Generation of a Culex Male Mosquito Sex-Separation RNAi Yeast Strain Using Cas-CLOVER and Super PiggyBac Engineering in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
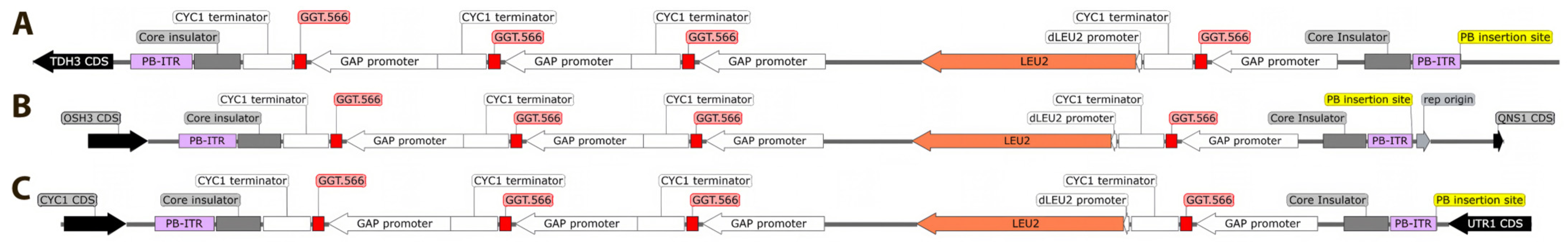
2.1. Yeast Strain Construction
2.2. Evaluation of GGT.566 Expression
2.3. Whole Genome Sequencing (WGS) of Yeast Strains
2.4. Pilot Fermentations
2.5. Mosquito Assays
3. Results
3.1. Production and Evaluation of Culex Sex-Separation Yeast Strains
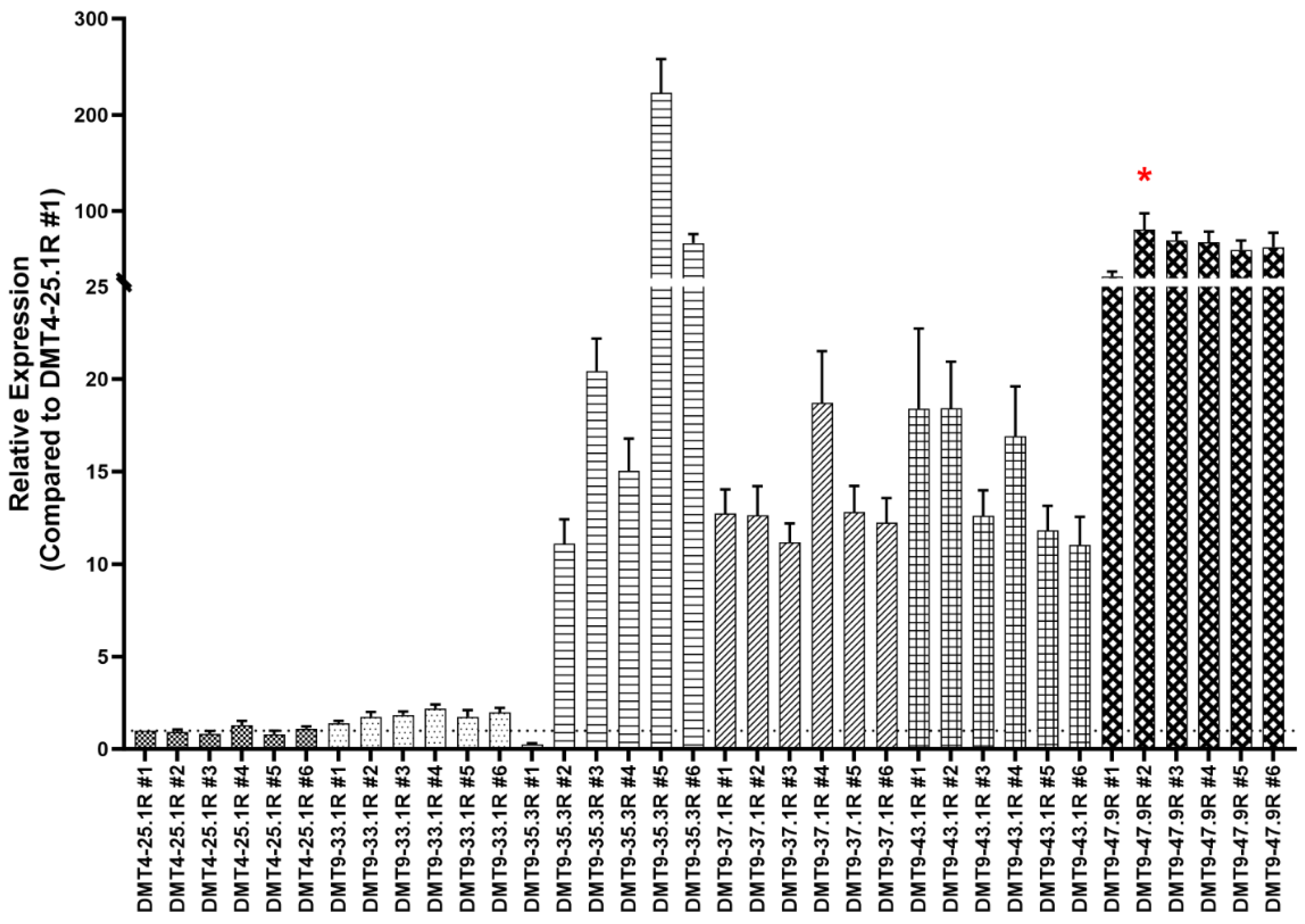
3.1.1. Generation of a Robust Yeast Strain with Multiple Integrations of the GGT.566 shRNA Expression Cassette
3.1.2. WGS Reveals the GGT.566 Expression Cassette Integration Sites
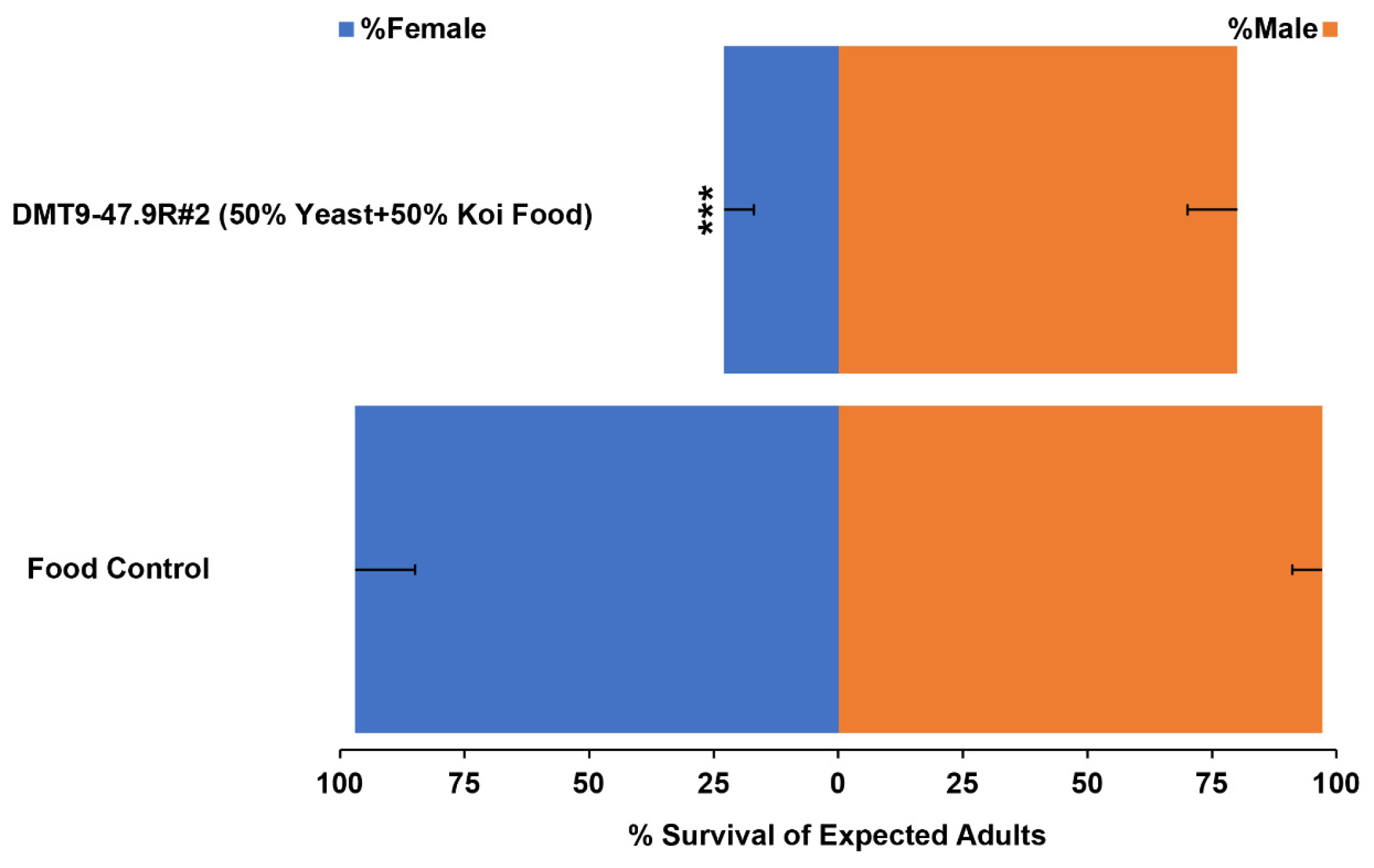
3.2. Assessment of Female-Specific Yeast Larvicide Activity
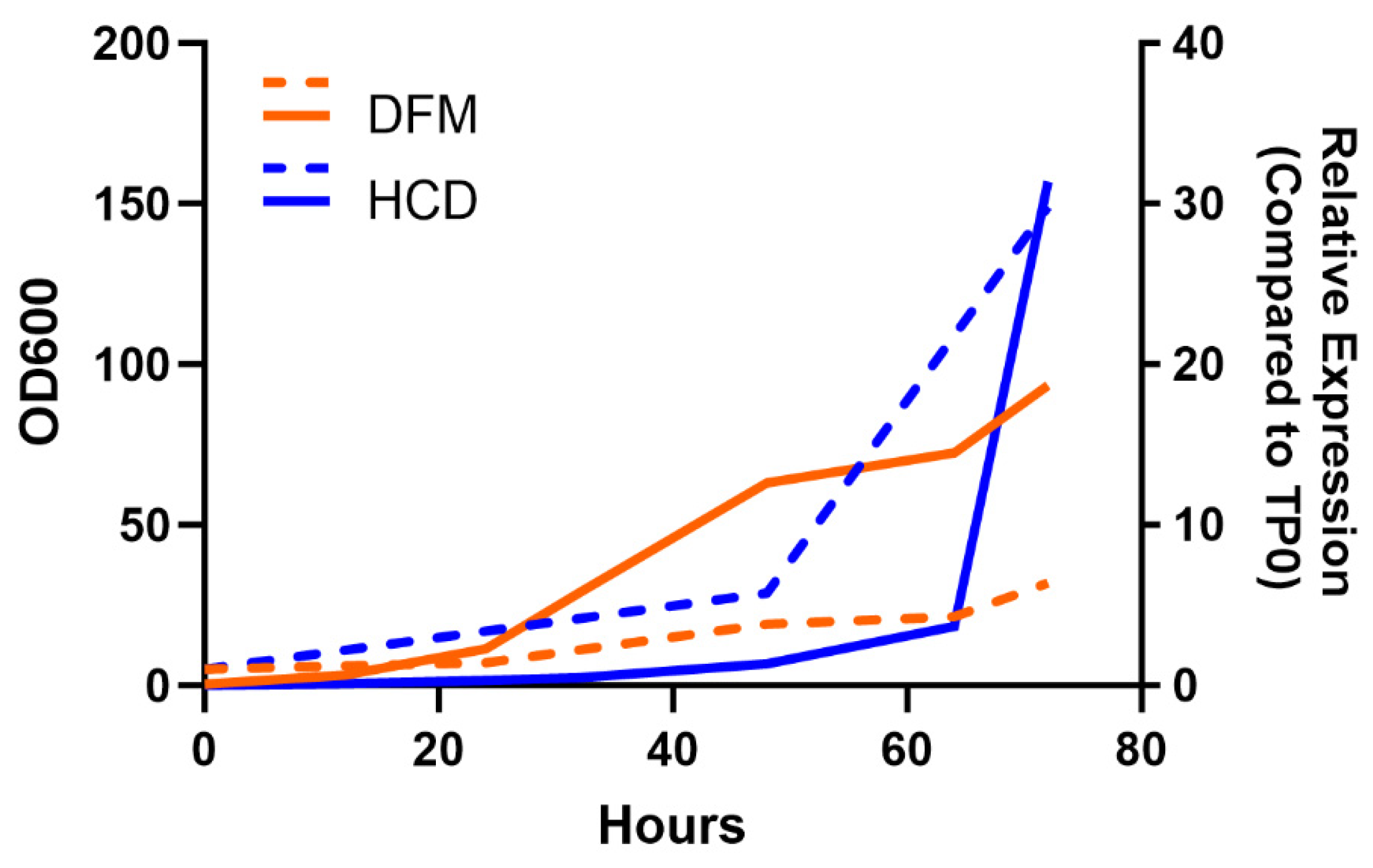
3.3. Performance of Strains in Pilot Fermentations
3.4. Evaluation of the Larvicidal Activity of Yeast Produced during the Scaled Fermentation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Center for Disease Control. Mosquitoes and Diseases: A–Z. Available online: https://www.cdc.gov/mosquitoes/about/diseases.html (accessed on 15 January 2020).
- Knipling, E.F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 1955, 48, 459–462. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, D.; Li, Y.; Yang, C.; Wu, Y.; Liang, X.; Liang, Y.; Pan, X.; Hu, L.; Sun, Q.; et al. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 2019, 572, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Kittayapong, P.; Ninphanomchai, S.; Limohpasmanee, W.; Chansang, C.; Chansang, U.; Mongkalangoon, P. Combined sterile insect technique and incompatible insect technique: The first proof-of-concept to suppress Aedes aegypti vector populations in semi-rural settings in Thailand. PLoS Negl. Trop. Dis. 2019, 13, e0007771. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.E.; Clarke, D.W.; Criswell, V.; Desnoyer, M.; Cornel, D.; Deegan, B.; Gong, K.; Hopkins, K.C.; Howell, P.; Hyde, J.S.; et al. Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat. Biotechnol. 2020, 38, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.O.; McKemey, A.R.; Garziera, L.; Lacroix, R.; Donnelly, C.A.; Alphey, L.; Malavasi, A.; Capurro, M.L. Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes. PLoS Negl. Trop. Dis. 2015, 9, e0003864. [Google Scholar] [CrossRef] [PubMed]
- Wise de Valdez, M.R.; Nimmo, D.; Betz, J.; Gong, H.F.; James, A.A.; Alphey, L.; Black, W.C., IV. Genetic elimination of dengue vector mosquitoes. Proc. Natl. Acad. Sci. USA 2011, 108, 4772–4775. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.M.; Ohm, J.R.; Rasgon, J.L. Gene drive for mosquito control: Where did it come from and where are we headed? Int. J. Environ. Res. Public Health 2017, 14, 1006. [Google Scholar] [CrossRef] [PubMed]
- Adelman, Z.N.; Tu, Z. Control of mosquito-borne infectious diseases: Sex and gene drive. Trends Parasitol. 2016, 32, 219–229. [Google Scholar] [CrossRef]
- Papathanos, P.A.; Bourtzis, K.; Tripet, F.; Bossin, H.; Virginio, J.F.; Capurro, M.L.; Pedrosa, M.C.; Guindo, A.; Sylla, L.; Coulibaly, M.B.; et al. A perspective on the need and current status of efficient sex separation methods for mosquito genetic control. Parasit. Vectors 2018, 11, 654. [Google Scholar] [CrossRef]
- Lutrat, C.; Giesbrecht, D.; Marois, E.; Whyard, S.; Baldet, T.; Bouyer, J. Sex sorting for pest control: It’s raining men! Trends. Parasitol. 2019, 35, 649–662. [Google Scholar] [CrossRef]
- Baylor College of Medicine. Emerging Infections and Biodefense. Available online: https://www.bcm.edu/departments/molecular-virology-and-microbiology/emerging-infections-and-biodefense/mosquitoes (accessed on 16 October 2023).
- Duman-Scheel, M. Saccharomyces cerevisiae (baker’s yeast) as an interfering RNA expression and delivery system. Curr. Drug Targets 2019, 20, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Hapairai, L.K.; Mysore, K.; Chen, Y.; Harper, E.I.; Scheel, M.P.; Lesnik, A.M.; Sun, L.; Severson, D.W.; Wei, N.; Duman-Scheel, M. Lure-and-kill yeast interfering RNA larvicides targeting neural genes in the human disease vector mosquito Aedes aegypti. Sci. Rep. 2017, 7, 13223. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.; Sun, L.; Li, P.; Roethele, J.B.; Misenti, J.K.; Kosmach, J.; Igiede, J.; Duman-Scheel, M. A conserved female-specific requirement for the GGT gene in mosquito larvae facilitates RNAi-mediated sex separation in multiple species of disease vector mosquitoes. Pathogens 2022, 11, 169. [Google Scholar] [CrossRef] [PubMed]
- Demeetra Ag. Bio. Cleaner Gene Editing with Cas-CLOVER. Available online: https://demeetra.com/cas-clover-the-clean-alternative-to-crispr-cas9-for-bioprocess/ (accessed on 1 July 2023).
- Brizzee, C.; Mysore, K.M.; Njoroge, T.M.; McConnell, S.; Hamid-Adiamoh, A.; Stewart, A.T.M.; Kinder, J.T.; Crawford, J.; Duman-Scheel, M. Targeting mosquitoes through generation of an insecticidal RNAi yeast strain using Cas-CLOVER and Super PiggyBac engineering in Saccharomyces cerevisiae. J. Fungi 2023, 9, 1056. [Google Scholar] [CrossRef]
- Waterham, H.R.; Digan, M.E.; Koutz, P.J.; Lair, S.V.; Cregg, J.M. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 1997, 186, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.A.; Morse, N.J.; Markham, K.A.; Wagman, A.M.; Gupta, A.; Alper, H.S. Short synthetic terminators for improved heterologous Gene Expression in Yeast. ACS Synth. Biol. 2015, 4, 824–832. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Maghini, D.G.; Moss, E.L.; Vance, S.E.; Bhatt, A.S. Improved high-molecular-weight DNA extraction, nanopore sequencing and metagenomic assembly from the human gut microbiome. Nat. Protoc. 2021, 16, 458–471. [Google Scholar] [CrossRef]
- Nanopore Technologies. Flongle. Available online: https://nanoporetech.com/products/flongle (accessed on 23 July 2023).
- Geneious. Geneious by Dotmatics. Available online: https://www.geneious.com (accessed on 1 July 2023).
- National Library of Medicine. Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 1 July 2023).
- Michigan State University. Bioeconomy Institute. Available online: https://bioeconomy.msu.edu/lansing-facility/ (accessed on 1 February 2021).
- van Hoek, P.; de Hulster, E.; van Dijken, J.P.; Pronk, J.T. Fermentative capacity in high-cell-density fed-batch cultures of baker’s yeast. Biotechnol. Bioeng. 2000, 68, 517–523. [Google Scholar] [CrossRef]
- MR4. Methods in Anopheles Research; CDC: Atlanta, GA, USA, 2015.
- Mysore, K.; Hapairai, L.K.; Wei, N.; Realey, J.S.; Scheel, N.D.; Severson, D.W.; Duman-Scheel, M. Preparation and use of a yeast shRNA delivery system for gene silencing in mosquito larvae. Methods Mol. Biol. 2019, 1858, 213–231. [Google Scholar] [CrossRef]
- Mysore, K.; Sun, L.; Roethele, J.B.; Li, P.; Igiede, J.; Misenti, J.K.; Duman-Scheel, M. A conserved female-specific larval requirement for MtnB function facilitates sex separation in multiple species of disease vector mosquitoes. Parasit. Vectors 2021, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D. Rejoice—RNAi for yeast. Science 2009, 326, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Crook, N.C.; Schmitz, A.C.; Alper, H.S. Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth. Biol. 2014, 3, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Harvey-Samuel, T.A.T.; Sutton, J.; Niebuhr, C.N.; Asigau, S.; Parker, P.; Sinkins, S.; Alphey, L. Culex quinquefasciatus: Status as a threat to island avifauna and options for genetic control. CABI Agric. Biosci. 2021, 2, 9. [Google Scholar] [CrossRef]
- WHO. Vector Alert: Anopheles stephensi Invasion and Spread in Africa and Sri Lanka. Available online: https://www.who.int/publications/i/item/9789240067714 (accessed on 15 April 2023).
- Li, M.A.; Pettitt, S.J.; Eckert, S.; Ning, Z.; Rice, S.; Cadiñanos, J.; Yusa, K.; Conte, N.; Bradley, A. The piggy bac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol. Cell. Biol. 2013, 33, 1317–1330. [Google Scholar] [CrossRef]
- Mitra, R.; Fain-Thornton, J.; Craig, N.L. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008, 27, 1097–1109. [Google Scholar] [CrossRef]
- He, L.; Huang, Y.; Tang, X. RNAi-based pest control: Production, application and the fate of dsRNA. Front. Bioeng. Biotechnol. 2022, 10, 1080576. [Google Scholar] [CrossRef]
- Marrone, P.G. Status of the biopesticide market and prospects for new bioherbicides. Pest. Manag. Sci. 2023. [Google Scholar] [CrossRef]
- Traber, G.M.; Yu, A.M. RNAi-based therapeutics and novel RNA bioengineering technologies. J. Pharmacol. Exp. Ther. 2023, 384, 133–154. [Google Scholar] [CrossRef]
- Corydon, I.J.; Fabian-Jessing, B.K.; Jakobsen, T.S.; Jorgensen, A.C.; Jensen, E.G.; Askou, A.L.; Aagaard, L.; Corydon, T.J. 25 years of maturation: A systematic review of RNAi in the clinic. Mol. Ther. Nucleic. Acids. 2023, 33, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Environmental Protection Agency. Final Regulatory Decision for New Active Ingredients Double-Stranded Ribonucleic Acid Transcript Comprising a DvSnj7 Inverted Repeat Sequence Derived from Western Corn Rootworm (Diabrotica virgifera virgifera) and Bacillus thuringiensis cry3Bbl Protein and the Genetic Material (Vector PV-ZMIR10871) Necessary for Their Production in MON 87411 Corn (OECD Unique Identifier: MON-87411-9). Available online: https://www.regulations.gov/document/EPA-HQ-OPP-2014-0293-0398 (accessed on 15 October 2015).
- Sparmann, A.; Vogel, J. RNA-based medicine: From molecular mechanisms to therapy. EMBO J. 2023, 42, e114760. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. FDA Approves First-of-Its Kind Targeted RNA-Based Therapy to Treat a Rare Disease. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-targeted-rna-based-therapy-treat-rare-disease (accessed on 20 November 2023).
- Food and Drug Administration. FDA Approves Givosiran for Acute Hepatic Porphyria. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-givosiran-acute-hepatic-porphyria#:~:text=On%20November%2020%2C%202019%2C%20the,enrolling%2094%20patients%20with%20AHP (accessed on 20 November 2023).
- Food and Drug Administration. FDA Approves First Drug to Treat Rare Metabolic Disorder. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-treat-rare-metabolic-disorder (accessed on 20 November 2023).
- Food and Drug Administration. FDA Approves Add-on Therapy to Lower Cholesterol among Certain High-Risk Adults. Available online: https://www.fda.gov/drugs/news-events-human-drugs/fda-approves-add-therapy-lower-cholesterol-among-certain-high-risk-adults (accessed on 20 November 2023).
- Erickson, B.E. RNAi pesticide moves closer to US approval. C&EN 2023, 101, 17. [Google Scholar]


| shRNA Type | Strain | Genotype | Original Auxotrophy | Restored Genotype |
|---|---|---|---|---|
| GTT.566 | DMT9-47.9R #2 | MATa, ura3∆0, leu2∆0, Piggybac (leu2d/PTDH3-shRNA_566-TCYC1, PTDH3-shRNA_566-TCYC1, PTDH3-shRNA_566-TCYC1), CEN/ARS (URA3/SPBase_Sc-CO) | Uracil | MATa, Piggybac (leu2d/PTDH3-shRNA_566-TCYC1, PTDH3-shRNA_566-TCYC1, PTDH3-shRNA_566-TCYC1), CEN/ARS (URA3/SPBase_Sc-CO) |
| Control | DMT4-347.1R a | MATa, ura3∆0, leu2∆0, his3∆0, trp1∆0, PiggyBac (LEU2/PTDH3-shRNA_Ctrl-TCYC1), 2um (URA3/SPBase_Sc-CO), PiggyBac (HIS3/PTDH3-shRNA_Ctrl-TCYC1), CEN/ARS (URA3/SPBase_Sc-CO) PiggyBac (trp1d/PTDH3-shRNA_Ctrl-TCYC1), CEN/ARS (URA3/SPBase_Sc-CO) | Uracil | MATa, PiggyBac (LEU2/PTDH3-shRNA_Ctrl-TCYC1), 2um (URA3/SPBase_Sc-CO), PiggyBac (HIS3/PTDH3-shRNA_Ctrl-TCYC1), CEN/ARS (URA3/SPBase_Sc-CO) PiggyBac (trp1d/PTDH3-shRNA_Ctrl-TCYC1), CEN/ARS (URA3/SPBase_Sc-CO) |
| Strain | Integrations | GGT.566 Copies | Total Copies | Genomic Integration Site | 5′ Flanking Sequence (60 bp) | 3′ Flanking Sequence (60 bp) |
|---|---|---|---|---|---|---|
| DMT9-47.9R #2 | 3 | 3 | 9 | Chromosome VII (883,906) | AAGTTCTTGGTGTTTTAAAACTAAAAA-AAAGACTAACTATAAAAGTAGAATTTAAGAAGT | GAAATAGATTTACAGAATTACAATCAATACCTACCGTCTTATATACTTATTAGTCAAGT |
| Chromosome VIII (245,747) | ACTATATACAACATGAAGGACTTTATAAATTCTTGATCAATCTTTCACACTAATTTTATA | CAGTTCTACACTATTTTTGTTTGCTTGACTGTAAAAACATTCTATTTTATGATTAGTTGC | ||||
| Chromosome X (526,821) | AGGAGTTAGACAACCTGAAGTCTAGGTCCCTATTTATTTTTTTTAATAGTTATGTTAGTA | GAACGTTATTTATATTTCAAATTTTTCTTTTTTTTCTGTACAAACGCGTGTACGCATGTA |
| Treatment | Male:Female a | p-Value |
|---|---|---|
| Food Control | 1:1 | 1 |
| DMT4-347.1R (50% Yeast + 50% Koi Food) | 1:1 | 0.86 |
| DMT9-47.9R #2 (50% Yeast + 50% Koi Food) | 5:1 | 2.1 × 10−9 |
| Treatment | Male/Female a | p-Value |
|---|---|---|
| Food Control | 1:1 | 0.72 |
| DMT9-47.9R #2 (50% Yeast + 50% Koi Food) | 3.5:1 | 8.4 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brizzee, C.; Mysore, K.; Njoroge, T.M.; McConnell, S.; Crawford, J.; Duman-Scheel, M. Generation of a Culex Male Mosquito Sex-Separation RNAi Yeast Strain Using Cas-CLOVER and Super PiggyBac Engineering in Saccharomyces cerevisiae. Fermentation 2023, 9, 999. https://doi.org/10.3390/fermentation9120999
Brizzee C, Mysore K, Njoroge TM, McConnell S, Crawford J, Duman-Scheel M. Generation of a Culex Male Mosquito Sex-Separation RNAi Yeast Strain Using Cas-CLOVER and Super PiggyBac Engineering in Saccharomyces cerevisiae. Fermentation. 2023; 9(12):999. https://doi.org/10.3390/fermentation9120999
Chicago/Turabian StyleBrizzee, Corey, Keshava Mysore, Teresia M. Njoroge, Seth McConnell, Jack Crawford, and Molly Duman-Scheel. 2023. "Generation of a Culex Male Mosquito Sex-Separation RNAi Yeast Strain Using Cas-CLOVER and Super PiggyBac Engineering in Saccharomyces cerevisiae" Fermentation 9, no. 12: 999. https://doi.org/10.3390/fermentation9120999
APA StyleBrizzee, C., Mysore, K., Njoroge, T. M., McConnell, S., Crawford, J., & Duman-Scheel, M. (2023). Generation of a Culex Male Mosquito Sex-Separation RNAi Yeast Strain Using Cas-CLOVER and Super PiggyBac Engineering in Saccharomyces cerevisiae. Fermentation, 9(12), 999. https://doi.org/10.3390/fermentation9120999






