Exploring the Microbial Diversity of Botswana’s Traditional Sourdoughs
Abstract
1. Introduction
2. Materials and Methods
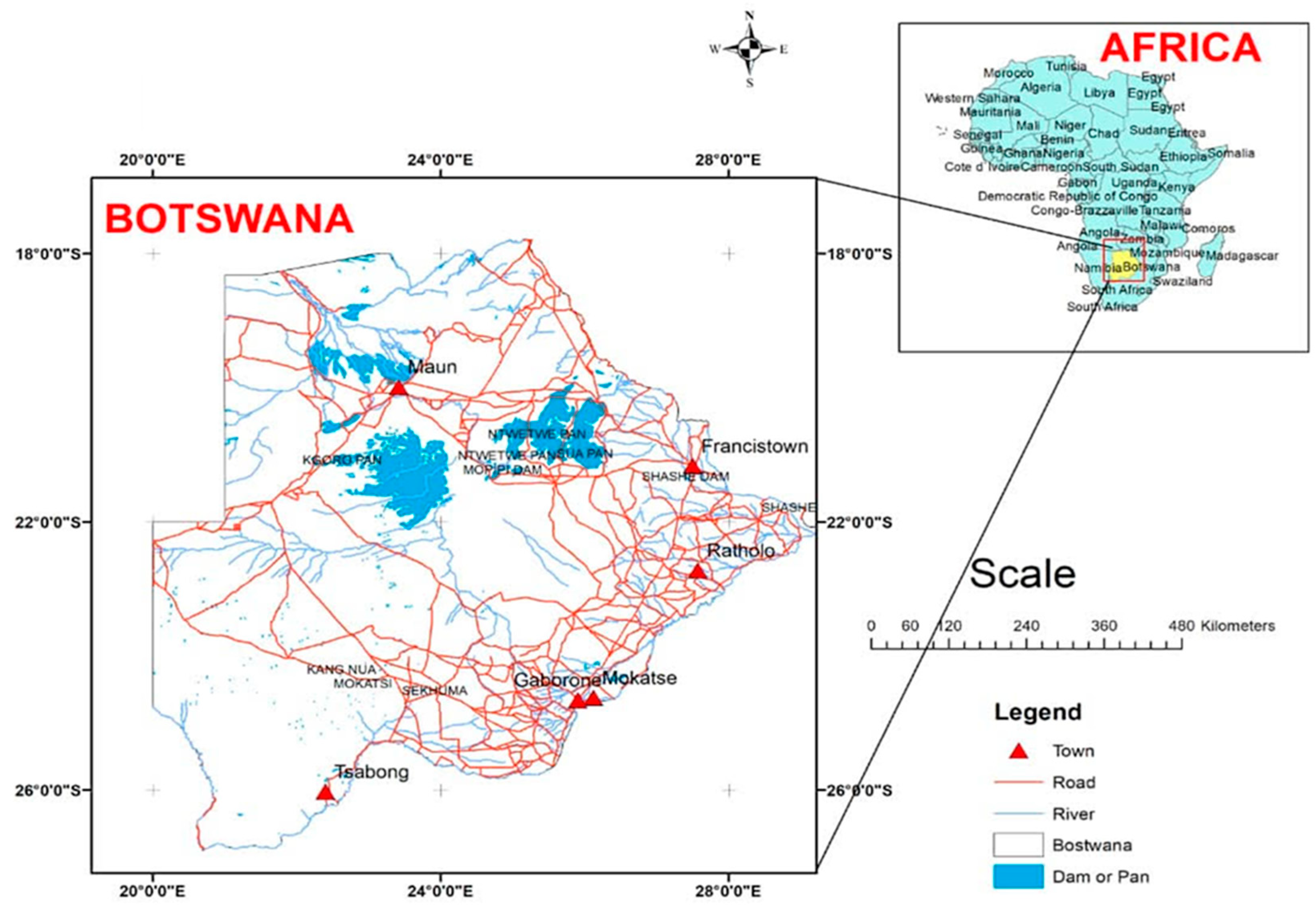
2.1. Sample Collection and Preparation
2.2. Culture Media
Isolation of Micro-Organisms from Sourdough Samples
2.3. Molecular Identification of Sourdough Isolates
2.3.1. Genomic DNA Extraction
2.3.2. Polymerase Chain Reaction and Sequencing
2.3.3. Phylogenetic Analysis of Sourdough Isolates
2.4. In Silico PCR-RFLP (Polymerase Chain Reaction–Restriction Fragment Length Polymorphism)
3. Results and Discussion
3.1. Microbial Communities Found in Traditional Sourdough in Botswana
3.2. Sourdoughs of Botswana Harbour Phylogenetically Diverse Yeasts
3.3. Sourdoughs of Botswana Harbour Phylogenetically Diverse LAB
3.4. Sourdoughs of Botswana Harbour Diverse AAB
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulp, K. Baker’s yeast and sourdough technologies in the production of U.S. bread products. In Handbook of Dough Fermentations; CRC Press: Boca Raton, FL, USA, 2003; pp. 101–148. [Google Scholar]
- Aboaba, O.; Obakpolor, E. The leavening ability of baker’s yeast on dough prepared with composite flour (wheat/cassava). Afr. J. Food Sci. 2010, 4, 325–329. [Google Scholar]
- Schieberle, P.; Grosch, W. Potent odorants of the wheat bread crumb Differences to the crust and effect of a longer dough fermentation. Z. Lebensm.-Unters. Forsch. 1991, 192, 130–135. [Google Scholar] [CrossRef]
- Chavan, R.S.; Chavan, S.R. Sourdough technology—A traditional way for wholesome foods: A review. Compr. Rev. Food Sci. Food Saf. 2011, 10, 169–182. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G. Sourdough/lactic acid bacteria. In Gluten-Free Cereal Products and Beverages; Elsevier: Amsterdam, The Netherlands, 2008; pp. 267–288. [Google Scholar]
- Katsi, P.; Kosma, I.S.; Michailidou, S.; Argiriou, A.; Badeka, A.V.; Kontominas, M.G. Characterization of artisanal spontaneous sourdough wheat bread from central Greece: Evaluation of physico-chemical, microbiological, and sensory properties in relation to conventional yeast leavened wheat bread. Foods 2021, 10, 635. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; Daniel, H.-M.; De Vuyst, L. Taxonomy and biodiversity of sourdough yeasts and lactic acid bacteria. In Handbook on Sourdough Biotechnology; Springer: New York, NY, USA, 2012; pp. 105–154. [Google Scholar]
- De Vuyst, L.; Harth, H.; Van Kerrebroeck, S.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Van Kerrebroeck, S.; Leroy, F. Microbial ecology and process technology of sourdough fermentation. Adv. Appl. Microbiol. 2017, 100, 49–160. [Google Scholar] [PubMed]
- Arena, M.P.; Russo, P.; Spano, G.; Capozzi, V. From microbial ecology to innovative applications in food quality improvements: The case of sourdough as a model matrix. J 2020, 3, 9–19. [Google Scholar] [CrossRef]
- Sekwati-Monang, B.; Gänzle, M.G. Microbiological and chemical characterisation of ting, a sorghum-based sourdough product from Botswana. Int. J. Food Microbiol. 2011, 150, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Landis, E.A.; Oliverio, A.M.; McKenney, E.A.; Nichols, L.M.; Kfoury, N.; Biango-Daniels, M.; Shell, L.K.; Madden, A.A.; Shapiro, L.; Sakunala, S. The diversity and function of sourdough starter microbiomes. eLife 2021, 10, e61644. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- Comasio, A.; Verce, M.; Van Kerrebroeck, S.; De Vuyst, L. Diverse microbial composition of sourdoughs from different origins. Front. Microbiol. 2020, 11, 1212. [Google Scholar] [CrossRef] [PubMed]
- Aouine, M.; Misbah, A.; Elabed, S.; Haggoud, A.; Mohammed, I.H.; Koraichi, S.I. Isolation and characterization of potential starter yeasts from traditional Moroccan sourdoughs. Microbiol. Biotechnol. Lett. 2021, 49, 501–509. [Google Scholar] [CrossRef]
- Gül, H.; Özçelik, S.; Sağdıç, O.; Certel, M. Sourdough bread production with lactobacilli and S. cerevisiae isolated from sourdoughs. Process Biochem. 2005, 40, 691–697. [Google Scholar] [CrossRef]
- Fujaroen, D.; Pathom-Aree, W. Application of chemical dyes as colour indicator for selective isolation of acetic acid bacteria. Res. J. Microbiol. 2007, 2, 885–888. [Google Scholar]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; De Vries, M.; Verkleij, G.; Crous, P.; Boekhout, T. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bartkiene, E.; Özogul, F.; Rocha, J.M. Bread sourdough lactic acid bacteria—Technological, antimicrobial, toxin-degrading, immune system-, and faecal microbiota-modelling biological agents for the preparation of food, nutraceuticals and feed. Foods 2022, 11, 452. [Google Scholar] [CrossRef]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.-M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef]
- Martorana, A.; Giuffrè, A.M.; Capocasale, M.; Zappia, C.; Sidari, R. Sourdoughs as a source of lactic acid bacteria and yeasts with technological characteristics useful for improved bakery products. Eur. Food Res. Technol. 2018, 244, 1873–1885. [Google Scholar] [CrossRef]
- Carbonetto, B.; Nidelet, T.; Guezenec, S.; Perez, M.; Segond, D.; Sicard, D. Interactions between Kazachstania humilis yeast species and lactic acid bacteria in sourdough. Microorganisms 2020, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.-M.; Moons, M.-C.; Huret, S.; Vrancken, G.; De Vuyst, L. Wickerhamomyces anomalus in the sourdough microbial ecosystem. Antonie Leeuwenhoek 2011, 99, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Coda, R.; Cassone, A.; Rizzello, C.G.; Nionelli, L.; Cardinali, G.; Gobbetti, M. Antifungal activity of Wickerhamomyces anomalus and Lactobacillus plantarum during sourdough fermentation: Identification of novel compounds and long-term effect during storage of wheat bread. Appl. Environ. Microbiol. 2011, 77, 3484–3492. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Sapone, A.; Zevallos, V.; Schuppan, D. Nonceliac gluten sensitivity. Gastroenterology 2015, 148, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; Piubeli, F.; Bonfá, M.; García, M.; Durrant, L.; Mellado, E. Analysis and characterization of cultivable extremophilic hydrolytic bacterial community in heavy-metal-contaminated soils from the Atacama Desert and their biotechnological potentials. J. Appl. Microbiol. 2012, 113, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Calvert, M.D.; Madden, A.A.; Nichols, L.M.; Haddad, N.M.; Lahne, J.; Dunn, R.R.; McKenney, E.A. A review of sourdough starters: Ecology, practices, and sensory quality with applications for baking and recommendations for future research. PeerJ 2021, 9, e11389. [Google Scholar] [CrossRef]
- Yassunaka Hata, N.N.; Surek, M.; Sartori, D.; Vassoler Serrato, R.; Aparecida Spinosa, W. Role of acetic acid bacteria in food and beverages. Food Technol. Biotechnol. 2023, 61, 85–103. [Google Scholar] [CrossRef]
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of acetic acid bacteria and their role in vinegar and fermented beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18, 587–625. [Google Scholar] [CrossRef]
- Sevgili, A.; Can, C.; Ceyhan, D.I.; Erkmen, O. Molecular identification of LAB and yeasts from traditional sourdoughs and their impacts on the sourdough bread quality characteristics. Curr. Res. Food Sci. 2023, 6, 100479. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Robinson, H.A. The Geographic Distributions of Saccharomyces cerevisiae and Saccharomyces paradoxus, and the Potential to Detect Past Yeast Populations with Ancient DNA. Ph.D. Thesis, The University of Manchester, Manchester, UK, 2016. [Google Scholar]
- Sampaio, J.P.; Gonçalves, P. Biogeography and ecology of the genus Saccharomyces. In Yeasts in Natural Ecosystems; Springer: Cham, Switzerland, 2017; pp. 131–153. [Google Scholar]
- Pearman, J.K.; Biessy, L.; Thomson-Laing, G.; Waters, S.; Vandergoes, M.J.; Howarth, J.D.; Rees, A.; Moy, C.; Pochon, X.; Wood, S.A. Local factors drive bacterial and microeukaryotic community composition in lake surface sediment collected across an altitudinal gradient. FEMS Microbiol. Ecol. 2020, 96, fiaa070. [Google Scholar] [CrossRef]
- Akamine, I.T.; Mansoldo, F.R.; Cardoso, V.S.; de Souza Dias, E.P.; Vermelho, A.B.J.F. Hydrolase Activities of Sourdough Microorganisms. Fermentation 2023, 9, 703. [Google Scholar] [CrossRef]
- Katina, K.; Heiniö, R.-L.; Autio, K.; Poutanen, K. Optimization of sourdough process for improved sensory profile and texture of wheat bread. LWT-Food Sci. Technol. 2006, 39, 1189–1202. [Google Scholar] [CrossRef]
- Lutter, L.; Jõudu, I.; Andreson, H. Volatile organic compounds and their generation in sourdough. Agron. Res. 2023, 21, 504–536. [Google Scholar]
- Alpers, T.; Kerpes, R.; Frioli, M.; Nobis, A.; Hoi, K.I.; Bach, A.; Jekle, M.; Becker, T. Impact of storing condition on staling and microbial spoilage behavior of bread and their contribution to prevent food waste. Foods 2021, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Islam, R.; Hasan, S.; Zzaman, W.; Rana, M.R.; Ahmed, S.; Roy, M.; Sayem, A.; Matin, A.; Raposo, A. A comprehensive review on bio-preservation of bread: An approach to adopt wholesome strategies. Foods 2022, 11, 319. [Google Scholar] [CrossRef]
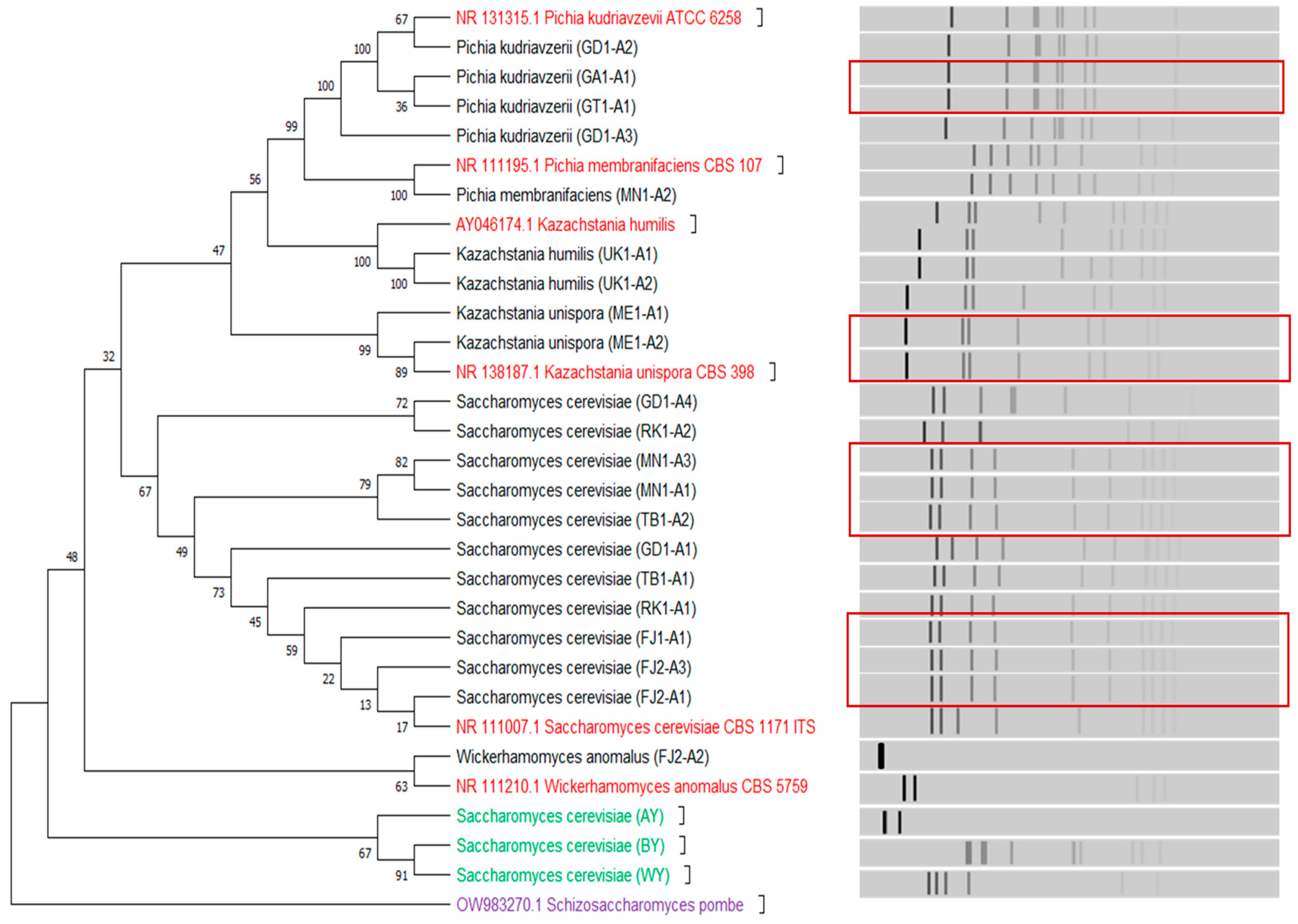
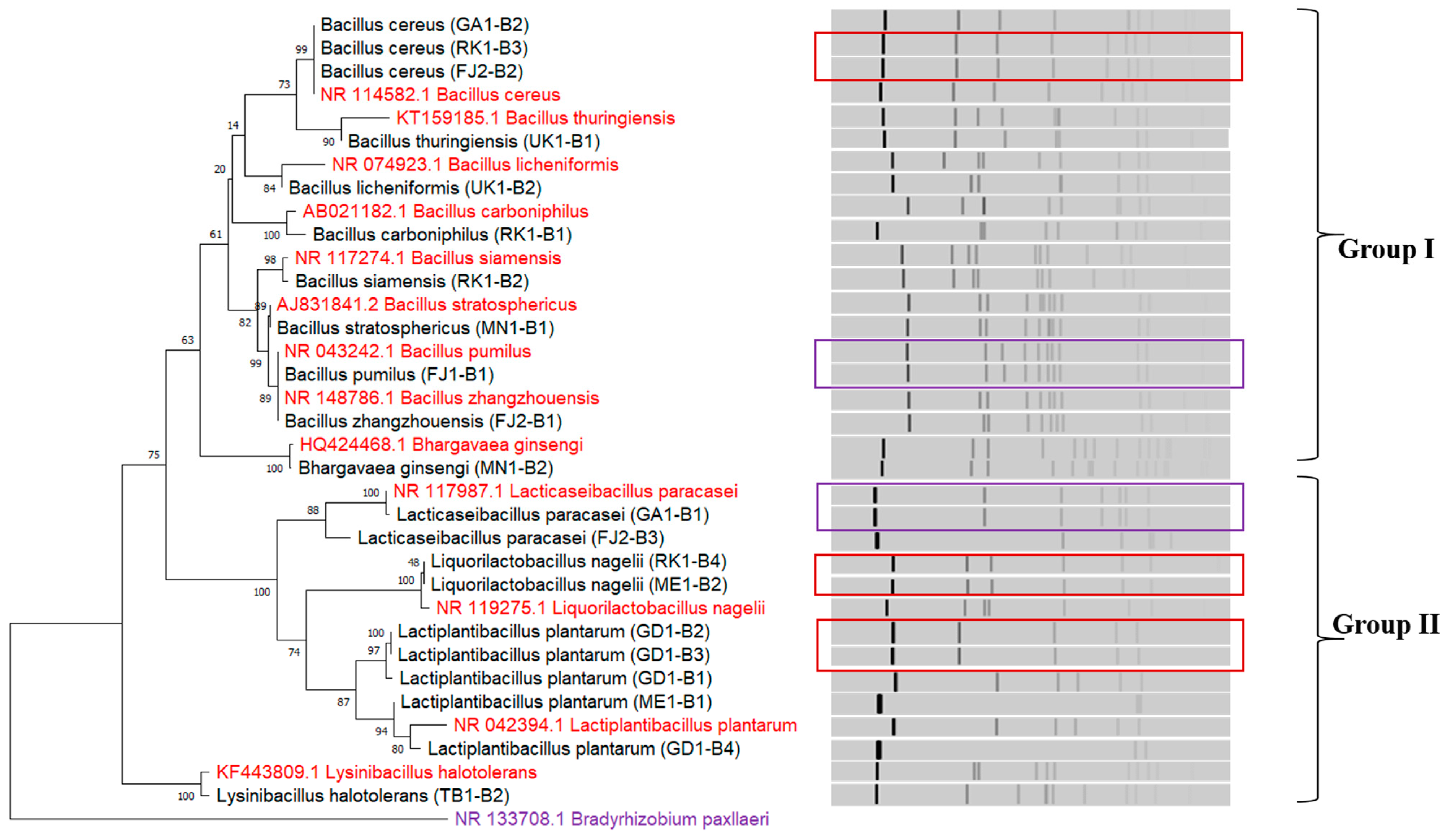

| Sample Name | Yeast Isolate Code | Scientific Name | LAB Isolate Code | Scientific Name | AAB Isolate Code | Scientific Name |
|---|---|---|---|---|---|---|
| FJ1 | FJ1-A1 | Saccharomyces cerevisiae | None | FJ1-C1 | Acetobacter pasteurianus | |
| FJ2 | FJ2-A1 | Saccharomyces cerevisiae | ||||
| FJ2-A2 | Wickerhamomyces anomalus | None | ||||
| FJ2-A3 | Saccharomyces cerevisiae | FJ2-B3 | Lacticaseibacillus paracasei | |||
| GA1 | GA1-A1 | Pichia kudriavzerii | GA1-B1 | Lacticaseibacillus paracasei | GA1-C1 | Acetobacter pasteurianus |
| GD1 | GD1-A1 | Saccharomyces cerevisiae | GD1-B1 | Lactiplantibacillus plantarum | GD1-C1 | Acetobacter indonesiensis |
| GD1-A2 | Pichia kudriavzerii | GD1-B2 | Lactiplantibacillus plantarum | |||
| GD1-A3 | Pichia kudriavzerii | GD1-B3 | Lactiplantibacillus plantarum | |||
| GD1-A4 | Saccharomyces cerevisiae | GD1-B4 | Lactiplantibacillus plantarum | |||
| RK1 | RK1-A1 | Saccharomyces cerevisiae | ||||
| RK1-A2 | Saccharomyces cerevisiae | None | ||||
| RK1-B4 | Liquorilactobacillus nageli | |||||
| MN1 | MN1-A1 | Saccharomyces cerevisiae | ||||
| MN1-A2 | Pichia membranifaciens | None | None | |||
| MN1-A3 | Saccharomyces cerevisiae | |||||
| GT1 | GT1-A1 | Pichia kudriavzerii | None | None | ||
| TB1 | TB1-A1 | Saccharomyces cerevisiae | None | None | ||
| TB1-A2 | Saccharomyces cerevisiae | |||||
| ME1 | ME1-A1 | Kazachstania unispora | ME1-B1 | Lactiplantibacillus plantarum | None | |
| ME1-A2 | Kazachstania unispora | ME1-B2 | Liquorilactobacillus nageli | |||
| UK1 | UK1-A1 | Kazachstania humilis | None | UK1-C1 | Acetobacter malorum | |
| UK1-A2 | Kazachstania humilis |
| Sample Name | Bacillus and Other Bacteria Codes | Scientific Name |
|---|---|---|
| FJ1 | FJ1-B1 | Bacillus pumilis |
| FJ2 | FJ2-B1 | Bacillus zhangzhouensis |
| FJ2-B2 | Bacillus cereus | |
| GA1 | None | |
| GA1-B2 | Bacillus cereus | |
| GD1 | None | |
| RK1 | RK1-B1 | Bacillus carboniphilus |
| RK1-B2 | Bacillus siamensis | |
| RK1-B3 | Bacillus cereus | |
| MN1 | MN1-B1 | Bacillus stratosphericus |
| MN1-B2 | Bhargavaea ginsengi | |
| GT1 | GT1-B1 | Bacillus cereus |
| TB1 | TB1-B1 | Lysinibacillus halotolerans |
| ME1 | None | |
| UK1 | UK1-B1 | Bacillus thuringiensis |
| UK1-B2 | Bacillus licheniformis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semumu, T.; Zhou, N.; Kebaneilwe, L.; Loeto, D.; Ndlovu, T. Exploring the Microbial Diversity of Botswana’s Traditional Sourdoughs. Fermentation 2024, 10, 417. https://doi.org/10.3390/fermentation10080417
Semumu T, Zhou N, Kebaneilwe L, Loeto D, Ndlovu T. Exploring the Microbial Diversity of Botswana’s Traditional Sourdoughs. Fermentation. 2024; 10(8):417. https://doi.org/10.3390/fermentation10080417
Chicago/Turabian StyleSemumu, Thandiwe, Nerve Zhou, Lebani Kebaneilwe, Daniel Loeto, and Thando Ndlovu. 2024. "Exploring the Microbial Diversity of Botswana’s Traditional Sourdoughs" Fermentation 10, no. 8: 417. https://doi.org/10.3390/fermentation10080417
APA StyleSemumu, T., Zhou, N., Kebaneilwe, L., Loeto, D., & Ndlovu, T. (2024). Exploring the Microbial Diversity of Botswana’s Traditional Sourdoughs. Fermentation, 10(8), 417. https://doi.org/10.3390/fermentation10080417






