Abstract
A potentially novel probiotic strain, YY-112, was previously isolated and identified as Lactiplantibacillus pentosus using 16S rDNA sequencing. The whole genome analysis showed that strain YY-112 has the potential to metabolize carbohydrates in the gastrointestinal environment and to regulate immunity. Further, comparative genomics analysis revealed that strain YY-112 was Lactiplantibacillus plantarum (L. plantarum) with more unique genes. The results of in vivo tests showed that L. plantarum YY-112 had no adverse effects and restored the damaged spleen and intestinal barrier of immunocompromised mice. L. plantarum YY-112 recovered the normal levels of lymphocytes, serum cytokines (Interferon-γ, tumor necrosis factor α, and interleukin-6), immunoglobulins (Ig) (IgA, IgM, and IgG) and lipids (albumin, cholesterol, triglyceride). Additionally, L. plantarum YY-112 might indirectly enhance the immune system by improving the intestinal microbiota structure. These results supported the potential of L. plantarum YY-112 as a probiotic to regulate the immune system of hosts.
1. Introduction
Probiotics can regulate the intestinal microbial structure, while intestinal microbiota produces short-chain fatty acids (SCFAs) by metabolizing dietary fiber to regulate host immunity and strengthen the intestinal barrier [1]. The primary representatives of probiotics are mostly lactic acid bacteria (LAB), including Lactiplantibacillus plantarum (L. plantarum), which provide significant health-promoting effects on the host intestinal barrier and immune system [2,3,4]. L. plantarum 23-1 improved intestinal inflammation and barrier function in obese mice through the Toll-like receptor 4/nuclear factor kappa-B signaling pathway [5]. L. plantarum 1201 increased D-mannose and alpha-tocopherol by restoring intestinal microbiota structure, thereby relieving colitis [6]. Nevertheless, LAB is highly strain-specific, so their biological effects cannot be judged at the species level [7]. This is why researchers are constantly screening for new probiotic strains with potential probiotic properties.
However, when it comes to discovering, developing and implementing novel LAB, traditional methods are labor- and time-intensive, which cannot meet the needs of strain safety risk assessment well at a high-throughput scale [8]. These conditions are being revolutionized by the development of next-generation sequencing technology, which offers significant advantages in terms of speed, cost, quality, and precision [9]. Furthermore, some strains have 99% similarity in 16S rRNA gene sequences, such as L. plantarum and Lactiplantibacillus pentosus. Their similar phenotypes made them easily confused and mistaken, requiring whole-genome sequencing for further differentiation [10].
The immune system is an organization of immune organs, cells, and molecules functioning as a defense against infection and disease [11]. However, many conditions impair host immune function, such as viral infections, long-term administration of chemotherapeutic drugs, and gastrointestinal immunodeficiency [12]. Encouragingly, the exploitation of probiotics to modulate the immune system without side effects is feasible. Lactobacillus reuteri can secrete indole-3-aldehyde to exert antitumor immunity [13]. Lactobacillus casei NCU011054 could enhance host intestinal immunity by modulating the Toll-like receptors/nuclear factor kappa-B pathway and improving the intestinal microbiota [14]. L. plantarum modulates the production of T cells and IgA, enhances phagocytic activity and expression of tumor necrosis factor α (TNF-α), interleukin (IL)-6, Toll-like receptor 2, nitric oxide, and inducible NO synthase in macrophages [15]. However, fewer studies have started with whole-genome analysis and utilized immunocompromised mouse models to gain a preliminary understanding of the effects of novel L. plantarum on the immune systems of normal and immunocompromised mice.
Previously, we isolated a strain YY-112 from waxberry (Myrica rubra), tolerated to acids, bile salts, artificial digestive fluids, etc. Its exopolysaccharide (EPS) was proven to exert beneficial effects on intestinal microbiota and facilitate SCFA production [16]. It seems that strain YY-112 is a potential probiotic strain with EPS that is beneficial for the intestinal barrier. Strain YY-112 was preliminarily identified as L. pentosus via 16S rRNA gene sequencing. However, 16S rRNA gene sequence similarity analysis could not distinguish the phylogenetic relationship between different cladistic LAB with recent in-depth research [10]. Here, we analyzed the genome sequence of strain YY-112 for its immune-related properties. Strain YY-112 was reclassified using comparative genome analysis and analyzed in comparison with other L. plantarum. Further, we evaluated the mitigating effects of L. plantarum YY-112 on cyclophosphamide (CP)-induced immunocompromised, including the immune environment, immune organs, and intestinal microbiota. The work will enrich the whole-genome sequence of L. plantarum and contribute to understanding the immunomodulatory potential of L. plantarum YY-112.
2. Materials and Methods
2.1. Whole-Genome Sequencing of YY-112
The strain YY-112 (CGMCC no.18492) was isolated from fresh waxberry in Zhejiang Province of China and stored at −80 °C with 25% (v/v) glycerol. Before whole-genome sequencing, the strain was cultured in MRS broth medium at 37 °C for 18 h twice. Then, it was centrifuged at 5000× g for 10 min at 4 °C and washed 3 times with phosphate-buffered saline. The strain was sequenced using PacBio Sequel and Illumina NovaSeq platforms. Additionally, the assembly of a complete sequence was performed using HGAP v.4, CANU v1.7.1, and pilon v1. 18 software.
2.2. Genome Annotation of YY-112
Coding sequences (CDSs) prediction was performed using GeneMarkS v4.32 software for the whole-genome sequence. tRNAscan-SE v2.0, Barrnap v0.9, and Rfam v14.1 software were used to predict tRNAs, rRNAs, and the remaining non-coding RNAs, respectively. Then, virulence factor-related genes and CAZy enzyme genes were annotated through the use of Virulence Factors of Pathogenic Bacteria (VFDB) and Carbohydrate-active enZYmes Database (CAZy) v6 databases. The functional annotation of CDS was conducted using the Clusters of Orthologous Groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Finally, genome circle mapping was performed using CGView and Photoshop CS software. In addition, the complete sequences of strain YY-112 have been deposited in the NCBI database (BioSample: SAMN35562085).
2.3. Comparative Genomics Analysis
Utilizing the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 26 June 2023), eight strains of L. plantarum and eight strains of L. pentosus were selected. Their genome sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov; accessed on 26 June 2023) and analyzed in comparison with strain YY-112. A pan-genomic analysis to understand the characteristics of bacterial genome dynamics and to analyze the dynamic changes of bacterial genomes during the evolutionary process [10]. The relationships between the species were assessed at the genome-wide level using average nucleotide identity (ANI).
2.4. Establishment of Immunocompromised Mouse Model and Interventions
Shanghai Slac Laboratory Animal Co., Ltd. provided a total of 60 male BALB/c mice (7 weeks old, 20–22 g body weight (BW)) with a license key SCXK 2017-0005 (Shanghai, China). Mice were housed under a constant temperature (22 ± 1 °C) and a 12 h light/dark cycle with unlimited access to standard laboratory chow and water. The relative humidity was controlled at 55 ± 5%. The experimental protocol was approved by the Ethics Committee of the Zhejiang Academy of Agricultural Sciences, which was carried out in accordance with the ethical treatment guidelines for experimental animals. Permission number: 2019DLSY1945.
The experimental design was based on a previous study conducted by Xu et al. [17]. After seven days of acclimatization, all mice were randomly divided into five groups (n = 12). Immunocompromised mice were continuously injected intraperitoneally with 80 mg/kg/d of CP, followed by another injection 14 days later and 21 days later, respectively. Two normal control groups, one with saline (labeled NC) and the other with 1.0 × 109 CFU/mL L. plantarum YY-112 (labeled NLAB). Another three groups using immunocompromised mouse model under different disposals were as follows: one experimental group was administered with 1.0 × 109 CFU/mL L. plantarum YY-112 (labeled LAB); while one positive control group was administered with 40 mg/kg levamisole (LMS) (labeled PC) and one model group was administered with normal saline (labeled MC). The gavage volume of mice in each group was 0.1 mL/10 g BW/day for 28 days.
During this experiment, their BW was weighed every day. Mice blood was sampled from eyeballs before being sacrificed via cervical dislocation after fasting for 12 h. Blood samples were used for the determination of biochemical indices. In addition, the thymus, kidney, liver, and spleen were, respectively, weighed to determine their organ coefficients (calculated as organ weight/BW). Feces in their colonic tissues were sampled and stored (−8 °C) for the following 16S rRNA sequencing analysis. Their splenic and distal colonic tissue were fixed with 4% paraformaldehyde for histopathological examination.
2.5. Determination of Blood Biochemical Indices
The level of whole-blood lymphocytes (LYMs) was immediately measured after sampling using a self-automated blood cell analyzer (BC-2800 Vet, Shenzhen Mindray Animal Medical Technology Co., Ltd., Shenzhen, China). Other biochemical indices, including serum albumin (ALB), cholesterol (CHO), triglyceride (TG), immunoglobulin (IgA, IgG, and IgM), and inflammatory factor (Interferon-γ (IFN-γ), TNF-α, and IL-6) were, respectively, analyzed through related commercial enzyme-linked immunosorbent assay kits (Xiamen Huijia Biotechnology Co., Ltd., Xiamen, China) based on the instructions.
2.6. Histopathological Analysis
The 4% paraformaldehyde-fixed colon and spleen were stained with hematoxylin and eosin (H&E) after paraffin embedding. Histopathological analysis was performed using a fluorescent microscope (ZEISS, Carl Zeiss Vision, Oberkochen, Germany) [14]. Immunofluorescence staining was performed to detect the expression of Mucin 2 (MUC2), Occludin, and Zona Occludens 1 (ZO-1) in the colon. After dewaxing and rehydrating the paraffin sections, the antigen was recovered by heating in citrate buffer solution, followed by treatment with 3% H2O2 for 10 min. Following phosphate-buffered saline washing, the sections were treated with a 5% BSA solution (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) for 30 min. Tissues were incubated overnight at 4 °C with MUC2, ZO-1 (Thermo Fisher Scientific, Shanghai, China) and Occludin (Abcam, Cambridge, UK) antibodies and then incubated with Fluorescein (FITC)-conjugated Affinipure Goat Anti-Rabbit IgG (H + L) (Proteintech Group, Rosemont, IL, USA) for 1 h. Images were taken under the reverse fluorescent microscope (ZEISS, Carl Zeiss Vision, Oberkochen, Germany) [18].
2.7. Intestinal Microbiota Analysis
Mice intestinal microbiota was analyzed via 16S rRNA sequencing in a commercial sequencing company (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China), following the procedures of genomic DNA extraction, PCR amplification, cloning and sequencing. The genomic DNA was amplified using ABI GeneAmp and Reg 9700 (Applied Biosystems, Carlsbad, CA, USA). Detection and quantification were performed using the QuantiFluor™ -ST Blue Fluorescence Quantification System (Promega Corporation, Beijing, China). After library creation using the TruSeq Nano DNA Low Throughput Library Prep Kit (Illumina Inc., San Diego, CA, USA), paired sequencing was performed using the Illumina Novaseq 6000 platform. To generate representative sequence and abundance information of amplicon sequence variants (ASV) for several statistical or graphical analyses, optimized data were processed using sequence noise reduction techniques.
2.8. Statistical Analysis
All data of the results were presented as mean ± standard error of the mean (SEM). SPSS software (v. 19.0; SPSS Inc., Chicago, IL, USA) was used to statistically analyze, including ordinary one-way analysis of variance (ANOVA) and the unpaired t-test for mean comparisons. Results with p < 0.05 were considered statistically significant (marked with different letters). Additionally, GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA, USA) was used for data processing and figure drawing. With the help of the Majorbio cloud platform (https://cloud.majorbio.com; accessed on 27 June 2023), a comparative genomic analysis of 17 strains was performed, in addition to bioinformatics analysis of the intestinal microbiota using the Kruskal–Wallis H test or Welch’s t-test.
3. Results and Discussion
3.1. Subsection
3.1.1. Whole-Genome Sequencing Analysis of YY-112
Genome Features of YY-112
The data from second-generation sequencing underwent quality assessment (Figure S1), which indicated that the filtered data had superior average quality. The genome of YY-112 was 3,441,908 bp in total length, with 3284 open reading frames (ORFs), a 3,346,642 bp circular chromosome (44.53% G+C content; Accession: CP127090.1) (Figure 1A) and two circular plasmids: plasmid 1 (72,756 bp; 39.51% G+C content; Accession: CP127091.1) and plasmid 2 (22,510 bp; 36.20% G+C content; Accession: CP127092.1) (Figure 1B,C) (Table S1). Figure 1 depicts the genomic distribution of YY-112 from a macroscopic perspective.
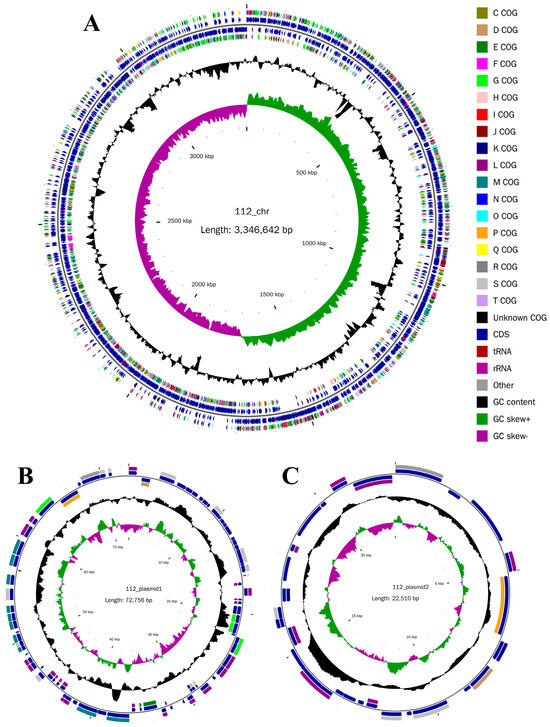
Figure 1.
Circular genome map of YY-112. (A) Chromosome; (B) Plasmid1; (C) Plasmid2. From inside to outside, the first circle represents the scale; the second circle represents GC skew (green represents GC skew+; purple represents GC skew-; GC skew = (G − C)/(G + C), which is used to measure the relative content of G and C. If G > C, the value of GC skew is positive, and G < C is negative); the third circle represents GC content; the fourth and seventh circles represent the COG to which each protein coding region (CDS) (nice blue) belongs; the fifth and sixth circles represent the position of CDS, tRNA, and rRNA on the genome.
Determination of Virulence Factors (VFs)
VFs include bacterial toxins, cell surface proteins that mediate bacterial attachment, cell surface carbohydrates and proteins that protect a bacterium, and hydrolytic enzymes that may contribute to the pathogenicity of the bacterium. The VFDB database mainly manages VFs of bacterial pathogens. However, the factors that induce virulence in some pathogens do not necessarily produce the same results for LAB [19]. Using the VFDB, we predicted eight related genes: five associated with immune modulation and antiphagocytosis (hasC, cpsI, glf, cpsA, and cpsF) and three with stress survival (clpP, clpE, and bsh) (Table S2).
Although genes associated with VFs were found in the genome of YY-112, they were not harmful. Specifically, the bsh gene is extensively present in LAB and encodes biliary salt hydrolase. Biliary salt hydrolase allowed better colonization and survival of host probiotic bacteria in the human gastrointestinal tract and might also be involved in decreasing blood cholesterol levels [20]. This might provide evidence at the genetic level for the ability of YY-112 to tolerate the harsh gastrointestinal environment [16]. The genes cpsI, cpsA, glf, and cpsF are associated with the biosynthesis of capsular polysaccharides (CPS). Bacterial polysaccharides can modulate host immunity by enhancing immunity or inducing immune tolerance. CPS-100 of L. plantarum IMB19 was a potent immunostimulatory molecule that stimulates cells to produce higher levels of IFN-γ, TNF-α, IL-6, and IL-12 [21]. The cps cluster of L. plantarum WCFS1 was associated with Toll-like receptor-dependent human nuclear factor kappa-B activation [22]. Furthermore, Liu et al. demonstrated that the modulatory ability of L. plantarum on the intestinal barrier was attributed to its CPS [23]. The VFs found in the YY-112 genome were likely to be common host interaction factors with non-specific or defensive functions [24]. These results indicated that the EPS-producing strain YY-112 potentially resists stress in the gastrointestinal environment and modulates host immunity.
Carbohydrate-Active Enzymes (CAZymes)
Carbohydrate-active enzymes are responsible for the biosynthesis and breakdown of carbohydrates and glycoconjugates. They are involved in many metabolic pathways and are of great health importance. A total of 135 ORF were found in the CAZymes of YY-112, including 59 glycoside hydrolases (GHs, 43.70%) genes, 39 glycosyltransferases (GTs, 28.89%) genes, 15 carbohydrate esterase (CEs, 11.11%) genes, 10 genes encoding auxiliary activities (AAs, 7.41%), 11 genes for enzymes related to carbohydrate-binding modules (CBMs, 8.15%), and one gene for polysaccharide lytic enzymes (PLs, 0.74%) (Table S3).
It has been demonstrated that GHs can hydrolyze or rearrange glycosidic linkages, while GTs are involved in the biosynthesis of disaccharides, oligosaccharides, and polysaccharides by catalyzing glycosidic linkages formation between them [25,26]. The numerous GHs (GH13, GH1, GH25, GH109, etc.) gave YY-112 the capacity to metabolize various carbohydrates efficiently [27]. Consequently, new glycosides and sapogenins were produced, which provide precursors for glycoconjugates such as EPS. The abundance of GTs (GT2, GT4, etc.) could assemble monosaccharide repeat units onto lipid carriers for reverse polymerization [26], given YY-112’s good ability to synthesize polysaccharides. Among them, YY-112 contained glucosyltransferases (EC 2.4.1.52, EC 2.4.1.157, EC 2.4.1.117), mannosyltransferases (EC 2.4.1.57, EC 2.4.1.83), galactosyltransferases (EC 2.4.1.-), and rhamnosyltransferases (EC 2.4.1.-). This should confirm the EPS-producing ability displayed by YY-112 in the earlier research (monosaccharide composition: glucose, mannose, glucosamine, galactose, and rhamnose). In particular, GHs and GTs were critical for surface structures recognized by immune receptors and cell molecular surface-mediated interactions between immune cells [28,29]. Indeed, we provided evidence for the previous findings at the genetic level and implicated that YY-112 can modulate the immune response.
COG and KEGG Database Annotation
To perform a comprehensive functional annotation for the inferred orthologs, the genes were classified via COG analysis. Based on the COG database, 2743 coding genes were specifically assigned to 19 categories (Figure 2A). Higher abundance of genes assigned to transcription (10.17%), carbohydrate transport and metabolism (9.70%), cell wall/membrane/envelope biogenesis (7.47%), amino acid transport and metabolism (7.29%) and replication, recombination and repair (7.25%). The carbohydrate transport and metabolism category were rich in phosphotransferase transport system (PTS) systems (21.80%). The PTS sugar transporter protein identified in the YY-112 genome showed broad substrate specificity, containing glucitol, sorbitol, mannose, fructose, sorbitol, galactitol, etc., allowing YY-112 to absorb a wide range of carbohydrates from the environment.
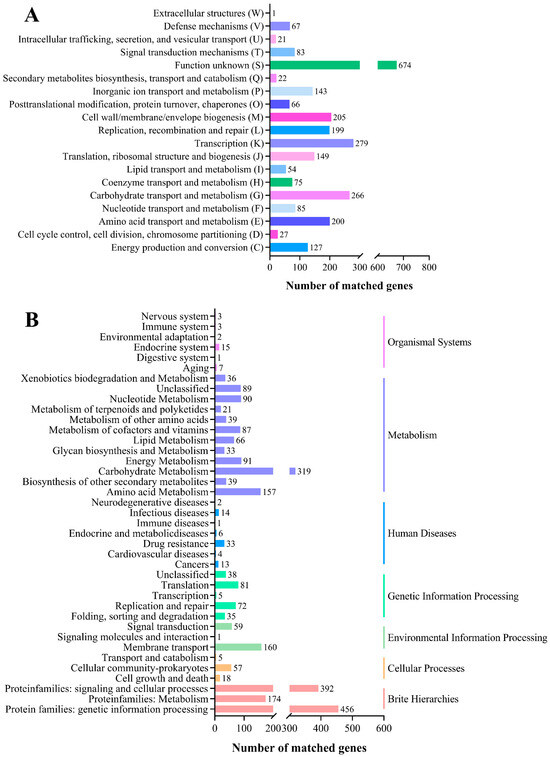
Figure 2.
Function classification of YY-112. (A) Histogram presentation of COG classification. The ordinate represents the COG classification entry whose abbreviations are shown in parentheses, and the abscissa represents the number of genes; (B) histogram presentation of the KEGG pathway. The ordinate is the name of the KEGG pathway, and the abscissa is the number of genes annotated to this pathway.
A network of molecular interactions and responses within the species was obtained through KEGG pathway annotation to clarify its underlying metabolic pathways. The genes with high abundance in these categories (Figure 2B) were consistent with previous studies [30]. It was shown that YY-112 could adapt well to challenging environmental conditions and possess a variety of metabolic pathways with excellent energy metabolism as well as material transport capabilities. Genes involved in propionate production were present in YY-112, including genes for the enzymes L-lactate dehydrogenase (EC:1.1.1.27), acetate kinase (EC:2.7.2.1), formate C-acetyltransferase (EC:2.3.1.54), and phosphate acetyltransferase (EC:2.3.1.8). These key enzymes constitute the complete metabolic pathway from 2-hydroxybutyrate to propionate. In addition, several genes encoding key enzymes of the acetate synthesis pathway were also identified, such as acylphosphatase (EC:3.6.1.7), acetate kinase (EC 2.7.2.1), pyruvate oxidase (EC:1.2.3.3), and pyruvate oxidase (EC:1.2.3.3). The process of acetate utilization is closely related to butyrate production that butyrate-producing bacteria can use acetate to produce butyrate in the intestine [31].
SCFAs are thought to play an important role in the regulation of the host immune system. According to Wu et al., Clostridium butyricum SLZX19-05 could increase propionate and butyrate production, thus improving intestinal health and weaning stress in early-weaned piglets [32]. Huang et al. indicated that L. plantarum DMDL 9010 increased the production of butyric acid and propionate that favored the regulation of intestinal immunity, thereby reducing colonic inflammation caused by dextran sulfate sodium [33]. Accordingly, we hypothesized that YY-112 had an excellent metabolic and environmental adaptation capacity. This allowed YY-112 to participate in multiple carbohydrate metabolism in the gastrointestinal tract to produce SCFAs, thereby enhancing host immunity.
Comparative Genome Analysis
Table S4 describes the genomic characteristics of the 17 strains used for comparative genomic analysis. The results showed that the genomic information of the strain YY-112 was closer to that of L. plantarum, including chromosome size and G+C content. ANI is a reliable method for estimating genetic correlations between genomes, where organisms belonging to one species usually show ≥95% ANI [34]. The ANI analysis results are shown in Figure 3A, and the ANI values between the strain YY-112 and eight L. plantarum strains were all greater than 0.989. Among them, the strain YY-112 had the highest ANI value of 0.993 with L. plantarum XJ25. Therefore, strain YY-112 has higher homology with L. plantarum, and strain YY-112 should be reclassified as L. plantarum rather than L. pentosus. Carbohydrate fermentation assays and 16S rRNA gene sequence information for L. plantarum and L. pentosus were very close. It was reported that both L. pentosus LTJ12 and L. pentosus KCA1 had been identified as L. plantarum, which was corrected based on whole-genome sequencing results in a follow-up study [10,35].
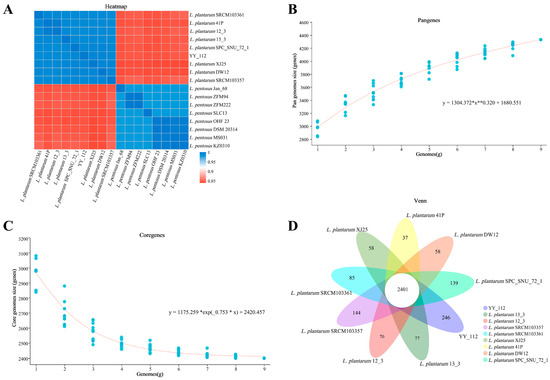
Figure 3.
Comparative genomic analysis of YY-112. (A) Heatmap of ANI matrix of YY-112, eight L. plantarum, and eight L. pentosus; (B,C) curves of YY-112 and eight L. plantarum pan-genome size (B) and core genome size change with the number of genomes, with the horizontal coordinate being the number of genomes and the vertical coordinate being the size of genomes. * represents the multiplication sign, and x raised to the power of 0.32 is conveniently written as x**0.32; (D) Venn diagram shows the number of core and unique genes.
Homologous genes were analyzed between eight selected L. plantarum strains and the strain YY-112. The pan-genome represents the genomic diversity of a species and includes both core and variable genes. Variable genes are absent in some individuals, including dispensable genes and unique genes, and core genes are present in all individuals [36]. As shown in Figure 3B,C, it can be concluded that the L. plantarum pangenome is open type. This represents that per additional genome sequenced increases the pan-genome size [36].
Further, the distribution of the pan-genome is displayed in a Venn diagram to visualize the number of core and unique genes in the sample (Figure 3D). The results showed that nine L. plantarum strains had 2401 homologous genes, while L. plantarum YY-112 had the highest number of unique homologous genes (246 genes). This suggested that L. plantarum YY-112 may have more physiological activities and more functional properties. Most of these unique genes have unknown functions and are partially related to transcriptional regulators (CLUSTER3879, CLUSTER3921, CLUSTER3968, CLUSTER3983, and CLUSTER4046), exopolysaccharide biosynthesis protein (CLUSTER3459) and capsular polysaccharide biosynthesis protein (CLUSTER4037), etc. Transcriptional regulators regulate gene transcription to maintain normal cellular processes and respond to various environmental stresses [37]. The EPS of L. plantarum YY-112 might distinguish it from other L. plantarum.
In particular, there are many unique genes related to transferase, including glycosyltransferase involved in cell wall biosynthesis (CLUSTER3948, CLUSTER3951, CLUSTER3997, and CLUSTER4010), mannosyltransferase OCH1 or related enzyme (CLUSTER4054), phosphotransferase system IIC components, glucose/maltose/N-acetylglucosamine-specific (CLUSTER4005), glycosyltransferase, GT2 family (CLUSTER3925), UDP-N-acetylglucosamine transferase subunit ALG13 (CLUSTER3903), and UDP-N-acetylglucosamine: LPS N-acetylglucosamine transferase (CLUSTER3864). The unique transferase contributed to the critical step in the metabolic pathway adjusted by L. plantarum YY-112 to efficiently influence the formation of metabolites in the gastrointestinal tract, thereby interfering with host immunity. In addition, immune receptor recognition and molecular surface mediation of immune cells might also differ from other L. plantarum [28,29]. However, the detailed mechanism needs to be further investigated.
In conclusion, whole-genome analysis showed that L. plantarum YY-112 has immunomodulatory potential, and an immunocompromised mouse model was established to assess its immune efficacy.
3.2. Direct Therapeutic Effect of L. plantarum YY-112 on Immune Status in Mice
3.2.1. Effects of L. plantarum YY-112 on BW and Organ Coefficients
The experimental design is shown in Figure 4A. The effects of L. plantarum YY-112 on the BW of CP-induced immunocompromised mice are shown in Figure S2A. Compared with the NC group, the BW significantly decreased after each CP injection of the MC group, especially at the end of the first injection on day 3 (p < 0.05). The results confirmed that CP affected the normal growth of mice and that CP can cause weight loss and imbalance of various leukocytes in peripheral blood in mice [38]. In addition, the BW in the NLAB group increased slightly than the NC group without significance, suggesting that the strain did no harm in terms of mouse growth. However, LMS and L. plantarum YY-112 did not restore the BW levels in immunocompromised mice, since there was no significant difference between the MC, PC, and LAB group. Their protective effect on immune mice might not be directly reflected in weight gain.
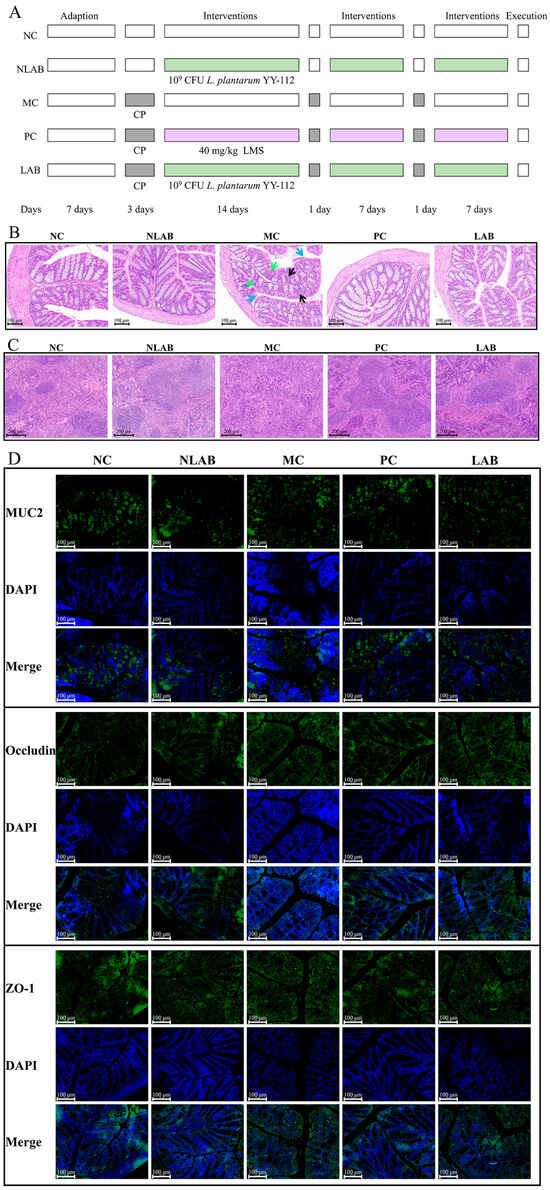
Figure 4.
Experimental design and effects of L. plantarum YY-112 on Colon and Spleen Histomorphology. (A) Schematic diagram of the animal experimental design; (B) representative images of H&E staining of the colon. Magnification: 300×. Blue arrows point to damaged epithelial cells, green arrows point to decreased goblet cells, and black arrows point to shortened crypts; (C) representative images of H&E staining of the spleen. Magnification: 200×; (D) immunofluorescence staining of MUC2, Occludin, and ZO-1. Magnification 400×. Mucin 2, MUC2; Zona Occludens 1, ZO-1. NC, normal saline; NLAB, 1.0 × 109 CFU/mL L. plantarum YY-112; MC, normal saline; PC, 40 mg/kg LMS; LAB, 1.0 × 109 CFU/mL L. plantarum YY-112.
The organ coefficients reflect their immune status to some extent. As shown in Figure S2B–E, there were no obvious changes in thymus, kidney, liver, and spleen coefficients between NC and NLAB groups, suggesting that L. plantarum YY-112 had no impaired effects on mouse organs. On the other hand, organ coefficients of the thymus, liver and spleen in the MC group were significantly higher than the NC group (p < 0.05), slightly higher than the PC and LAB groups. LMS and L. plantarum YY-112 could somewhat reduce the CP-induced increase in organ coefficients, revealing that they might relieve organ enlargement to some extent. Notably, L. plantarum YY-112 achieved a remission consistent with positive drug LMS. This was similar to previous findings that CP may have impaired host hematopoiesis, allowing extramedullary hematopoiesis to occur in the spleen, causing the onset of compensatory enlargement [39,40].
3.2.2. Effects of L. plantarum YY-112 on Mouse Colon and Spleen Histomorphology
To test the direct effect of L. plantarum YY-112 on immunity, we evaluated pathological tissue sections on the mouse colon and spleen. The histomorphology of the colon can be seen in Figure 4A. No abnormal histomorphometry changes occurred in the NLAB group compared to the NC group, showing that L. plantarum YY-112 did not cause colon damage. As for the MC groups, there existed fewer goblet cells and mucus, partially detached epithelial cells, and shortening of the crypts. Interestingly, after treatment with LSM and L. plantarum YY-112, the layers of the colon were clearer, the crypts were normalized, and mucus-producing goblet cells were increased.
It has been reported that the intestinal mucus layer and tight junction (TJ) proteins play a key role in maintaining the intestinal epithelial barrier. The main component of the intestinal mucus layer was MUC2. TJ proteins, such as ZO-1 and Occludin, are tightly apposed between adjacent cells to form a natural intestinal mechanical barrier [41]. As shown in Figure 4B, MUC2 was located in the crypt inner cup cells, whereas ZO-1 and Occludin were located in the cell membrane. The distribution of MUC2 expression in the other groups, excluding the MC group, exhibited a more predictable crypt-like pattern. Clear crypt structures and the MUC2 protein it encapsulates were observed in the LAB group compared to the MC group. In addition, the distribution and expression of Occludin and ZO-1 were not significantly altered in the MC group. Interestingly, however, the membrane protein network of TJ proteins in the crypts was well structured and dense after the L. plantarum YY-112 treatment. This was consistent with the results observed in H&E staining that L. plantarum YY-112 potentially enhances the intestinal chemical and mechanical barriers. However, additional research was required to ascertain whether L. plantarum YY-112 can enhance the expression of MUC2 and TJ proteins.
The histomorphology of the spleen can be seen in Figure 4A. In the NC group, the morphology of the red pulp and white pulp of the spleen was normal, and the boundary between them was clear. In the NLAB group, the spleen retained its normal white and red pulp structure, indicating that L. plantarum YY-112 had no adverse effect on the mouse spleen. The spleen is a secondary lymphoid organ with red pulps containing many myeloid cells and white pulps consisting of lymphoid tissue containing lymphocyte production centers [42]. The white pulp of the MC group was dysplastic with mixed red and had an uneven cell alignment. We could observe a decrease in LYMs and myeloid cells. This may be due to immunosuppression and damage to the spleen in mice, which was also observed by previous researchers [39]. Compared with the MC group, the boundary between the red and white pulp was clearer in the PC and LAB groups. In addition, we could visibly observe a larger area of white pulp. It revealed that LMS and L. plantarum YY-112 helped to increase LYMs in spleen tissue and restore spleen morphology, thus improving immune status.
In general, considering the results of H&E staining and immunofluorescence, L. plantarum YY-112 had no adverse effects on mice, improved the immune status of immunocompromised mice, promoted lymphocyte proliferation, protected spleen tissue, and strengthened intestinal chemical and mechanical barriers.
3.3. Modulation of the Immune Environment in Mice by L. plantarum YY-112
The hematological parameters, cytokine, and immunoglobulin levels were measured to confirm the modulatory effect of L. plantarum YY-112 on the immune environment in mice. We found that L. plantarum YY-112 did not cause significant changes in various indices of normal mice, indicating that it did not impair the immune environment of normal mice (Figure 5).
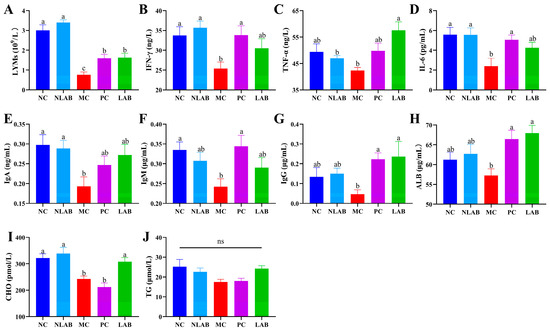
Figure 5.
Effects of L. plantarum YY-112 on immune environment. (A) LYMs; (B) IFN-γ; (C) TNF-α; (D) IL-6; (E) IgA; (F) IgM; (G) IgG; (H) ALB; (I) CHO; (J) TG. NC, normal saline; NLAB, 1.0 × 109 CFU/mL L. plantarum YY-112; MC, normal saline; PC, 40 mg/kg LMS; LAB, 1.0 × 109 CFU/mL L. plantarum YY-112. Lymphocytes, LYMs; interferon-γ, IFN-γ; tumor necrosis factor α, TNF-α; interleukin-6, IL-6; immunoglobulin A, IgA; immunoglobulin M, IgM; immunoglobulin G, IgG; albumin, ALB; cholesterol, CHO; triglyceride, TG. Values were expressed as mean ± SEM, assessed by one-way ANOVA with the Waller–Duncan test. The different letters (a, b, and c) represent significant differences between different groups (p < 0.05).
LYMs are white blood cells with essential immune functions and are the key performers of immune functions [43]. Compared with the NC group, there was a marked decrease in the number of LYMs in the peripheral blood of the MC group (p < 0.05) (Figure 5A). However, the levels of LYMs were significantly recovered in the PC and LAB groups compared with the MC group (p < 0.05). This was consistent with the results of H&E staining. It further illustrated that L. plantarum YY-112 might restore the CP-induced decrease in lymphocyte levels as to exert immunomodulatory effects.
Cytokines are critical in regulating and maintaining immune homeostasis in the body [43]. IFN-γ promotes T-cell differentiation of macrophages and enhances antimicrobial immunity [44]. TNF-α inhibits intracellular pathogens and controls inflammatory processes. IL-6 regulates the immune response and the differentiation of T and B cells [45]. As seen in Figure 5B–D, the serum levels of INF-γ, TNF-α, and IL-6 were decreased in mice after CP injection compared with the NC group. In contrast, both LMS and L. plantarum YY-112 were able to reverse this trend. Several researchers have been conducted to treat immune diseases with immunomodulatory cytokines, the results of which are consistent with our findings [14,45]. It was indicated that L. plantarum YY-112 could enhance host immunity by stimulating cytokine production.
Serum immunoglobulins, including immunoglobulins A, G and M (IgA, IgG and IgM), belong to the adaptive immune system. IgM provides a rapid immune response and participates in tissue homeostasis. IgG is the primary antibody in every immune response of the body, while IgA provides mainly mucosal immunity [46]. The serum levels of IgA, IgG, and IgM are shown in Figure 5E–G. Compared with the NC group, immunoglobulin levels decreased in the MC group, especially IgA and IgM levels (p < 0.05). Treatment with LMS and L. plantarum YY-112 recovered immunoglobulin levels, approaching those of normal mice. Similar results were observed in previous studies [47]. Therefore, L. plantarum YY-112 could attenuate the CP-induced decreased immunoglobulin levels, thus exerting immunomodulatory effects and improving immune function in mice.
The concentration of ALB reflects the anabolic and reserve functions of the liver. CHO and TG levels reflect liver lipid metabolism. The results showed that LMS and L. plantarum YY-112 could significantly recover ALB levels to bring them close to normal mice (p < 0.05), while LMS could not restore CHO levels. There was no significant difference in TG levels between the groups, but the same trend as the change in CHO content was observed. This suggested that L. plantarum YY-112 could regulate liver function in CP-injured mice. Although we did not observe a noticeable recovery in the BW of mice in the LAB group, the elevated ALB, CHO, and TG levels illustrated that L. plantarum YY-112 could regulate body fat levels, which might not yet be visually reflected in BW.
In conclusion, as an immunosuppressive agent, CP may disrupt DNA structure and immune cells, thus suppressing cellular and humoral immune levels [48]. However, L. plantarum YY-112 ameliorated CP-induced immunosuppression and recovered serum levels of LYMs, cytokines, immunoglobulins, and lipids. Herein, L. plantarum YY-112 could exert immunomodulatory effects by promoting the proliferation of lymphocytes to promote immune factor secretion.
3.4. Effects of L. plantarum YY-112 on the Intestinal Microbiota in Mice
Numerous studies have shown that LAB can modulate the host immune system by regulating the composition of the intestinal microbiota [49]. To minimize the impact of sequencing depth on subsequent data analysis, the number of sequences of all samples was drawn flat to 43,085 and divided into 3517 ASVs. The L. plantarum YY-112 treatment resulted in an increase in Chao1 and Shannon indices compared to the NC and MC groups (Figure 6A,B). L. plantarum YY-112 had the potential to promote the richness and diversity of intestinal microbiota. Figure 6C demonstrates the relative abundance of intestinal microbiota at the phylum level in mice. Firmicutes, Desulfobacterota, Proteobacteria, Actionobacteria, Verrucomicrobia, and Bacteroidetes, were the dominant phyla in the intestinal microbiota. Compared to the NC group, the MC group showed a decrease in the relative abundance of Firmicutes and an increase in that of Desulfobacterota, Proteobacteria, and Actionobacteria. Notably, L. plantarum YY-112 ameliorated the CP-induced alterations in the phylum level. At the genus level, the community composition of the MC and NC groups differed considerably (Figure 6D). In addition, a greater proportion of Akkermansia was observed in the NLAB group, which explains the greater relative abundance of Verrucomicrobiota in the NLAB group.
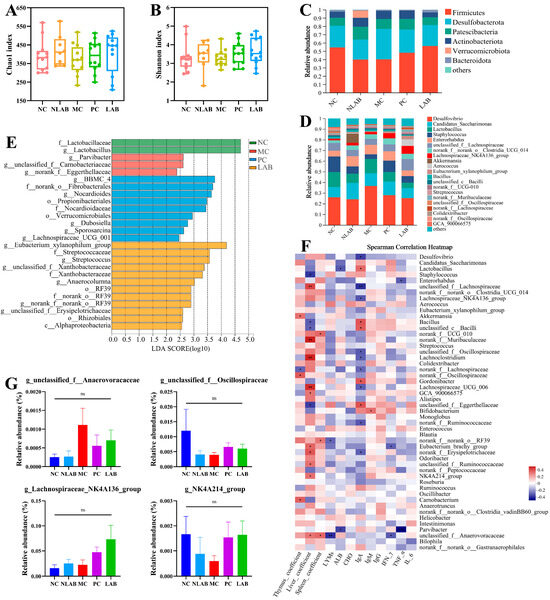
Figure 6.
Effect of L. plantarum YY-112 on intestinal microbiota. (A) Chao1 index; (B) Shannon index; (C,D) community composition at the phylum level (C) and genus level (D) of each group; (E) LEfSe analysis; (F) Spearman correlation analysis between different intestinal microbiota at genus level and immune-related indicators; (G) the relative abundances of unclassified_f__Anaerovoracaceae, unclassified_f_O-scillospiraceae, Lachnospiraceae_NK4A136_group, and NK4A214_group. Values were expressed as mean ± SEM and were analyzed by one-way ANOVA with the Tukey multiple comparison test using GraphPad Prism 8.0 Software. NC, normal saline; NLAB, 1.0 × 109 CFU/mL L. plantarum YY-112; MC, normal saline; PC, 40 mg/kg LMS; LAB, 1.0 × 109 CFU/mL L. plantarum YY-112. * p < 0.05 and ** p < 0.01, while ns means no significant difference (p > 0.05).
Species characteristics of intergroup differences were further analyzed using LEfSe analysis (LDA > 2; p < 0.05; Figure 6E). In the MC group, the biomarkers were Parvibacter, unclassified_f__Carnobacteriaceae, and norank_f__Eggerthellaceae. In the PC group, the levels of BBMC-4, norank_o__Fibrobacterales, and Nocardioides were significantly increased (LDA > 3.64). In the LAB group, Eubacterium_xylanophilum_group, Streptococcus, Xanthobacteraceae, and RF39 were significantly increased (LDA > 3.56).
Parvibacter is a member of the Coriobacteriaceae family, which has a significant role in detoxification [50]. Its increase in the MC group might be due to a protective response of the intestinal immune system to CP injection. In a previous study, researchers found an increase in the relative abundance of Carnobacteriaceae in the gut during accelerated mortality of Plutella xylostella [51]. Eggerthellaceae is more abundant in the intestinal microbiota of hosts with severe diseases [52]. Obviously, CP induced unfavorable changes in mice intestinal microbiota. Eubacterium xylanophilum is noted to ferment phytochemicals and produce SCFAs, such as butyrate, while butyrate has anti-obesity effects [53]. Streptococcus was reported to be positively correlated with IL-6, serum TG, and liver weight [54]. Those might explain why we failed to observe a significant increase in BW but an improvement in mice’s body condition. Xanthobacteraceae, enriched in the LAB group, shows great potential in carbon-related metabolism [55]. Anaerocolumna, belonging to Lachnospiraceae, is a widespread anaerobic bacterium in the anaerobic digestive system, capable of utilizing a wide range of carbohydrates and fermenting to produce acetate [56]. Order RF39 is the potential probiotic. Yu et al. assessed the correlation between long-term diet quality and gut microbes in 1920 adults and found that the relative abundance of RF39 was positively correlated with diet quality [57].
The above results showed that CP treatment-induced changes in the intestinal microbiota of mice were undesirable, while the intervention of L. plantarum YY-112 could improve the structure of the intestinal microbiota. In particular, L. plantarum YY-112 was significantly enriched in the Eubacterium_xylanophilum_group, Xanthobacteraceae, and Anaerocolumna, which favor fermentable carbohydrates. This echoed the findings of the whole-genome analysis that L. plantarum YY-112 intervened in host immunity through superior metabolic capacity to produce SCFAs. However, this conjecture needs to be supported by further data.
In addition, we assessed the correlation between intestinal microbiota (top 50) and biochemical indexes via the Spearman calculation method (Figure 6F). We found that some bacteria were closely associated with immune indicators. The relative abundance of the biomarker Parvibacter in the MC group was significantly but negatively correlated with ALB and TNF-α levels (p < 0.05). The relative abundance of unclassified_f__Anaerovoracaceae increasing under CP treatment was markedly but negatively correlated with the content of LYMs and IFN-γ (p < 0.05) (Figure 6G). The relative abundance of the biomarker Eubacterium_xylanophilum_group in the LAB group was positively correlated with the levels of IgM, IgG, and ALB, when Streptococcus positively correlated with TNF-α, IFN-γ, and ALB (p < 0.05). Following the intervention of L. plantarum YY-112, the relative abundance of unclassified_f_Oscillospiraceae, Lachnospiraceae_NK4A136_group, and NK4A214_group dramatically increased, positively correlating with IgM and IgG content (Figure 6G). The results indicated that L. plantarum YY-112 could regulate the host immune system by modulating intestinal microbiota consumption. In general, L. plantarum YY-112 might regulate host immune function in a favorable direction through modulated mice intestinal microbiota compositions.
4. Conclusions
Based on the results of whole-genome analysis, it could be concluded that L. plantarum YY-112 is a probiotic candidate with excellent metabolic capacity and potential to modulate host immunity. The present experiments in vivo suggested that L. plantarum YY-112 had no adverse effects on mice. It repaired the injured spleen and recovered the level of LYMs, cytokines, and immunoglobulins. According to histopathological analysis, the chemical and mechanical barriers of the intestine were protected by L. plantarum YY-112. Furthermore, the intervention of L. plantarum YY-112 improved the structure of the intestinal microbiota and showed more promise in fermenting carbohydrates to produce SCFAs. However, the specific mechanism by which L. plantarum YY-112 exerts its immune effects remains to be further explored, particularly the relationship between L. plantarum YY-112 and its metabolites with intestinal microbes and host immunity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9120996/s1, Figure S1: Quality control chart of partial next-generation sequencing data; Figure S2: Effects of L. plantarum YY-112 on BW and organ coefficients; Table S1: General genome features of L. plantarum YY-112; Table S2: Distribution of virulence factors in L. plantarum YY-112; Table S3: Distribution of CAZymes in L. plantarum YY-112; Table S4: Genomic characterization of strain YY-112, selected L. plantarum and L. pentosus.
Author Contributions
Conceptualization, Y.Y. and Y.G.; Data curation, M.L. and W.Z.; Formal analysis, M.L., W.Z., W.T. and J.L.; Funding acquisition, Y.Y. and Y.G.; Investigation, J.X., Y.Y. and Y.G.; Methodology, M.L., J.L. and Y.Y.; Project administration, Y.Y.; Resources, J.X., Y.Y. and Y.G.; Software, M.L., W.Z. and W.T.; Supervision, J.X., Y.Y. and Y.G.; Validation, W.Z., W.T. and J.L.; Visualization, M.L.; Writing—original draft, M.L.; Writing—review and editing, W.Z. and W.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Research and Development Project in Zhejiang Province (grant number 2020C02034), the National Natural Science Foundation of China (grant number 31972093) and Forestry Science and Technology Innovation in Jiangsu Province (grant number LYKJ [2022]12).
Institutional Review Board Statement
The animal study protocol was approved by the Laboratory Animal Welfare and Ethics Committee of Zhejiang Academy of Agricultural Sciences (Permission number: 2019DLSY1945).
Data Availability Statement
Data will be made available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chang, X.; Shen, Y.; Yun, L.; Wang, X.; Feng, J.; Yang, G.; Meng, X.; Zhang, J.; Su, X. The antipsychotic drug olanzapine altered lipid metabolism in the common carp (Cyprinus carpio L.): Insight from the gut microbiota-SCFAs-liver axis. Sci. Total Environ. 2023, 856, 159054. [Google Scholar] [CrossRef]
- Mayo, B.; Flórez, A.B. Lactic Acid Bacteria: Lactobacillus plantarum. In Encyclopedia of Dairy Sciences, 3rd ed.; McSweeney, P.L.H., McNamara, J.P., Eds.; Academic Press: Oxford, UK, 2022; pp. 206–217. [Google Scholar]
- DiBaise, J.K.; Lof, J.; Taylor, K.; Quigley, E.M. Lactobacillus plantarum 299V in the irritable bowel syndrome: A randomized, double-blind, placebo-controlled crossover study. Gastroenterology 2000, 118, A615. [Google Scholar] [CrossRef]
- Peñalva, R.; Martínez-López, A.L.; Gamazo, C.; Gonzalez-Navarro, C.J.; González-Ferrero, C.; Virto-Resano, R.; Brotons-Canto, A.; Vitas, A.I.; Collantes, M.; Peñuelas, I.; et al. Encapsulation of Lactobacillus plantarum in casein-chitosan microparticles facilitates the arrival to the colon and develops an immunomodulatory effect. Food Hydrocoll. 2023, 136, 108213. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, J.; Sun, R.; Wang, M.; Wang, K.; Li, Y.; Shang, H.; Hou, J.; Jiang, Z. Lactobacillus plantarum 23-1 improves intestinal inflammation and barrier function through the TLR4/NF-κB signaling pathway in obese mice. Food Funct. 2022, 13, 5971–5986. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ren, Z.; Huo, Y.; Yang, W.; Peng, L.; Lv, H.; Nie, L.; Wei, H.; Wan, C. Targeting the gut microbiota to investigate the mechanism of Lactiplantibacillus plantarum 1201 in negating colitis aggravated by a high-salt diet. Food Res. Int. 2022, 162, 112010. [Google Scholar] [CrossRef]
- Koduru, L.; Lakshmanan, M.; Lee, Y.Q.; Ho, P.-L.; Lim, P.-Y.; Ler, W.X.; Ng, S.K.; Kim, D.; Park, D.-S.; Banu, M.; et al. Systematic evaluation of genome-wide metabolic landscapes in lactic acid bacteria reveals diet- and strain-specific probiotic idiosyncrasies. Cell Rep. 2022, 41, 111735. [Google Scholar] [CrossRef]
- Peng, X.; Ed-Dra, A.; Yue, M. Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 2022, 2022, 2087174. [Google Scholar] [CrossRef]
- Akaçin, İ.; Ersoy, Ş.; Doluca, O.; Güngörmüşler, M. Comparing the significance of the utilization of next generation and third generation sequencing technologies in microbial metagenomics. Microbiol. Res. 2022, 264, 127154. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, C.; Xu, Q.; Li, Z.; Song, Y.; Zhou, S.; Guo, L.; Zhang, T.; Luo, X. Comparative Genomics Analysis Provides New Insights into High Ethanol Tolerance of Lactiplantibacillus pentosus LTJ12, a Novel Strain Isolated from Chinese Baijiu. Foods 2023, 12, 35. [Google Scholar] [CrossRef]
- Delves, P.J.; Roitt, I.M. The immune system. First of two parts. N. Engl. J. Med. 2000, 343, 37–49. [Google Scholar] [CrossRef]
- Panarelli, N.C.; Yantiss, R.K. Inflammatory and infectious manifestations of immunodeficiency in the gastrointestinal tract. Mod. Pathol. 2018, 31, 844–861. [Google Scholar] [CrossRef]
- Bender, M.J.; McPherson, A.C.; Phelps, C.M.; Pandey, S.P.; Laughlin, C.R.; Shapira, J.H.; Medina Sanchez, L.; Rana, M.; Richie, T.G.; Mims, T.S.; et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell 2023, 186, 1846–1862.e1826. [Google Scholar] [CrossRef]
- Min, F.; Hu, J.; Huang, T.; Huang, Y.; Nie, S.; Xiong, T.; Xie, M. Effects of Lactobacillus casei NCU011054 on immune response and gut microbiota of cyclophosphamide induced immunosuppression mice. Food Chem. Toxicol. 2023, 174, 113662. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, D.H.; Jo, S.; Cho, M.J.; Cho, Y.R.; Lee, Y.J.; Byun, S. Immunomodulatory functional foods and their molecular mechanisms. Exp. Mol. Med. 2022, 54, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhou, W.; Yang, Y.; Xing, J.; Xu, X.; Lin, Y. Potential prebiotic properties of exopolysaccharides produced by a novel Lactobacillus strain, Lactobacillus pentosus YY-112. Food Funct. 2021, 12, 9456–9465. [Google Scholar] [CrossRef]
- Xu, D.; Hu, J.; Zhong, Y.; Zhang, Y.; Liu, W.; Nie, S.; Xie, M. Effects of Rosa roxburghii & edible fungus fermentation broth on immune response and gut microbiota in immunosuppressed mice. Food Sci. Hum. Wellness 2024, 13, 154–165. [Google Scholar] [CrossRef]
- Li, S.; Chen, Y.; Zhang, Y.; Lv, H.; Luo, L.; Wang, S.; Guan, X. Polyphenolic Extracts of Coffee Cherry Husks Alleviated Colitis-Induced Neural Inflammation via NF-κB Signaling Regulation and Gut Microbiota Modification. J. Agric. Food. Chem. 2022, 70, 6467–6477. [Google Scholar] [CrossRef] [PubMed]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic resistance and virulence factors in lactobacilli: Something to carefully consider. Food Microbiol. 2022, 103, 103934. [Google Scholar] [CrossRef]
- Kusada, H.; Morinaga, K.; Tamaki, H. Identification of Bile Salt Hydrolase and Bile Salt Resistance in a Probiotic Bacterium Lactobacillus gasseri JCM1131T. Microorganisms 2021, 9, 1011. [Google Scholar] [CrossRef]
- Garcia-Vello, P.; Sharma, G.; Speciale, I.; Molinaro, A.; Castro, C.D. Structural features and immunological perception of the cell surface glycans of Lactobacillus plantarum: A novel rhamnose-rich polysaccharide and teichoic acids. Carbohydr. Polym. 2020, 233, 115857. [Google Scholar] [CrossRef]
- Remus, D.M.; van Kranenburg, R.; van Swam, I.I.; Taverne, N.; Bongers, R.S.; Wels, M.; Wells, J.M.; Bron, P.A.; Kleerebezem, M. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb. Cell Fact. 2012, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Strain-specific regulative effects of Lactobacillus plantarum on intestinal barrier dysfunction are associated with their capsular polysaccharides. Int. J. Biol. Macromol. 2022, 222, 1343–1352. [Google Scholar] [CrossRef]
- Sui, S.J.H.; Fedynak, A.; Hsiao, W.W.L.; Langille, M.G.I.; Brinkman, F.S.L. The Association of Virulence Factors with Genomic Islands. PLoS ONE 2009, 4, e8094. [Google Scholar] [CrossRef]
- Lammens, W.; Roy, K.L.; Schroeven, L.; Laere, A.V.; Rabijns, A.; Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef]
- Soumya, M.P.; Nampoothiri, K.M. An overview of functional genomics and relevance of glycosyltransferases in exopolysaccharide production by lactic acid bacteria. Int. J. Biol. Macromol. 2021, 184, 1014–1025. [Google Scholar] [CrossRef]
- Zhan, M.; Wang, L.; Xie, C.; Fu, X.; Zhang, S.; Wang, A.; Zhou, Y.; Xu, C.; Zhang, H. Succession of Gut Microbial Structure in Twin Giant Pandas During the Dietary Change Stage and Its Role in Polysaccharide Metabolism. Front. Microbiol. 2020, 11, 551038. [Google Scholar] [CrossRef] [PubMed]
- Lihong, Z.; Hongcai, M.; FakharEAlam, K.M.; Huachun, P.; Kewei, L.; Aoyun, L.; Quan, M.; Yaping, W.; Hailong, D.; Yuhua, B.; et al. Complete genome analysis of Lactobacillus fermentum YLF016 and its probiotic characteristics. Microb. Pathogen. 2021, 162, 105212. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, D.; Jia, X.; Liang, M.; Lu, Y.; Liu, J. Whole genome sequencing of Lactobacillus plantarum DMDL 9010 and its effect on growth phenotype under nitrite stress. LWT 2021, 149, 111778. [Google Scholar] [CrossRef]
- Duncan, S.H.; Barcenilla, A.; Stewart, C.S.; Pryde, S.E.; Flint, H.J. Acetate Utilization and Butyryl Coenzyme A (CoA):Acetate-CoA Transferase in Butyrate-Producing Bacteria from the Human Large Intestine. Appl. Environ. Microbiol. 2002, 68, 5186–5190. [Google Scholar] [CrossRef]
- Wu, J.; Wang, J.; Lin, Z.; Liu, C.; Zhang, Y.; Zhang, S.; Zhou, M.; Zhao, J.; Liu, H.; Ma, X. Clostridium butyricum alleviates weaned stress of piglets by improving intestinal immune function and gut microbiota. Food Chem. 2023, 405, 135014. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Wu, Y.P.; Jia, X.Z.; Lin, J.; Xiao, L.F.; Liu, D.M.; Liang, M.H. Lactiplantibacillus plantarum DMDL 9010 alleviates dextran sodium sulfate (DSS)-induced colitis and behavioral disorders by facilitating microbiota-gut-brain axis balance. Food Funct. 2022, 13, 411–424. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Anukam, K.C.; Macklaim, J.M.; Gloor, G.B.; Reid, G.; Siezen, R.J. Genome Sequence of Lactobacillus pentosus KCA1: Vaginal Isolate from a Healthy Premenopausal Woman. PLoS ONE 2013, 8, e59239. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Bayer, P.E.; Bhalla, P.L.; Batley, J.; Edwards, D. Pangenomics Comes of Age: From Bacteria to Plant and Animal Applications. Trends Genet. 2020, 36, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, Y.; Wang, J. Engineering of transcriptional regulators enhances microbial stress tolerance. Biotechnol. Adv. 2013, 31, 986–991. [Google Scholar] [CrossRef]
- Qi, Q.; Dong, Z.; Sun, Y.; Li, S.; Zhao, Z. Protective Effect of Bergenin against Cyclophosphamide-Induced Immunosuppression by Immunomodulatory Effect and Antioxidation in Balb/c Mice. Molecules 2018, 23, 2668. [Google Scholar] [CrossRef]
- Dong, Y.-J.; Lin, M.-Q.; Fang, X.; Xie, Z.-Y.; Luo, R.; Teng, X.; Li, B.; Li, B.; Li, L.-Z.; Jin, H.-Y.; et al. Modulating effects of a functional food containing Dendrobium officinale on immune response and gut microbiota in mice treated with cyclophosphamide. J. Funct. Foods 2022, 94, 105102. [Google Scholar] [CrossRef]
- Xiang, X.; Cao, N.; Chen, F.; Qian, L.; Wang, Y.; Huang, Y.; Tian, Y.; Xu, D.; Li, W. Polysaccharide of Atractylodes macrocephala Koidz (PAMK) Alleviates Cyclophosphamide-induced Immunosuppression in Mice by Upregulating CD28/IP3R/PLCγ-1/AP-1/NFAT Signal Pathway. Front. Pharmacol. 2020, 11, 529657. [Google Scholar] [CrossRef]
- Bai, Y.; Zeng, Z.; Xie, Z.; Chen, G.; Chen, D.; Sun, Y.; Zeng, X.; Liu, Z. Effects of polysaccharides from Fuzhuan brick tea on immune function and gut microbiota of cyclophosphamide-treated mice. J. Nutr. Biochem. 2022, 101, 108947. [Google Scholar] [CrossRef]
- Nagelkerke, S.Q.; Bruggeman, C.W.; den Haan, J.M.M.; Mul, E.P.J.; van den Berg, T.K.; van Bruggen, R.; Kuijpers, T.W. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-γ receptors. Blood Adv. 2018, 2, 941–953. [Google Scholar] [CrossRef]
- Li, X.; Li, Z.; Zhang, X.; Zeng, Q.; Huang, X.; Sheng, L.; Ahn, D.U.; Cai, Z. Restoration of immunity by whole egg was superior to egg white or egg yolk in a cyclophosphamide-induced immunocompromised mouse model. Food Biosci. 2022, 50, 102013. [Google Scholar] [CrossRef]
- Xiang, X.-W.; Zheng, H.-Z.; Wang, R.; Chen, H.; Xiao, J.-X.; Zheng, B.; Liu, S.-L.; Ding, Y.-T. Ameliorative Effects of Peptides Derived from Oyster (Crassostrea gigas) on Immunomodulatory Function and Gut Microbiota Structure in Cyclophosphamide-Treated Mice. Mar. Drugs 2021, 19, 456. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-E.; Lee, W.-K. Immune enhancing effects of Weissella cibaria JW15 on BALB/c mice immunosuppressed by cyclophosphamide. J. Funct. Foods 2018, 49, 518–525. [Google Scholar] [CrossRef]
- Khan, S.R.; van der Burgh, A.C.; Peeters, R.P.; van Hagen, P.M.; Dalm, V.A.S.H.; Chaker, L. Determinants of Serum Immunoglobulin Levels: A Systematic Review and Meta-Analysis. Front. Immunol. 2021, 12, 1103. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Sun, Y.; Bai, Y.; Xu, J.; Sun, J.; Han, L.; Sun, H.; Han, R. A polysaccharide from Pleurotus citrinopileatus mycelia enhances the immune response in cyclophosphamide-induced immunosuppressed mice via p62/Keap1/Nrf2 signal transduction pathway. Int. J. Biol. Macromol. 2023, 228, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiang, X.; Cao, N.; Chen, W.; Tian, Y.; Zhang, X.; Shen, X.; Jiang, D.; Xu, D.; Xu, S. Polysaccharide of atractylodes macrocephala koidz activated T lymphocytes to alleviate cyclophosphamide-induced immunosuppression of geese through novel_mir2/CD28/AP-1 signal pathway. Poult. Sci. 2021, 100, 101129. [Google Scholar] [CrossRef]
- Santiago-López, L.; Hernández-Mendoza, A.; Vallejo-Cordoba, B.; Wall-Medrano, A.; González-Córdova, A.F. Th17 immune response in inflammatory bowel disease: Future roles and opportunities for lactic acid bacteria and bioactive compounds released in fermented milk. Trends Food Sci. Technol. 2021, 112, 109–117. [Google Scholar] [CrossRef]
- Choi, B.S.-Y.; Varin, T.V.; St-Pierre, P.; Pilon, G.; Tremblay, A.; Marette, A. A polyphenol-rich cranberry extract protects against endogenous exposure to persistent organic pollutants during weight loss in mice. Food Chem. Toxicol. 2020, 146, 111832. [Google Scholar] [CrossRef]
- Li, S.; Xu, X.; De Mandal, S.; Shakeel, M.; Hua, Y.; Shoukat, R.F.; Fu, D.; Jin, F. Gut microbiota mediate Plutella xylostella susceptibility to Bt Cry1Ac protoxin is associated with host immune response. Environ. Pollut. 2021, 271, 116271. [Google Scholar] [CrossRef]
- Nagata, N.; Takeuchi, T.; Masuoka, H.; Aoki, R.; Ishikane, M.; Iwamoto, N.; Sugiyama, M.; Suda, W.; Nakanishi, Y.; Terada-Hirashima, J.; et al. Human Gut Microbiota and Its Metabolites Impact Immune Responses in COVID-19 and Its Complications. Gastroenterology 2023, 164, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.P.; Wilkens, L.R.; Shvetsov, Y.B.; Maskarinec, G.; Park, S.-Y.; Shepherd, J.A.; Boushey, C.J.; Hebert, J.R.; Wirth, M.D.; Ernst, T.; et al. Associations of the Dietary Inflammatory Index with total adiposity and ectopic fat through the gut microbiota, LPS, and C-reactive protein in the Multiethnic Cohort–Adiposity Phenotype Study. Am. J. Clin. Nutr. 2022, 115, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Xu, M.; Qiao, G.; Li, C.; Lin, L.; Zheng, G. Smilax china L. polyphenols alleviates obesity and inflammation by modulating gut microbiota in high fat/high sucrose diet-fed C57BL/6J mice. J. Funct. Foods 2021, 77, 104332. [Google Scholar] [CrossRef]
- Mai, Z.; Ye, M.; Wang, Y.; Foong, S.Y.; Wang, L.; Sun, F.; Cheng, H. Characteristics of Microbial Community and Function with the Succession of Mangroves. Front. Microbiol. 2021, 12, 764974. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fu, J.; Luo, Y.; Li, P.; Song, B.; Lv, Z.; Guo, Y. Effects of tannic acid on the immunity and intestinal health of broiler chickens with necrotic enteritis infection. J. Anim. Sci. Biotechnol. 2023, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Nguyen, S.M.; Yang, Y.; Xu, W.; Cai, H.; Wu, J.; Cai, Q.; Long, J.; Zheng, W.; Shu, X.-O. Long-term diet quality is associated with gut microbiome diversity and composition among urban Chinese adults. Am. J. Clin. Nutr. 2021, 113, 684–694. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).