Profile of PKS and NRPS Gene Clusters in the Genome of Streptomyces cellostaticus NBRC 12849T
Abstract
1. Introduction
2. Materials and Methods
2.1. Streptomyces cellostaticus NBRC 12849T Growth Conditions
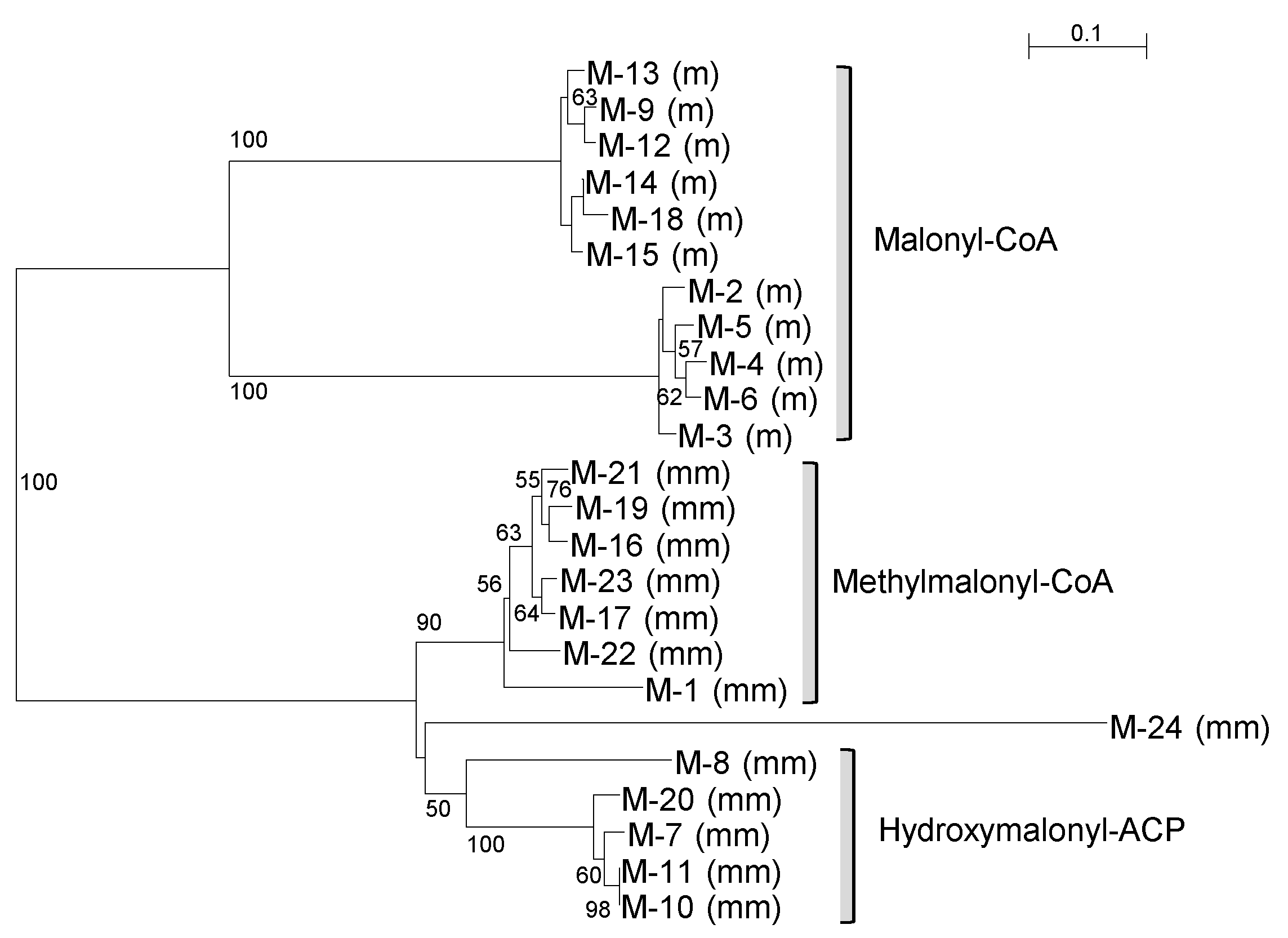
2.2. Phylogenetic Analysis
2.3. DNA Extraction
2.4. Whole Genome Sequencing
2.5. Annotation of PKS and NRPS Gene Clusters
2.6. Detection of Blasticidin A Produced by S. cellostaticus NBRC 12849T
3. Results
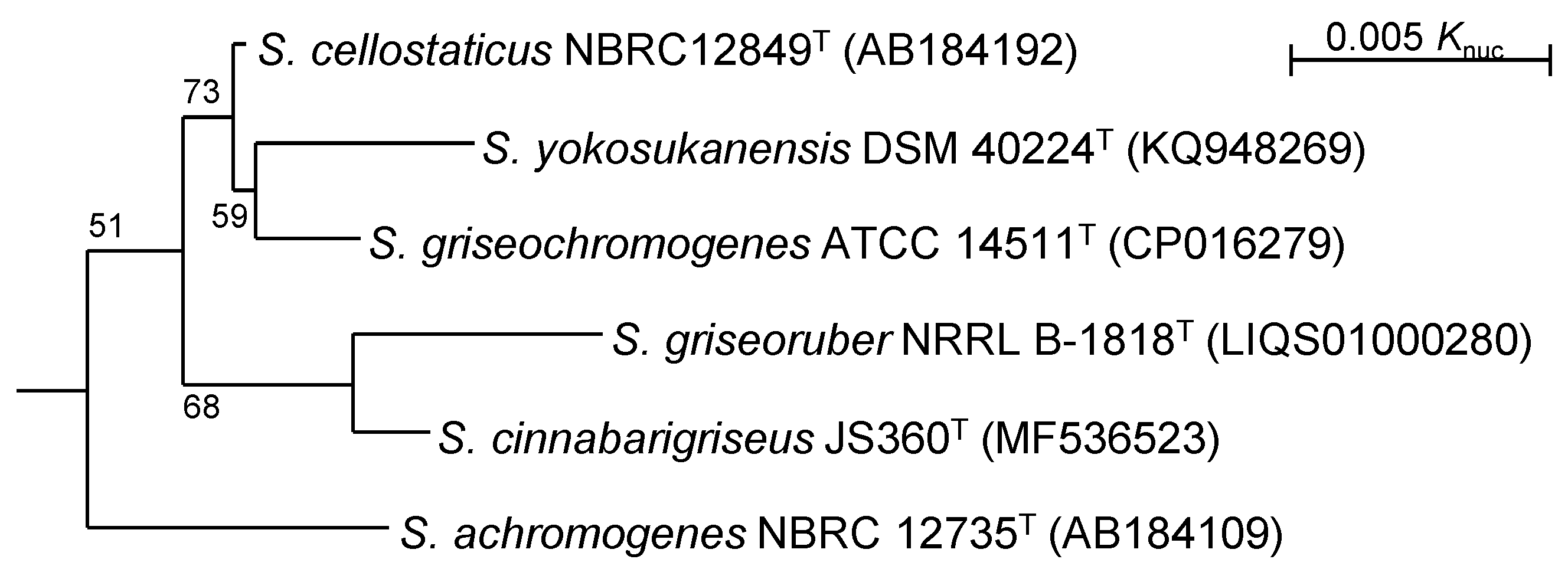
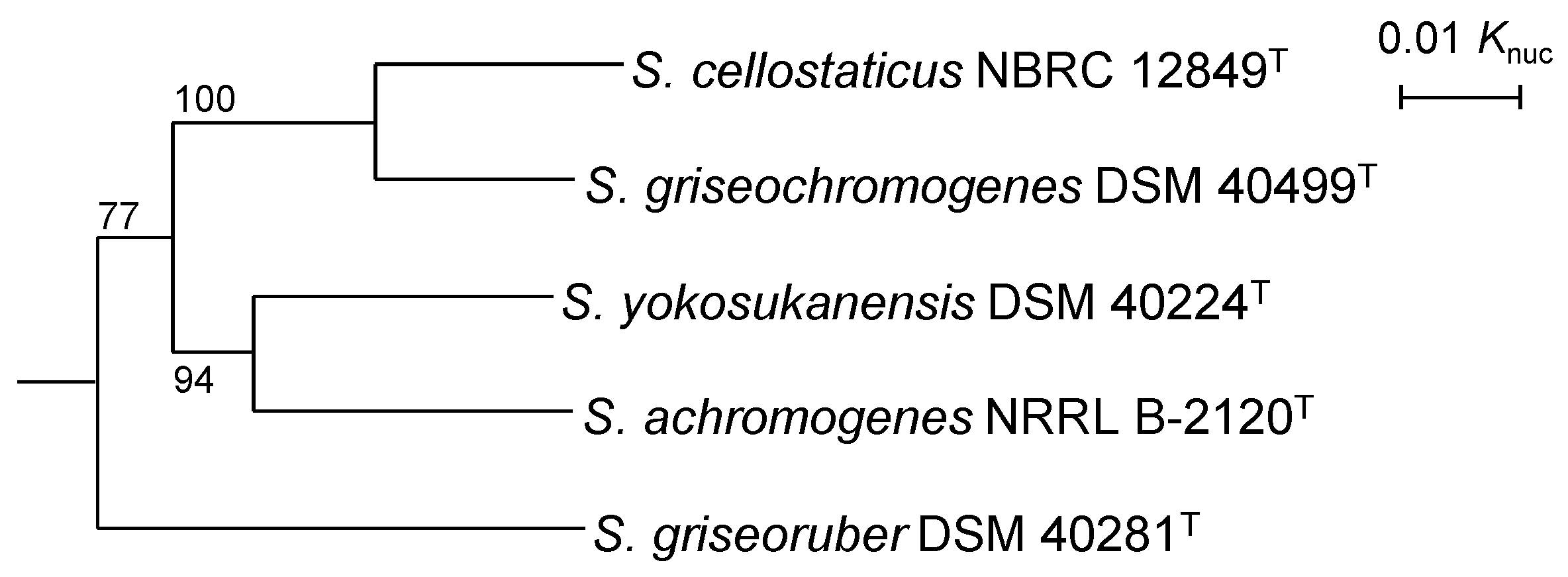
3.1. Phylogenetic Position of S. cellostaticus
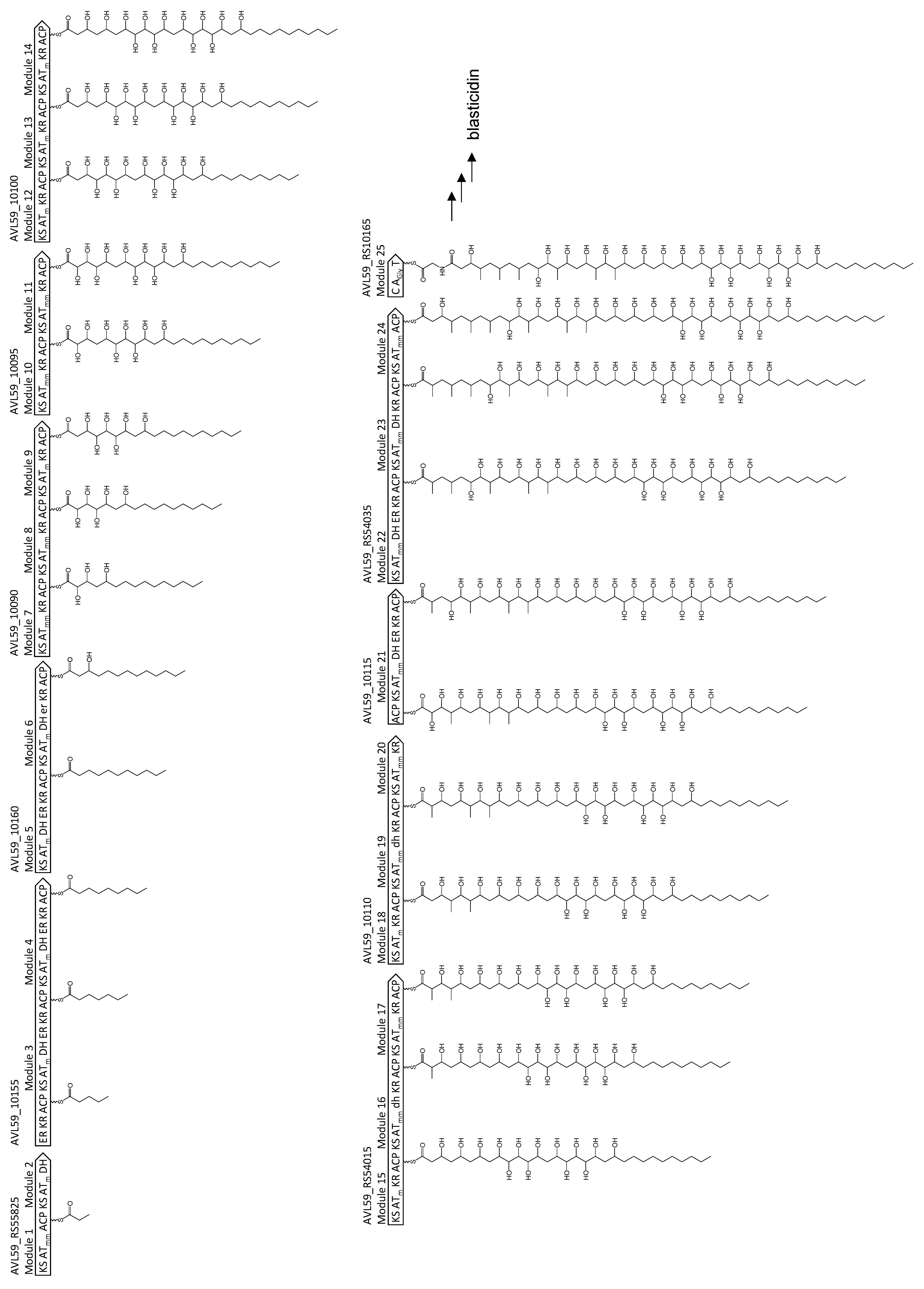
3.2. PKS and NRPS Gene Clusters in S. cellostaticus NBRC 12849T
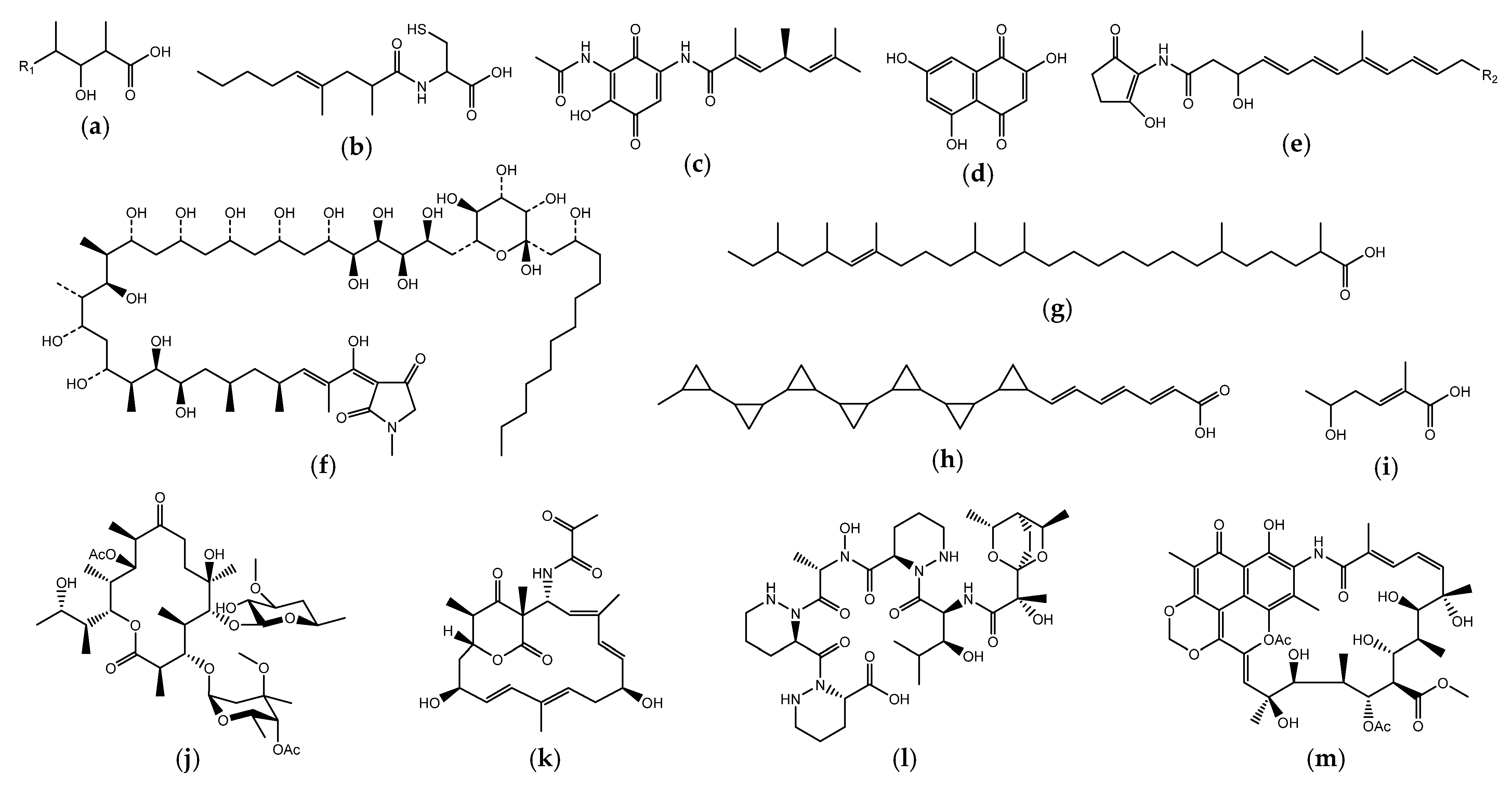
3.3. Annotation of Each Gene Cluster in S. cellostaticus NBRC 12849T
3.4. Identification of Blasticidin-Biosynthetic Gene Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H. Recent progress of reclassification of the genus Streptomyces. Microorganisms 2023, 11, 831. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006, 106, 3468–3496. [Google Scholar] [CrossRef]
- Hamada, S. A study of a new antitumor substance, cellostatin. I. On the isolation and some properties of cellostatin. Tohoku J. Exp. Med. 1958, 67, 173–179. [Google Scholar] [CrossRef][Green Version]
- Pessotti, R.C.; Hansen, B.L.; Reaso, J.N.; Ceja-Navarro, J.A.; El-Hifnawi, L.; Brodie, E.L.; Traxler, M.F. Multiple lineages of Streptomyces produce antimicrobials within passalid beetle galleries across eastern North America. eLife 2021, 10, e65091. [Google Scholar] [CrossRef]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Saito, H.; Miura, K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 1963, 72, 619–629. [Google Scholar] [CrossRef]
- Komaki, H.; Igarashi, Y.; Tamura, T. Taxonogenomic analysis of marine-derived Streptomyces sp. N11-50 and the profile of NRPS and PKS gene clusters. Hydrobiology 2023, 2, 382–394. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Greule, A.; Marolt, M.; Deubel, D.; Peintner, I.; Zhang, S.; Jessen-Trefzer, C.; De Ford, C.; Burschel, S.; Li, S.M.; Friedrich, T.; et al. Wide distribution of foxicin biosynthetic gene clusters in Streptomyces strains—An unusual secondary metabolite with various properties. Front. Microbiol. 2017, 8, 221. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T.; Igarashi, Y. Classification and secondary metabolite-biosynthetic gene clusters of marine Streptomyces strains including a lobophorin- and divergolide-producer. Hydrobiology 2023, 2, 151–161. [Google Scholar] [CrossRef]
- Kalan, L.; Gessner, A.; Thaker, M.N.; Waglechner, N.; Zhu, X.; Szawiola, A.; Bechthold, A.; Wright, G.D.; Zechel, D.L. A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem. Biol. 2013, 20, 1214–1224. [Google Scholar] [CrossRef]
- Sakuda, S.; Ono, M.; Ikeda, H.; Nakamura, T.; Inagaki, Y.; Kawachi, R.; Nakayama, J.; Suzuki, A.; Isogai, A.; Nagasawa, H. Blasticidin A as an inhibitor of aflatoxin production by Aspergillus parasiticus. J. Antibiot. 2000, 53, 1265–1271. [Google Scholar] [CrossRef][Green Version]
- Cruz-Morales, P.; Yin, K.; Landera, A.; Cort, J.R.; Young, R.P.; Kyle, J.E.; Bertrand, R.; Iavarone, A.T.; Acharya, S.; Cowan, A.; et al. Biosynthesis of polycyclopropanated high energy biofuels. Joule 2022, 6, 1590–1605. [Google Scholar] [CrossRef]
- Arakawa, K. Genetic and biochemical analysis of the antibiotic biosynthetic gene clusters on the Streptomyces linear plasmid. Biosci. Biotechnol. Biochem. 2014, 78, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Morgan, K.D.; Williams, D.E.; Patrick, B.O.; Remigy, M.; Banuelos, C.A.; Sadar, M.D.; Ryan, K.S.; Andersen, R.J. Incarnatapeptins A and B, nonribosomal peptides discovered using genome mining and 1H/15N HSQC-TOCSY. Org. Lett. 2020, 22, 4053–4057. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Li, Z.; Xu, W.; Tao, W.; Wu, J.; Yang, J.; Deng, Z.; Sun, Y. Functional analysis of cytochrome P450s involved in streptovaricin biosynthesis and generation of anti-MRSA analogues. ACS Chem. Biol. 2017, 12, 2589–2597. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Boyne, M.T., 2nd; Podevels, A.M.; Klimowicz, A.K.; Handelsman, J.; Kelleher, N.L.; Thomas, M.G. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc. Natl. Acad. Sci. USA 2006, 103, 14349–14354. [Google Scholar] [CrossRef] [PubMed]
- Sakuda, S.; Ikeda, H.; Nakamura, T.; Kawachi, R.; Kondo, T.; Ono, M.; Sakurada, M.; Inagaki, H.; Ito, R.; Nagasawa, H. Blasticidin A derivatives with highly specific inhibitory activity toward aflatoxin production in Aspergillus parasiticus. J. Antibiot. 2000, 53, 1378–1384. [Google Scholar] [CrossRef][Green Version]
- Komaki, H. Reclassification of Streptomyces costaricanus and Streptomyces phaeogriseichromatogenes as later heterotypic synonyms of Streptomyces murinus. Int. J. Syst. Evol. Microbiol. 2021, 71, 004638. [Google Scholar] [CrossRef]
- Oshima, K.; Hattori, M.; Shimizu, H.; Fukuda, K.; Nemoto, M.; Inagaki, K.; Tamura, T. Draft genome sequence of Streptomyces incarnatus NRRL8089, which produces the nucleoside antibiotic sinefungin. Genome Announc. 2015, 3, e00715-15. [Google Scholar] [CrossRef]


| Contig | Accession No. | Length (bp) | G + C Content |
|---|---|---|---|
| sequence01 | BNDU01000001.1 | 15,464 | 69.9% |
| sequence02 | BNDU01000002.1 | 520,187 | 70.9% |
| sequence03 | BNDU01000003.1 | 651,433 | 71.4% |
| sequence04 | BNDU01000004.1 | 1,755,540 | 71.3% |
| sequence05 | BNDU01000005.1 | 142,300 | 72.0% |
| sequence06 | BNDU01000006.1 | 6,225,385 | 70.8% |
| sequence07 | BNDU01000007.1 | 60,471 | 70.7% |
| sequence08 | BNDU01000008.1 | 211,191 | 69.5% |
| sequence09 | BNDU01000009.1 | 59,991 | 70.5% |
| sequence10 | BNDU01000010.1 | 101,974 | 72.9% |
| sequence11 | BNDU01000011.1 | 15,410 | 68.2% |
| sequence12 | BNDU01000012.1 | 123,546 | 69.9% |
| sequence13 | BNDU01000013.1 | 20,918 | 70.1% |
| Total | 9,903,810 | 70.9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komaki, H.; Tamura, T. Profile of PKS and NRPS Gene Clusters in the Genome of Streptomyces cellostaticus NBRC 12849T. Fermentation 2023, 9, 924. https://doi.org/10.3390/fermentation9110924
Komaki H, Tamura T. Profile of PKS and NRPS Gene Clusters in the Genome of Streptomyces cellostaticus NBRC 12849T. Fermentation. 2023; 9(11):924. https://doi.org/10.3390/fermentation9110924
Chicago/Turabian StyleKomaki, Hisayuki, and Tomohiko Tamura. 2023. "Profile of PKS and NRPS Gene Clusters in the Genome of Streptomyces cellostaticus NBRC 12849T" Fermentation 9, no. 11: 924. https://doi.org/10.3390/fermentation9110924
APA StyleKomaki, H., & Tamura, T. (2023). Profile of PKS and NRPS Gene Clusters in the Genome of Streptomyces cellostaticus NBRC 12849T. Fermentation, 9(11), 924. https://doi.org/10.3390/fermentation9110924





