. Interplay between Sulfur Assimilation and Biodesulfurization Activity in Rhodococcus qingshengii IGTS8: Insights into a Regulatory Role of the Reverse Transsulfuration Pathway. mBio 2022, 13, e0075422. [Google Scholar] [CrossRef]
- Matsui, T.; Noda, K.-i.; Tanaka, Y.; Maruhashi, K.; Kurane, R. Recombinant Rhodococcus sp. strain T09 can desulfurize DBT in the presence of inorganic sulfate. Curr. Microbiol. 2002, 45, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Abin-Fuentes, A.; Mohamed, M.E.-S.; Wang, D.I.; Prather, K.L. Exploring the mechanism of biocatalyst inhibition in microbial desulfurization. Appl. Environ. Microbiol. 2013, 79, 7807–7817. [Google Scholar] [CrossRef]
- Martínez, I.; Mohamed, M.E.-S.; Rozas, D.; García, J.L.; Díaz, E. Engineering synthetic bacterial consortia for enhanced desulfurization and revalorization of oil sulfur compounds. Metab. Eng. 2016, 35, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Al-Jamimi, H.A.; BinMakhashen, G.M.; Saleh, T.A. Multiobjectives optimization in petroleum refinery catalytic desulfurization using Machine learning approach. Fuel 2022, 322, 124088. [Google Scholar] [CrossRef]
- Gunam, I.B.W.; Sitepu, A.; Antara, N.S.; Triani, I.G.A.L.; Arnata, I.W.; Setiyo, Y. Bacterial desulfurization of dibenzothiophene by Pseudomonas sp. strain KWN5 immobilized in alginate beads. J. Teknol. 2021, 83, 107–115. [Google Scholar] [CrossRef]
- Dinamarca, M.A.; Rojas, A.; Baeza, P.; Espinoza, G.; Ibacache-Quiroga, C.; Ojeda, J. Optimizing the biodesulfurization of gas oil by adding surfactants to immobilized cell systems. Fuel 2014, 116, 237–241. [Google Scholar] [CrossRef]
- Shan, G.; Xing, J.; Zhang, H.; Liu, H. Biodesulfurization of dibenzothiophene by microbial cells coated with magnetite nanoparticles. Appl. Environ. Microbiol. 2005, 71, 4497–4502. [Google Scholar] [CrossRef]
- Li, S.; Ma, T. The Desulfurization Pathway in Rhodococcus. In Biology of Rhodococcus; Springer: Berlin/Heidelberg, Germany, 2019; pp. 203–229. [Google Scholar]
- Kaufman, E.N.; Harkins, J.B.; Rodriguez, M., Jr.; Tsouris, C.; Selvaraj, P.T.; Murphy, S.E. Development of an electro-spray bioreactor for crude oil processing. Fuel Process. Technol. 1997, 52, 127–144. [Google Scholar] [CrossRef]
- Konishi, M.; Kishimoto, M.; Tamesui, N.; Omasa, T.; Shioya, S.; Ohtake, H. The separation of oil from an oil–water–bacteria mixture using a hydrophobic tubular membrane. Biochem. Eng. J. 2005, 24, 49–54. [Google Scholar] [CrossRef]
- Peighami, R.; Motamedian, E.; Rasekh, B.; Yazdian, F. Investigating role of abiotic side and finding optimum abiotic condition for improving gas biodesulfurization using Thioalkalivibrio versutus. Sci. Rep. 2022, 12, 6260. [Google Scholar] [CrossRef]
- Grossman, M.; Lee, M.; Prince, R.; Garrett, K.; George, G.; Pickering, I. Microbial desulfurization of a crude oil middle-distillate fraction: Analysis of the extent of sulfur removal and the effect of removal on remaining sulfur. Appl. Environ. Microbiol. 1999, 65, 181–188. [Google Scholar] [CrossRef]
- Chang, J.H.; Rhee, S.K.; Chang, Y.K.; Chang, H.N. Desulfurization of diesel oils by a newly isolated dibenzothiophene-degrading Nocardia sp. strain CYKS2. Biotechnol. Prog. 1998, 14, 851–855. [Google Scholar] [CrossRef]
- Ishii, Y.; Kozaki, S.; Furuya, T.; Kino, K.; Kirimura, K. Thermophilic biodesulfurization of various heterocyclic sulfur compounds and crude straight-run light gas oil fraction by a newly isolated strain Mycobacterium phlei WU-0103. Curr. Microbiol. 2005, 50, 63–70. [Google Scholar] [CrossRef]
- Ma, C.Q.; Feng, J.H.; Zeng, Y.Y.; Cai, X.F.; Sun, B.P.; Zhang, Z.B.; Blankespoor, H.D.; Xu, P. Methods for the preparation of a biodesulfurization biocatalyst using Rhodococcus sp. Chemosphere 2006, 65, 165–169. [Google Scholar] [CrossRef]
- Konishi, J.; Ishii, Y.; Onaka, T.; Okumura, K.; Suzuki, M. Thermophilic carbon-sulfur-bond-targeted biodesulfurization. Appl. Environ. Microbiol. 1997, 63, 3164–3169. [Google Scholar] [CrossRef]
- Wang, J.; Davaadelger, B.; Salazar, J.K.; Butler, R.R.; Pombert, J.-F.; Kilbane, J.J.; Stark, B.C. Isolation and characterization of an interactive culture of two Paenibacillus species with moderately thermophilic desulfurization ability. Biotechnol. Lett. 2015, 37, 2201–2211. [Google Scholar] [CrossRef]
- Derikvand, P.; Etemadifar, Z. Sulfur removal from dibenzothiophene by newly isolated Paenibacillus validus strain PD2 and process optimization in aqueous and biphasic (model-oil) systems. Pol. J. Microbiol. 2015, 64, 5. [Google Scholar] [CrossRef]
- Onaka, T.; Konishi, J.; Ishii, Y.; Maruhashi, K. Desulfurization characteristics of thermophilic Paenibacillus sp. strain A11-2 against asymmetrically alkylated dibenzothiophenes. J. Biosci. Bioeng. 2001, 92, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, M.; Mu, T.; Liu, J.; Chen, L.; Miao, D.; Xing, J. Organic layer characteristics and microbial utilization of the biosulfur globules produced by haloalkaliphilic Thioalkalivibrio versutus D301 during biological desulfurization. Extremophiles 2022, 26, 27. [Google Scholar] [CrossRef]
- Konishi, J.; Ishii, Y.; Onaka, T.; Ohta, Y.; Suzuki, M.; Maruhashi, K. Purification and characterization of dibenzothiophene sulfone monooxygenase and FMN-dependent NADH oxidoreductase from the thermophilic bacterium Paenibacillus sp. strain A11-2. J. Biosci. Bioeng. 2000, 90, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Awasthi, G.; Singh, B. Extremophiles: A novel source of industrially important enzymes. Biotechnology 2011, 10, 121–135. [Google Scholar] [CrossRef]
- Siddiqui, K.S. Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar] [CrossRef]
- Siddiqui, K.S. Defying the activity–stability trade-off in enzymes: Taking advantage of entropy to enhance activity and thermostability. Crit. Rev. Biotechnol. 2017, 37, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Hokmabadi, M.; Khosravinia, S.; Mahdavi, M.A.; Gheshlaghi, R. Enhancing the biodesulphurization capacity of Rhodococcus sp. FUM94 in a biphasic system through optimization of operational factors. J. Appl. Microbiol. 2022, 132, 3461–3475. [Google Scholar] [CrossRef]
- Kirimura, K.; Furuya, T.; Nishii, Y.; Ishii, Y.; Kino, K.; Usami, S. Biodesulfurization of dibenzothiophene and its derivatives through the selective cleavage of carbon-sulfur bonds by a moderately thermophilic bacterim Bacillus subtilis WU-S2B. J. Biosci. Bioeng. 2001, 91, 262–266. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, W.-J.; Cai, Y.-B.; Zhang, Y.; Li, W. Elucidation of 2-hydroxybiphenyl effect on dibenzothiophene desulfurization by Microbacterium sp. strain ZD-M2. Bioresour. Technol. 2008, 99, 6928–6933. [Google Scholar] [CrossRef]
- Hou, Y.; Kong, Y.; Yang, J.; Zhang, J.; Shi, D.; Xin, W. Biodesulfurization of dibenzothiophene by immobilized cells of Pseudomonas stutzeri UP-1. Fuel 2005, 84, 1975–1979. [Google Scholar] [CrossRef]
- Chang, J.H.; Kim, Y.J.; Lee, B.H.; Cho, K.S.; Ryu, H.W.; Chang, Y.K.; Chang, H.N. Production of a desulfurization biocatalyst by two-stage fermentation and its application for the treatment of model and diesel oils. Biotechnol. Prog. 2001, 17, 876–880. [Google Scholar] [CrossRef]
- Alves, L.; Marques, S.; Matos, J.; Tenreiro, R.; Gírio, F.M. Dibenzothiophene desulfurization by Gordonia alkanivorans strain 1B using recycled paper sludge hydrolyzate. Chemosphere 2008, 70, 967–973. [Google Scholar] [CrossRef]
- Guobin, S.; Jianmin, X.; Huaiying, Z.; Huizhou, L. Deep desulfurization of hydrodesulfurized diesel oil by Pseudomonas delafieldii R-8. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 2005, 80, 420–424. [Google Scholar]
- Guobin, S.; Huaiying, Z.; Jianmin, X.; Guo, C.; Wangliang, L.; Huizhou, L. Biodesulfurization of hydrodesulfurized diesel oil with Pseudomonas delafieldii R-8 from high density culture. Biochem. Eng. J. 2006, 27, 305–309. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Z.; Feng, J.; Cai, X.; Xu, P. Biodesulfurization of DBT in tetradecane and crude oil by a facultative thermophilic bacterium Mycobacterium goodii X7B. J. Biotechnol. 2007, 127, 222–228. [Google Scholar] [CrossRef]
- Maghsoudi, S.; Vossoughi, M.; Kheirolomoom, A.; Tanaka, E.; Katoh, S. Biodesulfurization of hydrocarbons and diesel fuels by Rhodococcus sp. strain P32C1. Biochem. Eng. J. 2001, 8, 151–156. [Google Scholar] [CrossRef]
- Labana, S.; Pandey, G.; Jain, R. Desulphurization of dibenzothiophene and diesel oils by bacteria. Lett. Appl. Microbiol. 2005, 40, 159–163. [Google Scholar] [CrossRef]
- Ayala, M.; Tinoco, R.; Hernandez, V.; Bremauntz, P.; Vazquez-Duhalt, R. Biocatalytic oxidation of fuel as an alternative to biodesulfurization. Fuel Process. Technol. 1998, 57, 101–111. [Google Scholar] [CrossRef]
- Yu, B.; Xu, P.; Shi, Q.; Ma, C. Deep desulfurization of diesel oil and crude oils by a newly isolated Rhodococcus erythropolis strain. Appl. Environ. Microbiol. 2006, 72, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Grossman, M.; Lee, M.; Prince, R.; Minak-Bernero, V.; George, G.; Pickering, I. Deep desulfurization of extensively hydrodesulfurized middle distillate oil by Rhodococcus sp. strain ECRD-1. Appl. Environ. Microbiol. 2001, 67, 1949–1952. [Google Scholar] [CrossRef] [PubMed]
- Gunam, I.B.W.; Yaku, Y.; Hirano, M.; Yamamura, K.; Tomita, F.; Sone, T.; Asano, K. Biodesulfurization of alkylated forms of dibenzothiophene and benzothiophene by Sphingomonas subarctica T7b. J. Biosci. Bioeng. 2006, 101, 322–327. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, W.-J.; Chen, J.-M.; Cai, Y.-B.; Li, W. Desulfurization of various organic sulfur compounds and the mixture of DBT+ 4, 6-DMDBT by Mycobacterium sp. ZD-19. Bioresour. Technol. 2008, 99, 3630–3634. [Google Scholar] [CrossRef]
- Oda, S.; Ohta, H. Biodesulfurization of dibenzothiophene with Rhodococcus erythropolis ATCC 53968 and its mutant in an interface bioreactor. J. Biosci. Bioeng. 2002, 94, 474–477. [Google Scholar] [CrossRef]
- Mohamed Mel, S.; Al-Yacoub, Z.H.; Vedakumar, J.V. Biocatalytic desulfurization of thiophenic compounds and crude oil by newly isolated bacteria. Front. Microbiol. 2015, 6, 112. [Google Scholar] [CrossRef]
- Hwan Chang, J.; Keun Chang, Y.; Cho, K.-S.; Nam Chang, H. Desulfurization of model and diesel oils by resting cells of Gordona sp. Biotechnol. Lett. 2000, 22, 193–196. [Google Scholar] [CrossRef]
- Chacón-Patiño, M.L.; Nelson, J.; Rogel, E.; Hench, K.; Poirier, L.; Lopez-Linares, F.; Ovalles, C. Vanadium and nickel distributions in Pentane, In-between C5-C7 Asphaltenes, and heptane asphaltenes of heavy crude oils. Fuel 2021, 292, 120259. [Google Scholar] [CrossRef]
- Chacón-Patiño, M.L.; Nelson, J.; Rogel, E.; Hench, K.; Poirier, L.; Lopez-Linares, F.; Ovalles, C. Vanadium and nickel distributions in selective-separated n-heptane asphaltenes of heavy crude oils. Fuel 2022, 312, 122939. [Google Scholar] [CrossRef]
- Sousa, J.P.M.; Neves, R.P.P.; Sousa, S.F.; Ramos, M.J.; Fernandes, P.A. Reaction Mechanism and Determinants for Efficient Catalysis by DszB, a Key Enzyme for Crude Oil Bio-desulfurization. ACS Catal. 2020, 10, 9545–9554. [Google Scholar] [CrossRef]
- Visscher, P.T.; Taylor, B.F. Aerobic and anaerobic degradation of a range of alkyl sulfides by a denitrifying marine bacterium. Appl. Environ. Microbiol. 1993, 59, 4083–4089. [Google Scholar] [CrossRef]
- Visscher, P.T.; Taylor, B.F. A new mechanism for the aerobic catabolism of dimethyl sulfide. Appl. Environ. Microbiol. 1993, 59, 3784–3789. [Google Scholar] [CrossRef]
- Kilbane, J.J.; Jackowski, K. Biocatalytic detoxification of 2-chloroethyl ethyl sulfide. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 1996, 65, 370–374. [Google Scholar] [CrossRef]
- Lee, T.-S.; Chan, S.-H.; Weigand, W.; Bentley, W. A metabolic model for thiodiglycol degradation: Capacity constraint leads to byproduct accumulation. Bioprocess Biosyst. Eng. 2001, 24, 33–38. [Google Scholar]
- Jenisch-Anton, A.; Adam, P.; Michaelis, W.; Connan, J.; Herrmann, D.; Rohmer, M.; Albrecht, P. Molecular evidence for biodegradation of geomacromolecules. Geochim. Cosmochim. Acta 2000, 64, 3525–3537. [Google Scholar] [CrossRef]
- Van Hamme, J.D.; Fedorak, P.M.; Foght, J.M.; Gray, M.R.; Dettman, H.D. Use of a novel fluorinated organosulfur compound to isolate bacteria capable of carbon-sulfur bond cleavage. Appl. Environ. Microbiol. 2004, 70, 1487–1493. [Google Scholar] [CrossRef] [PubMed]
- Murgich, J.; Abanero, J.A.; Strausz, O.P. Molecular recognition in aggregates formed by asphaltene and resin molecules from the Athabasca oil sand. Energy Fuels 1999, 13, 278–286. [Google Scholar] [CrossRef]
- Omori, T.; Saiki, Y.; Kasuga, K.; Kodama, T. Desulfurization of alkyl and aromatic sulfides and sulfonates by dibenzothiophene-desulfurizing Rhodococcus sp. strain SY1. Biosci. Biotechnol. Biochem. 1995, 59, 1195–1198. [Google Scholar] [CrossRef]
- Ahmad, A.; Chauhan, A.K.; Javed, S.; Kumar, A. Desulfurization of thianthrene by a Gordonia sp. IITR100. Biotechnol. Lett. 2014, 36, 2209–2214. [Google Scholar] [CrossRef]
- Adlakha, J.; Singh, P.; Ram, S.K.; Kumar, M.; Singh, M.; Singh, D.; Sahai, V.; Srivastava, P. Optimization of conditions for deep desulfurization of heavy crude oil and hydrodesulfurized diesel by Gordonia sp. IITR100. Fuel 2016, 184, 761–769. [Google Scholar] [CrossRef]
- Kirkwood, K.; Ebert, S.; Foght, J.; Fedorak, P.; Gray, M. Bacterial biodegradation of aliphatic sulfides under aerobic carbon-or sulfur-limited growth conditions. J. Appl. Microbiol. 2005, 99, 1444–1454. [Google Scholar] [CrossRef]
- Kirkwood, K.M.; Foght, J.M.; Gray, M.R. Selectivity among organic sulfur compounds in one-and two-liquid-phase cultures of Rhodococcus sp. strain JVH1. Biodegradation 2007, 18, 473–480. [Google Scholar] [CrossRef]
- Kargi, F. Biological oxidation of thianthrene, thioxanthene and dibenzothiophene by the thermophilic organism Sulfolobus acidocaldarius. Biotechnol. Lett. 1987, 9, 478–482. [Google Scholar] [CrossRef]
- Schreiner, R.P.; Stevens, S.E., Jr.; Tien, M. Oxidation of thianthrene by the ligninase of Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1988, 54, 1858–1860. [Google Scholar] [CrossRef]
- Mohebali, G.; Ball, A.S. Biocatalytic desulfurization (BDS) of petrodiesel fuels. Microbiology 2008, 154, 2169–2183. [Google Scholar] [CrossRef]
- Yang, J.; Marison, I.W. Two-stage process design for the biodesulphurisation of a model diesel by a newly isolated Rhodococcus globerulus DAQ3. Biochem. Eng. J. 2005, 27, 77–82. [Google Scholar] [CrossRef]
- Lee, K.; Levy, E.M. Bioremediation: Waxy crude oils stranded on low-energy shorelines. In Proceedings of the International Oil Spill Conference, San Diego, CA, USA, 4–7 March 1991; pp. 541–547. [Google Scholar]
- Gallagher, J.R.; Olson, E.S.; Stanley, D.C. Microbial desulfurization of dibenzothiophene: A sulfur-specific pathway. FEMS Microbiol. Lett. 1993, 107, 31–35. [Google Scholar] [CrossRef]
- Monticello, D.J. Continuous process for biocatalytic desulfurization of sulfur-bearing heterocyclic molecules. Biotechnol. Adv. 1997, 15, 89. [Google Scholar]
- Kayser, K.J.; Bielaga-Jones, B.A.; Jackowski, K.; Odusan, O.; Kilbane II, J.J. Utilization of organosulphur compounds by axenic and mixed cultures of Rhodococcus rhodochrous IGTS8. Microbiology 1993, 139, 3123–3129. [Google Scholar] [CrossRef]
- Wang, P.; Krawiec, S. Desulfurization of dibenzothiophene to 2-hydroxybiphenyl by some newly isolated bacterial strains. Arch. Microbiol. 1994, 161, 266–271. [Google Scholar] [CrossRef]
- Kropp, K.G.; Goncalves, J.A.; Andersson, J.T.; Fedorak, P.M. Bacterial transformations of benzothiophene and methylbenzothiophenes. Environ. Sci. Technol. 1994, 28, 1348–1356. [Google Scholar] [CrossRef]
- Gilbert, S.C.; Morton, J.; Buchanan, S.; Oldfield, C.; McRoberts, A. Isolation of a unique benzothiophene-desulphurizing bacterium, Gordona sp. strain 213E (NCIMB 40816), and characterization of the desulphurization pathway. Microbiology 1998, 144, 2545–2553. [Google Scholar] [CrossRef]
- Tanaka, Y.; Matsui, T.; Konishi, J.; Maruhashi, K.; Kurane, R. Biodesulfurization of benzothiophene and dibenzothiophene by a newly isolated Rhodococcus strain. Appl. Microbiol. Biotechnol. 2002, 59, 325–328. [Google Scholar]
- Tanaka, Y.; Onaka, T.; Matsui, T.; Maruhashi, K.; Kurane, R. Desulfurization of benzothiophene by the gram-negative bacterium, Sinorhizobium sp. KT55. Curr. Microbiol. 2001, 43, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Hirasawa, K.; Konishi, J.; Tanaka, Y.; Maruhashi, K.; Kurane, R. Microbial desulfurization of alkylated dibenzothiophene and alkylated benzothiophene by recombinant Rhodococcus sp. strain T09. Appl. Microbiol. Biotechnol. 2001, 56, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Kirimura, K.; Furuya, T.; Sato, R.; Ishii, Y.; Kino, K.; Usami, S. Biodesulfurization of naphthothiophene and benzothiophene through selective cleavage of carbon-sulfur bonds by Rhodococcus sp. strain WU-K2R. Appl. Environ. Microbiol. 2002, 68, 3867–3872. [Google Scholar] [CrossRef]
- Khairy, H.; Wübbeler, J.H.; Steinbüchel, A. Biodegradation of the organic disulfide 4,4′-dithiodibutyric acid by Rhodococcus spp. Appl. Environ. Microbiol. 2015, 81, 8294–8306. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Tao, F.; Li, F.; Hou, J.; Tang, H.; Ma, C.; Xu, P. Complete genome sequence of Mycobacterium goodii X7B, a facultative thermophilic biodesulfurizing bacterium with industrial potential. J. Biotechnol. 2015, 212, 56–57. [Google Scholar] [CrossRef]
- Jaishankar, J.; Singh, P.; Srivastava, P. Draft genome sequence of a biodesulfurizing bacterium, Gordonia sp. strain IITR100. Genome Announc. 2017, 5, e00230-17. [Google Scholar] [CrossRef]
- Akhtar, N.; Ghauri, M.A.; Anwar, M.A.; Heaphy, S. Phylogenetic characterization and novelty of organic sulphur metabolizing genes of Rhodococcus spp.(Eu-32). Biotechnol. Lett. 2015, 37, 837–847. [Google Scholar] [CrossRef]
- Takada, M.; Nomura, N.; Okada, H.; Nakajima-Kambe, T.; Nakahara, T.; Uchiyama, H. De-repression and comparison of oil–water separation activity of the dibenzothiophene desulfurizing bacterium, Mycobacterium sp. G3. Biotechnol. Lett. 2005, 27, 871–874. [Google Scholar] [CrossRef]
- Wang, J.; Butler III, R.R.; Wu, F.; Pombert, J.-F.; Kilbane, J.J.; Stark, B.C. Enhancement of microbial biodesulfurization via genetic engineering and adaptive evolution. PLoS ONE 2017, 12, e0168833. [Google Scholar] [CrossRef]
- Watanabe, K.; Noda, K.-i.; Maruhashi, K. Enhanced desulfurization in a transposon-mutant strain of Rhodococcus erythropolis. Biotechnol. Lett. 2003, 25, 1299–1304. [Google Scholar] [CrossRef]
- Reichmuth, D.S.; Blanch, H.W.; Keasling, J.D. Dibenzothiophene biodesulfurization pathway improvement using diagnostic GFP fusions. Biotechnol. Bioeng. 2004, 88, 94–99. [Google Scholar] [CrossRef]
- Li, G.-q.; Li, S.-s.; Zhang, M.-l.; Wang, J.; Zhu, L.; Liang, F.-l.; Liu, R.-l.; Ma, T. Genetic rearrangement strategy for optimizing the dibenzothiophene biodesulfurization pathway in Rhodococcus erythropolis. Appl. Environ. Microbiol. 2008, 74, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Liu, Y.; Luo, Q.; Su, F.; Xu, Y.; Li, F.; Yu, B.; Ma, C.; Xu, P. Novel organic solvent-responsive expression vectors for biocatalysis: Application for development of an organic solvent-tolerant biodesulfurizing strain. Bioresour. Technol. 2011, 102, 9380–9387. [Google Scholar] [CrossRef]
- Aliebrahimi, S.; Raheb, J.; Ebrahimipour, G.; Bardania, H.; Nurollah, M.; Aghajani, Z. Designing a new recombinant indigenous Klebsiella oxytoca ISA4 by cloning of dsz genes. Energy Sources Part A Recovery Util. Environ. Eff. 2015, 37, 2056–2063. [Google Scholar] [CrossRef]
- Pan, J.; Wu, F.; Wang, J.; Yu, L.; Khayyat, N.H.; Stark, B.C.; Kilbane II, J.J. Enhancement of desulfurization activity by enzymes of the Rhodococcus dsz operon through coexpression of a high sulfur peptide and directed evolution. Fuel 2013, 112, 385–390. [Google Scholar] [CrossRef]
- Aggarwal, S.; Karimi, I.; Lee, D.Y. Reconstruction of a genome-scale metabolic network of Rhodococcus erythropolis for desulfurization studies. Mol. BioSystems 2011, 7, 3122–3131. [Google Scholar] [CrossRef]
- Zhuo, Y.; Han, Y.; Qu, Q.; Li, J.; Zhong, C.; Peng, D. Characteristics of low H2S concentration biogas desulfurization using a biotrickling filter: Performance and modeling analysis. Bioresour. Technol. 2019, 280, 143–150. [Google Scholar] [CrossRef]
- Feng, S.; Lin, X.; Tong, Y.; Huang, X.; Yang, H. Biodesulfurization of sulfide wastewater for elemental sulfur recovery by isolated Halothiobacillus neapolitanus in an internal airlift loop reactor. Bioresour. Technol. 2018, 264, 244–252. [Google Scholar] [CrossRef]
- Martínez, I.; Mohamed, M.E.-S.; Santos, V.E.; García, J.L.; García-Ochoa, F.; Díaz, E. Metabolic and process engineering for biodesulfurization in Gram-negative bacteria. J. Biotechnol. 2017, 262, 47–55. [Google Scholar] [CrossRef]
- Sar, T.; Ozturk, M.; Stark, B.C.; Akbas, M.Y. Improvement in desulfurization of dibenzothiophene and dibenzothiophene sulfone by Paenibacillus strains using immobilization or nanoparticle coating. J. Appl. Microbiol. 2022, 133, 1040–1051. [Google Scholar] [CrossRef]
- Kilbane, J.J. Biodesulfurization: How to Make it Work? Arab. J. Sci. Eng. 2017, 42, 1–9. [Google Scholar] [CrossRef]
- Mol, A.R.; Pruim, S.D.; de Korte, M.; Meuwissen, D.J.M.; van der Weijden, R.D.; Klok, J.B.M.; Keesman, K.J.; Buisman, C.J.N. Removal of small elemental sulfur particles by polysulfide formation in a sulfidic reactor. Water Res. 2022, 227, 119296. [Google Scholar] [CrossRef] [PubMed]
- Murarka, P.; Srivastava, P. Characterization of DNA binding and ligand binding properties of the TetR family protein involved in regulation of dsz operon in Gordonia sp. IITR100. Int. J. Biol. Macromol. 2019, 141, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Martín-Cabello, G.; Terrón-González, L.; Santero, E. Characterization of a dszEABC operon providing fast growth on dibenzothiophene and construction of broad-host-range biodesulfurization catalysts. Environ. Microbiol. 2022, 24, 1946–1963. [Google Scholar] [CrossRef] [PubMed]
- Keshav, A.; Murarka, P.; Srivastava, P. Bending is required for activation of dsz operon by the TetR family protein (DszGR). Gene 2022, 810, 146061. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).