A Review of Basic Bioinformatic Techniques for Microbial Community Analysis in an Anaerobic Digester
Abstract
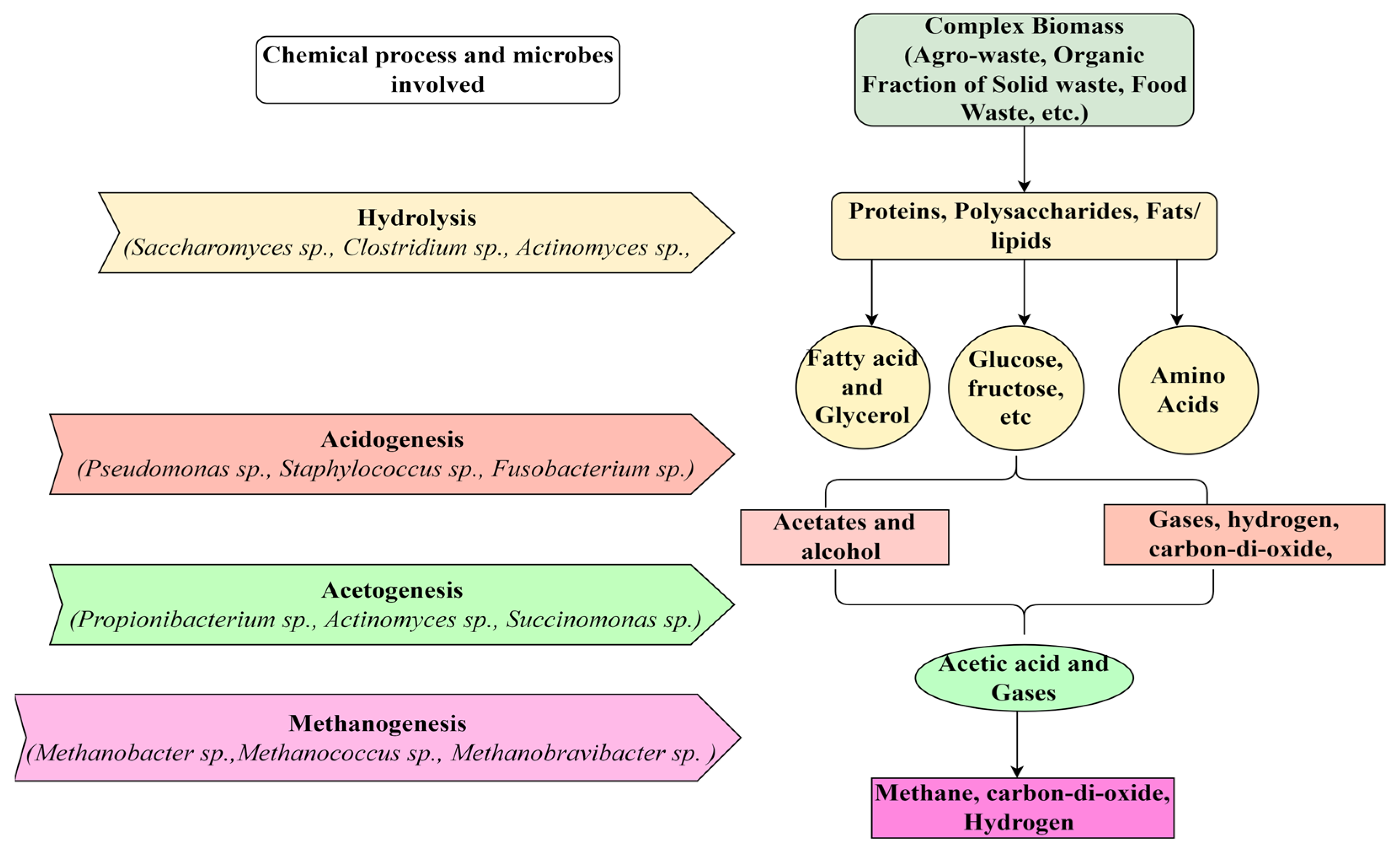
1. Introduction
2. Methods of DNA Isolation and DNA Sequencing
3. Analysis of Anaerobic Microbial Communities Using Bioinformatic Tools
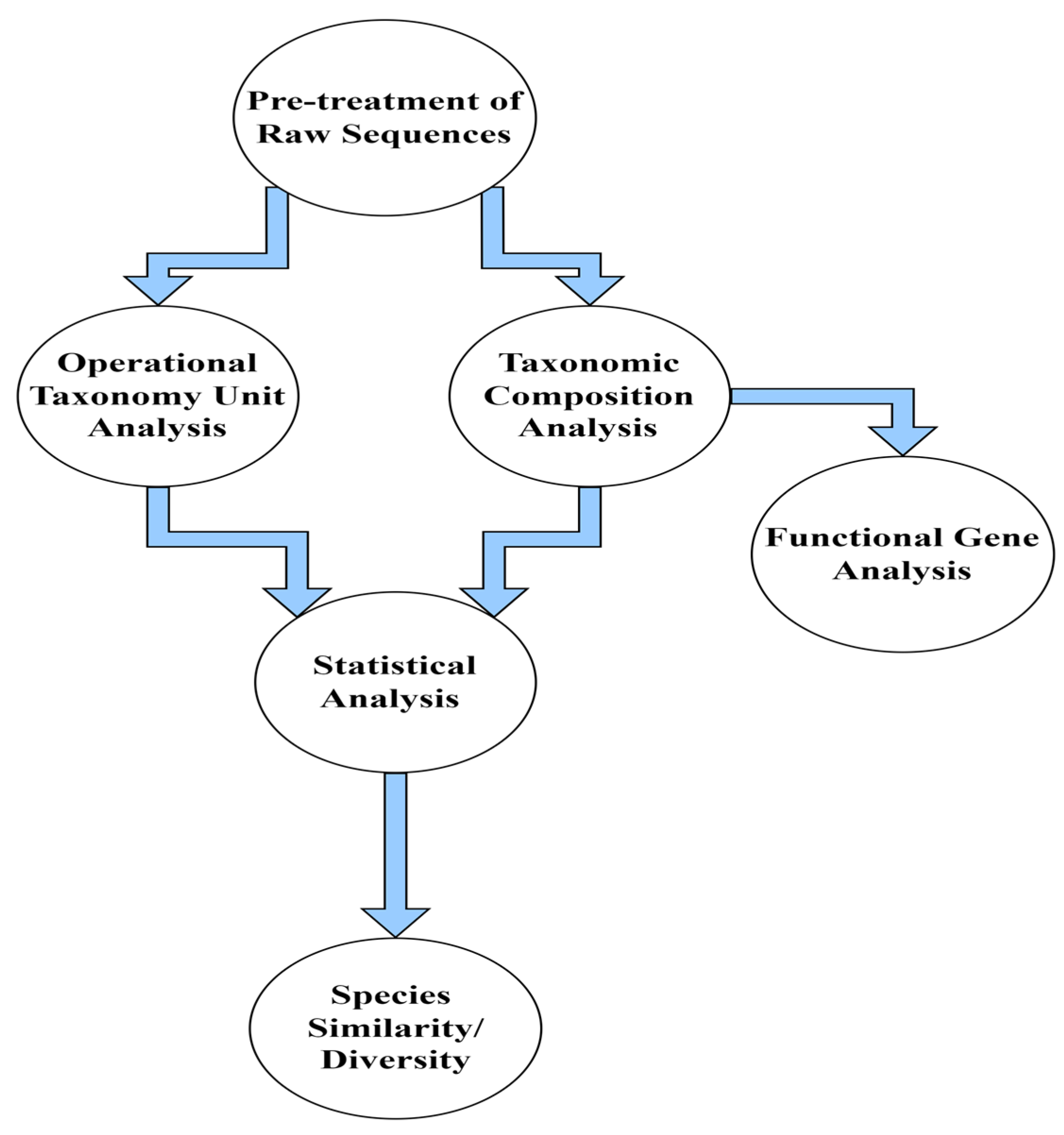
3.1. Pre-Processing of Raw Sequences
3.2. Analysis of OTU Clustering
3.3. Analysis of Alpha Diversity
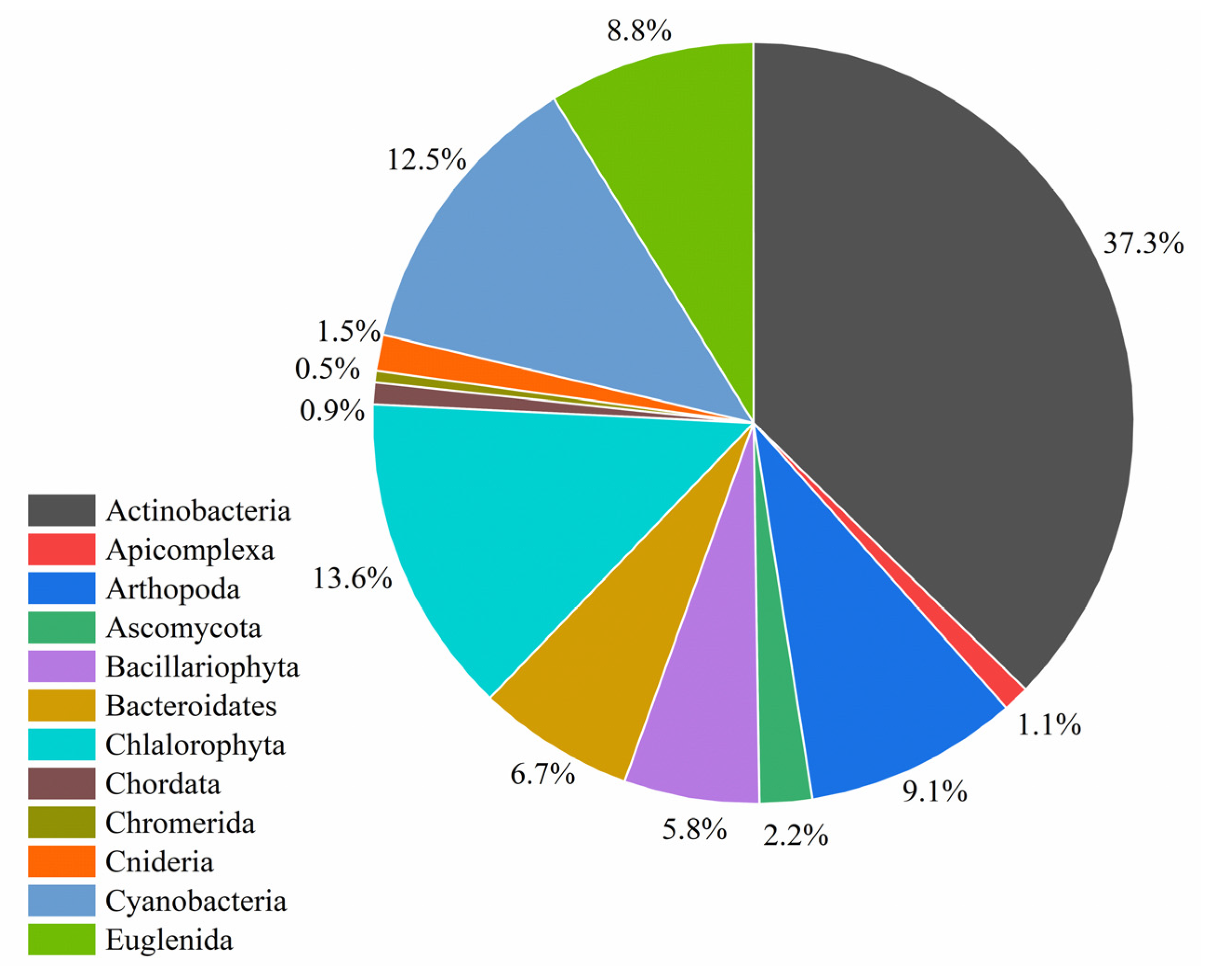
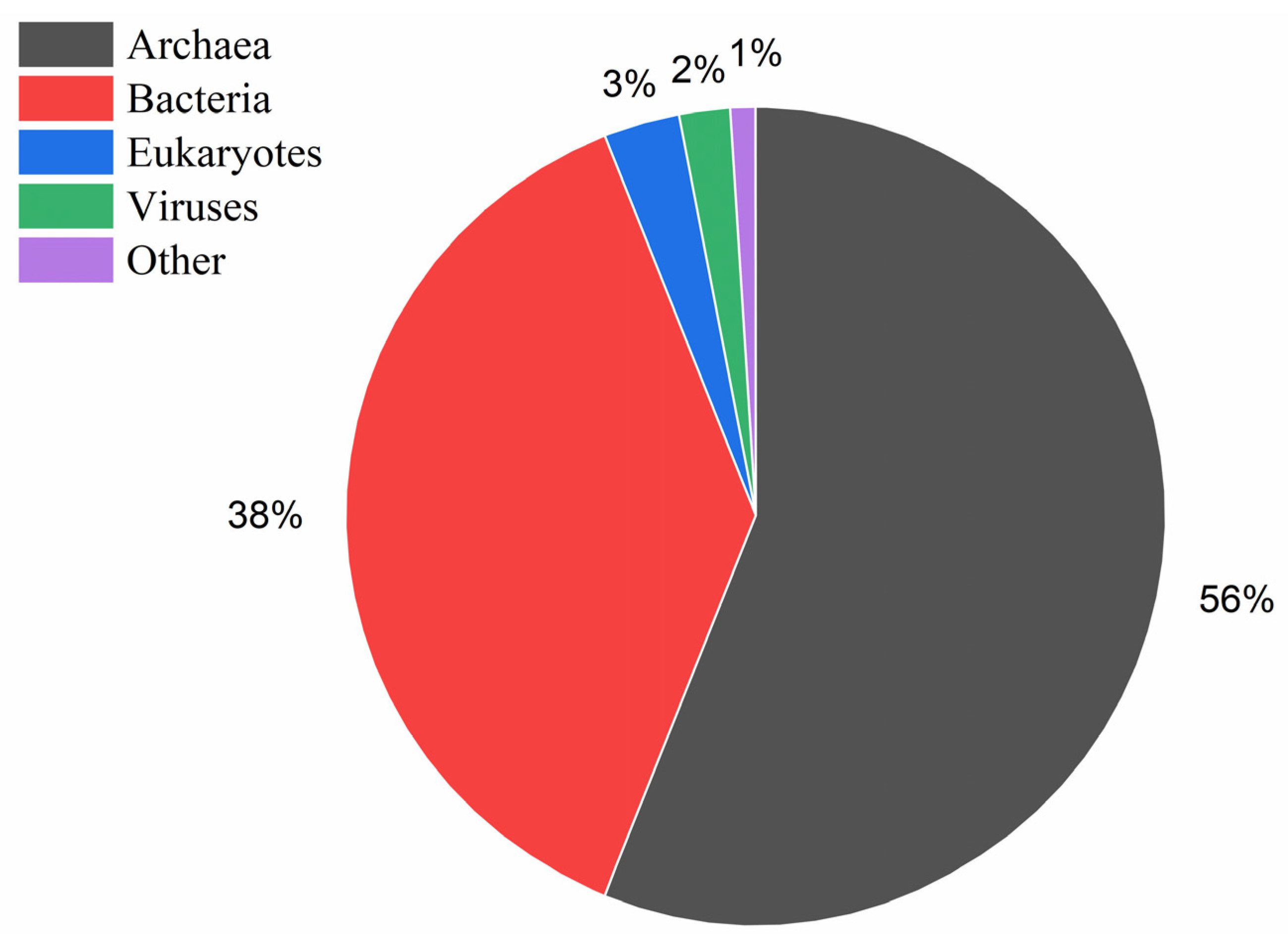
3.4. Metataxonomic Analysis
3.5. Study of the Similarities/Differences in Microbial Taxonomic Compositions
3.6. Statistical Multivariate Analysis
3.7. Metatranscriptomics and Application
4. Application of the Analysis of Anaerobic Microbial Community on Biogas Production
4.1. Development of Improved AD Processes
4.2. Development on Metabolomics Analysis
4.3. Development in Identification of Antibiotic-Resistant Genes
4.4. Creation of the Modelling and Optimization of Microbial Population Dynamics
5. Restrictions and Predictions of Bioinformatics Analysis on Microbial Metagenomics
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ju, F.; Lau, F.; Zhang, T. Linking Microbial Community, Environmental Variables, and Methanogenesis in Anaerobic Biogas Digesters of Chemically Enhanced Primary Treatment Sludge. Environ. Sci. Technol. 2017, 51, 3982–3992. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Qiu, Y.; Ren, L.; Jiang, B. Microbial Characteristics in Anaerobic Digestion Process of Food Waste for Methane Production–A Review. Bioresour. Technol. 2018, 248, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Hassa, J.; Maus, I.; Off, S.; Pühler, A.; Scherer, P.; Klocke, M.; Schlüter, A. Metagenome, Metatranscriptome, and Metaproteome Approaches Unraveled Compositions and Functional Relationships of Microbial Communities Residing in Biogas Plants. Appl. Microbiol. Biotechnol. 2018, 102, 5045–5063. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Lee, J.; Han, G.; Hwang, S. Comprehensive Analysis of Microbial Communities in Full-Scale Mesophilic and Thermophilic Anaerobic Digesters Treating Food Waste-Recycling Wastewater. Bioresour. Technol. 2018, 259, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Lu, F.; Jiang, Q.; Jiang, C.; Kashif, M.; Shen, P. Assessment of Multiple Anaerobic Co-Digestions and Related Microbial Community of Molasses with Rice-Alcohol Wastewater. Energies 2020, 13, 4866. [Google Scholar] [CrossRef]
- Björnsson, L.; Murto, M.; Mattiasson, B. Evaluation of Parameters for Monitoring an Anaerobic Co-Digestion Process. Appl. Microbiol. Biotechnol. 2001, 54, 844–849. [Google Scholar] [CrossRef]
- Holm-Nielsen, J.B.; Lomborg, C.J.; Oleskowicz-Popiel, P.; Esbensen, K.H. On-Line near Infrared Monitoring of Glycerol-Boosted Anaerobic Digestion Processes: Evaluation of Process Analytical Technologies. Biotechnol. Bioeng. 2008, 99, 302–313. [Google Scholar] [CrossRef]
- Boe, K.; Batstone, D.J.; Steyer, J.-P.; Angelidaki, I. State Indicators for Monitoring the Anaerobic Digestion Process. Water Res. 2010, 44, 5973–5980. [Google Scholar] [CrossRef]
- Molina, F.; Ruiz-Filippi, G.; Garcia, C.; Lema, J.M.; Roca, E. Pilot-Scale Validation of a New Sensor for On-Line Analysis of Volatile Fatty Acids and Alkalinity in Anaerobic Wastewater Treatment Plants. Environ. Eng. Sci. 2009, 26, 641–649. [Google Scholar] [CrossRef]
- Fernández, A.; Huang, S.; Seston, S.; Xing, J.; Hickey, R.; Criddle, C.; Tiedje, J. How Stable Is Stable? Function versus Community Composition. Appl. Environ. Microbiol. 1999, 65, 3697–3704. [Google Scholar] [CrossRef]
- Li, L.; He, Q.; Ma, Y.; Wang, X.; Peng, X. Dynamics of Microbial Community in a Mesophilic Anaerobic Digester Treating Food Waste: Relationship between Community Structure and Process Stability. Bioresour. Technol. 2015, 189, 113–120. [Google Scholar] [CrossRef]
- Hashemi, S.; Hashemi, S.; Lien, K.; Lamb, J. Molecular Microbial Community Analysis as an Analysis Tool for Optimal Biogas Production. Microorganisms 2021, 9, 1162. [Google Scholar] [CrossRef]
- Narihiro, T.; Sekiguchi, Y. Microbial Communities in Anaerobic Digestion Processes for Waste and Wastewater Treatment: A Microbiological Update. Curr. Opin. Biotechnol. 2007, 18, 273–278. [Google Scholar] [CrossRef]
- Zhang, L.; Kuroki, A.; Loh, K.-C.; Seok, J.K.; Dai, Y.; Tong, Y.W. Highly Efficient Anaerobic Co-Digestion of Food Waste and Horticultural Waste Using a Three-Stage Thermophilic Bioreactor: Performance Evaluation, Microbial Community Analysis, and Energy Balance Assessment. Energy Convers. Manag. 2020, 223, 113290. [Google Scholar] [CrossRef]
- Zhen, F.; Luo, X.; Xing, T.; Sun, Y.; Kong, X.; Li, W. Performance Evaluation and Microbial Community Analysis of Microaerobic Pretreatment on Thermophilic Dry Anaerobic Digestion. Biochem. Eng. J. 2021, 167, 107873. [Google Scholar] [CrossRef]
- Laserson, J.; Jojic, V.; Koller, D. Genovo: De Novo Assembly for Metagenomes. J. Comput. Biol. 2011, 18, 429–443. [Google Scholar] [CrossRef]
- De Vrieze, J.; Saunders, A.M.; He, Y.; Fang, J.; Nielsen, P.H.; Verstraete, W.; Boon, N. Ammonia and Temperature Determine Potential Clustering in the Anaerobic Digestion Microbiome. Water Res. 2015, 75, 312–323. [Google Scholar] [CrossRef]
- Rincón, B.; Borja, R.; González, J.M.; Portillo, M.C.; Sáiz-Jiménez, C. Influence of Organic Loading Rate and Hydraulic Retention Time on the Performance, Stability and Microbial Communities of One-Stage Anaerobic Digestion of Two-Phase Olive Mill Solid Residue. Biochem. Eng. J. 2008, 40, 253–261. [Google Scholar] [CrossRef]
- Luo, G.; Fotidis, I.; Angelidaki, I. Comparative Analysis of Taxonomic, Functional, and Metabolic Patterns of Microbiomes from 14 Full-Scale Biogas Reactors by Metagenomic Sequencing and Radioisotopic Analysis. Biotechnol. Biofuels 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Zhang, J.; Mao, L.; Zhang, L.; Loh, K.-C.; Dai, Y.; Tong, Y. Metagenomic Insight into the Microbial Networks and Metabolic Mechanism in Anaerobic Digesters for Food Waste by Incorporating Activated Carbon. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, T.; Wang, T.; Fang, Z. Microbial Structures, Functions, and Metabolic Pathways in Wastewater Treatment Bior actors Revealed Using High-Throughput Sequencing. Environ. Sci. Technol. 2012, 46, 13244–13252. [Google Scholar] [CrossRef]
- Regueiro, L.; Spirito, C.; Usack, J.; Hospodsky, D.; Werner, J.; Angenent, L. Comparing the Inhibitory Thresholds of Dairy Manure Co-Digesters after Prolonged Acclimation Periods: Part 2–Correlations between Microbiomes and Environment. Water Res. 2015, 87, 458–466. [Google Scholar] [CrossRef]
- Genitsaris, S.; Monchy, S.; Denonfoux, J.; Ferreira, S.; Kormas, K.; Sime-Ngando, T.; Viscogliosi, E.; Christaki, U. Marine Microbial Community Structure Assessed from Combined Metagenomic Analysis and Ribosomal Amplicon Deep-Sequencing. Mar. Biol. Res. 2015, 12, 1–13. [Google Scholar] [CrossRef]
- Krakat, N.; Anjum, R.; Demirel, B.; Schröder, P. Methodological Flaws Introduce Strong Bias into Molecular Analysis of Microbial Populations. J. Appl. Microbiol. 2016, 122, 364–377. [Google Scholar] [CrossRef]
- Alvarez-Silva, M.C.; Álvarez-Yela, A.C.; Gómez-Cano, F.; Zambrano, M.M.; Husserl, J.; Danies, G.; Restrepo, S.; González-Barrios, A.F. Compartmentalized Metabolic Network Reconstruction of Microbial Communities to Determine the Effect of Agricultural Intervention on Soils. PLoS ONE 2017, 12, e0181826. [Google Scholar] [CrossRef]
- Badhai, J.; Ghosh, T.S.; Das, S.K. Taxonomic and Functional Characteristics of Microbial Communities and Their Correlation with Physicochemical Properties of Four Geothermal Springs in Odisha, India. Front. Microbiol. 2015, 6, 1166. [Google Scholar] [CrossRef]
- Kumar, G.R.; Chowdhary, N. Biotechnological and Bioinformatics Approaches for Augmentation of Biohydrogen Production: A Review. Renew. Sustain. Energy Rev. 2016, 56, 1194–1206. [Google Scholar] [CrossRef]
- Mukherjee, A.; Chettri, B.; Langpoklakpam, J.; Basak, P.; Prasad, A.; Mukherjee, A.; Bhattacharyya, M.; Singh, A.; Chattopadhyay, D. Bioinformatic Approaches Including Predictive Metagenomic Profiling Reveal Characteristics of Bacterial Response to Petroleum Hydrocarbon Contamination in Diverse Environments. Sci. Rep. 2017, 7, 1–22. [Google Scholar] [CrossRef]
- Zhang, L.; Loh, K.-C.; Lim, J.W.; Zhang, J. Bioinformatics Analysis of Metagenomics Data of Biogas-Producing Microbial Communities in Anaerobic Digesters: A Review. Renew. Sustain. Energy Rev. 2019, 100, 110–126. [Google Scholar] [CrossRef]
- Suhartini, S.; Heaven, S.; Banks, C. Comparison of Mesophilic and Thermophilic Anaerobic Digestion of Sugar Beet Pulp: Performance, Dewaterability and Foam Control. Bioresour. Technol. 2013, 152, 202–211. [Google Scholar] [CrossRef]
- Riggio, S.; Hernandez Shek, M.; Torrijos, M.; Vives, G.; Esposito, G.; van Hullebusch, E.; Steyer, J.-P.; Escudié, R. Comparison of the Mesophilic and Thermophilic Anaerobic Digestion of Spent Cow Bedding in Leach-Bed Reactors. Bioresour. Technol. 2017, 234, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Munk, B.; Guebitz, G.M.; Lebuhn, M. Influence of Nitrogen-Rich Substrates on Biogas Production and on the Methanogenic Community under Mesophilic and Thermophilic Conditions. Anaerobe 2017, 46, 146–154. [Google Scholar] [CrossRef]
- Ju, F.; Zhang, T. Experimental Design and Bioinformatics Analysis for the Application of Metagenomics in Environmental Sciences and Biotechnology. Environ. Sci. Technol. 2015, 49, 12628–12640. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Yi, J.; Dai, L.; Dai, X. Evaluation of Several DNA Extraction Methods for Obtaining Total Community DNA from Anaerobic Digestion Sludge. Procedia Environ. Sci. 2013, 18, 856–863. [Google Scholar] [CrossRef]
- Bareither, C.; Wolfe, G.; Mcmahon, K.; Benson, C. Microbial Diversity and Dynamics during Methane Production from Municipal Solid Waste. Waste Manag. 2013, 33, 1982–1992. [Google Scholar] [CrossRef]
- Lee, S.-H.; Kang, H.-J.; Lee, Y.; Lee, T.; Han, K.; Choi, Y.; Park, H.-D. Monitoring Bacterial Community Structure and Variability in Time Scale in Full-Scale Anaerobic Digesters. J. Environ. Monit. JEM 2012, 14, 1893–1905. [Google Scholar] [CrossRef]
- Sun, L.; Liu, T.; Müller, B.; Schnürer, A. The Microbial Community Structure in Industrial Biogas Plants Influences the Degradation Rate of Straw and Cellulose in Batch Tests. Biotechnol. Biofuels 2016, 9, 1–20. [Google Scholar] [CrossRef]
- Han, G.; Shin, S.G.; Lee, J.; Shin, J.; Hwang, S. A Comparative Study on the Process Efficiencies and Microbial Community Structures of Six Full-Scale Wet and Semi-Dry Anaerobic Digesters Treating Food Wastes. Bioresour. Technol. 2017, 245, 869–875. [Google Scholar] [CrossRef]
- Wei, Z.; Jin, D.; Deng, Y. Bioinformatics Tools and Applications in the Study of Environmental Microbial Metagenomics. Microbiol. China 2015, 42, 890–901. [Google Scholar] [CrossRef]
- Chen, S.; Cheng, H.; Wyckoff, K.; He, Q. Linkages of Firmicutes and Bacteroidetes Populations to Methanogenic Process Performance. J. Ind. Microbiol. Biotechnol. 2016, 43, 771–781. [Google Scholar] [CrossRef]
- Carabeo-Pérez, A.; Guerra-Rivera, G.; Ramos-Leal, M.; Jiménez-Hernández, J. Metagenomic Approaches: Effective Tools for Monitoring the Structure and Functionality of Microbiomes in Anaerobic Digestion Systems. Appl. Microbiol. Biotechnol. 2019, 103, 9379–9390. [Google Scholar] [CrossRef] [PubMed]
- Hardegen, J.; Latorre-Pérez, A.; Vilanova, C.; Günther, T.; Porcar, M.; Luschnig, O.; Simeonov, C.; Abendroth, C. Methanogenic Community Shifts during the Transition from Sewage Mono-Digestion to Co-Digestion of Grass Biomass. Bioresour. Technol. 2018, 265, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.; Bongcam-Rudloff, E.; Müller, B. Abundance Tracking by Long-Read Nanopore Sequencing of Complex Microbial Communities in Samples from 20 Different Biogas/Wastewater Plants. Appl. Sci. 2020, 10, 7518. [Google Scholar] [CrossRef]
- Garcia, S.L.; Jangid, K.; Whitman, W.B.; Das, K.C. Transition of Microbial Communities during the Adaption to Anaerobic Digestion of Carrot Waste. Bioresour. Technol. 2011, 102, 7249–7256. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Lou, Z.; Zhang, D.; Shan, A.; Yuan, H.; Zhu, N.; Zhang, K. Variations of Organic Matters and Microbial Community in Thermophilic Anaerobic Digestion of Waste Activated Sludge with the Addition of Ferric Salts. Bioresour. Technol. 2015, 179, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Bae, J.; Choi, O.; Ju, D.; Lee, J.; Sung, H.; Park, S.; Sang, B.-I.; Um, Y. A Pilot Scale Two-Stage Anaerobic Digester Treating Food Waste Leachate (FWL): Performance and Microbial Structure Analysis Using Pyrosequencing. Process Biochem. 2014, 49, 301–308. [Google Scholar] [CrossRef]
- Cho, K.; Shin, S.G.; Kim, W.; Lee, J.; Lee, C.; Hwang, S. Microbial Community Shifts in a Farm-Scale Anaerobic Digester Treating Swine Waste: Correlations between Bacteria Communities Associated with Hydrogenotrophic Methanogens and Environmental Conditions. Sci. Total Environ. 2017, 601–602, 167–176. [Google Scholar] [CrossRef]
- Lee, J.; Han, G.; Shin, S.G.; Koo, T.; Cho, K.; Kim, W.; Hwang, S. Seasonal Monitoring of Bacteria and Archaea in a Full-Scale Thermophilic Anaerobic Digester Treating Food Waste-Recycling Wastewater: Correlations between Microbial Community Characteristics and Process Variables. Chem. Eng. J. 2016, 300, 291–299. [Google Scholar] [CrossRef]
- Leite, A.; Janke, L.; Harms, H.; Richnow, H.; Nikolausz, M. Lessons Learned from the Microbial Ecology Resulting from Different Inoculation Strategies for Biogas Production from Waste Products of the Bioethanol/Sugar Industry. Biotechnol. Biofuels 2016, 9, 1–16. [Google Scholar] [CrossRef]
- Li, L.; He, Q.; Ma, Y.; Wang, X.; Peng, X. A Mesophilic Anaerobic Digester for Treating Food Waste: Process Stability and Microbial Community Analysis Using Pyrosequencing. Microb. Cell Factories 2016, 15, 1–11. [Google Scholar] [CrossRef]
- Yu, Z.; Wen, X.; Xu, M.; Huang, X. Characteristics of Extracellular Polymeric Substances and Bacterial Communities in an Anaerobic Membrane Bioreactor Coupled with Online Ultrasound Equipment. Bioresour. Technol. 2012, 117, 333–340. [Google Scholar] [CrossRef]
- Ho, D.; Jensen, P.; Batstone, D. Effects of Temperature and Hydraulic Retention Time on Acetotrophic Pathways and Performance in High-Rate Sludge Digestion. Environ. Sci. Technol. 2014, 48, 6468–6476. [Google Scholar] [CrossRef]
- Lebuhn, M.; Hanreich, A.; Klocke, M.; Schlueter, A.; Bauer, C.; Pérez, C. Towards Molecular Biomarkers for Biogas Production from Lignocellulose-Rich Substrates. Anaerobe 2014, 29, 10–21. [Google Scholar] [CrossRef]
- Ziganshin, A.M.; Liebetrau, J.; Pröter, J.; Kleinsteuber, S. Microbial Community Structure and Dynamics during Anaerobic Digestion of Various Agricultural Waste Materials. Appl. Microbiol. Biotechnol. 2013, 97, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Oosterkamp, M.; Méndez-García, C.; Kim, C.-H.; Bauer, S.; Ibáñez, A.; Zimmerman, S.; Hong, P.; Cann, I.; Mackie, R. Lignocellulose-Derived Thin Stillage Composition and Efficient Biological Treatment with a High-Rate Hybrid Anaerobic Bioreactor System. Biotechnol. Biofuels 2016, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Belostotskiy, D.E.; Ziganshina, E.E.; Siniagina, M.; Boulygina, E.A.; Miluykov, V.A.; Ziganshin, A.M. Impact of the Substrate Loading Regime and Phosphoric Acid Supplementation on Performance of Biogas Reactors and Microbial Community Dynamics during Anaerobic Digestion of Chicken Wastes. Bioresour. Technol. 2015, 193, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Ziganshina, E.; Belostotskiy, D.; Ilinskaya, O.; Boulygina, E.; Grigoryeva, T.; Ziganshin, A. Effect of the Organic Loading Rate Increase and the Presence of Zeolite on Microbial Community Composition and Process Stability During Anaerobic Digestion of Chicken Wastes. Microb. Ecol. 2015, 70, 948–960. [Google Scholar] [CrossRef]
- Razaviarani, V.; Buchanan, I.D. Anaerobic Co-Digestion of Biodiesel Waste Glycerin with Municipal Wastewater Sludge: Microbial Community Structure Dynamics and Reactor Performance. Bioresour. Technol. 2015, 182, 8–17. [Google Scholar] [CrossRef]
- Solli, L.; Håvelsrud, O.E.; Horn, S.J.; Rike, A.G. A Metagenomic Study of the Microbial Communities in Four Parallel Biogas Reactors. Biotechnol. Biofuels 2014, 7, 146. [Google Scholar] [CrossRef]
- Maspolim, Y.; Zhou, Y.; Guo, C.; Xiao, K.; Ng, W.J. Determination of the Archaeal and Bacterial Communities in Two-Phase and Single-Stage Anaerobic Systems by 454 Pyrosequencing. J. Environ. Sci. 2015, 36, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Cardinali-Rezende, J.; Rojas-Ojeda, P.; Nascimento, A.; Sanz, J. Proteolytic Bacterial Dominance in a Full-Scale Municipal Solid Waste Anaerobic Reactor Assessed by 454 Pyrosequencing Technology. Chemosphere 2016, 146, 519–525. [Google Scholar] [CrossRef]
- Azizi, A.; Kim, W.; Lee, J.H. Comparison of Microbial Communities during the Anaerobic Digestion of Gracilaria under Mesophilic and Thermophilic Conditions. World J. Microbiol. Biotechnol. 2016, 32, 158. [Google Scholar] [CrossRef]
- Wirth, R.; Kovács, E.; Maróti, G.; Bagi, Z.; Rákhely, G.; Kovacs, K. Characterization of a Biogas—Producing Microbial Community by Short-Read next Generation DNA Sequencing. Biotechnol. Biofuels 2012, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.A.; Romero, H.; Perotti, N.I. Two Amplicon Sequencing Strategies Revealed Different Facets of the Prokaryotic Community Associated with the Anaerobic Treatment of Vinasses from Ethanol Distilleries. Bioresour. Technol. 2014, 153, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, S.; Treu, L.; Kougias, P.; De Francisci, D.; Valle, G.; Angelidaki, I. Metagenomic Analysis and Functional Characterization of the Biogas Microbiome Using High Throughput Shotgun Sequencing and a Novel Binning Strategy. Biotechnol. Biofuels 2016, 9, 1–17. [Google Scholar] [CrossRef]
- Begmatov, S.; Dorofeev, A.; Kadnikov, V.; Beletsky, A.; Pimenov, N.; Ravin, N.; Mardanov, A. The Structure of Microbial Communities of Activated Sludge of Large-Scale Wastewater Treatment Plants in the City of Moscow. Sci. Rep. 2022, 12, 3458. [Google Scholar] [CrossRef]
- Krause, L.; Diaz, N.N.; Edwards, R.A.; Gartemann, K.-H.; Krömeke, H.; Neuweger, H.; Pühler, A.; Runte, K.J.; Schlüter, A.; Stoye, J.; et al. Taxonomic Composition and Gene Content of a Methane-Producing Microbial Community Isolated from a Biogas Reactor. J. Biotechnol. 2008, 136, 91–101. [Google Scholar] [CrossRef]
- Mitra, S.; Rupek, P.; Richter, D.; Urich, T.; Jack, G.; Meyer, F.; Wilke, A.; Huson, D. Functional Analysis of Metagenomes and Metatranscriptomes Using SEED and KEGG. BMC Bioinform. 2011, 12 (Suppl. 1), S21. [Google Scholar] [CrossRef]
- Shaw, G.T.-W.; Liu, A.-C.; Weng, C.-Y.; Chou, C.-Y.; Wang, D. Inferring Microbial Interactions in Thermophilic and Mesophilic Anaerobic Digestion of Hog Waste. PLoS ONE 2017, 12, e0181395. [Google Scholar] [CrossRef]
- Guo, X.; Wang, C.; Sun, F.; Zhu, W.; Wu, W.-X. A Comparison of Microbial Characteristics between the Thermophilic and Mesophilic Anaerobic Digesters Exposed to Elevated Food Waste Loadings. Bioresour. Technol. 2013, 152, 420–428. [Google Scholar] [CrossRef]
- Lu, X.; Rao, S.; Shen, Z.; Lee, P.K.H. Substrate Induced Emergence of Different Active Bacterial and Archaeal Assemblages during Biomethane Production. Bioresour. Technol. 2013, 148, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D.; Rao, S.; Lu, X.; Lee, P. Effects of Sludge Inoculum and Organic Feedstock on Active Microbial Communities and Methane Yield during Anaerobic Digestion. Front. Microbiol. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pope, P.B.; Vivekanand, V.; Eijsink, V.G.H.; Horn, S.J. Microbial Community Structure in a Biogas Digester Utilizing the Marine Energy Crop Saccharina Latissima. 3 Biotech 2013, 3, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Kröber, M.; Bekel, T.; Diaz, N.N.; Goesmann, A.; Jaenicke, S.; Krause, L.; Miller, D.; Runte, K.J.; Viehöver, P.; Pühler, A.; et al. Phylogenetic Characterization of a Biogas Plant Microbial Community Integrating Clone Library 16S-RDNA Sequences and Metagenome Sequence Data Obtained by 454-Pyrosequencing. J. Biotechnol. 2009, 142, 38–49. [Google Scholar] [CrossRef]
- Yun, Y.-M.; Kim, D.-H.; Cho, S.-K.; Shin, H.-S.; Jung, K.-W.; Kim, H.-W. Mitigation of Ammonia Inhibition by Internal Dilution in High-Rate Anaerobic Digestion of Food Waste Leachate and Evidences of Microbial Community Response. Biotechnol. Bioeng. 2016, 113, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wang, N.; Wei, D.; Liang, C.; Chen, X.; Liu, L.; Shi, J. Bacterial Community Compositions and Nitrogen Metabolism Function in a Cattle Farm Wastewater Treatment Plant Revealed by Illumina High-Throughput Sequencing. Environ. Sci. Pollut. Res. 2021, 28, 40895–40907. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; Hara, B.; Henry, M.; Stevens, H. The Vegan Package. 2007. Available online: https://www.researchgate.net/publication/228975085 (accessed on 22 October 2022).
- Morris, E.K.; Caruso, T.; Buscot, F.; Fischer, M.; Hancock, C.; Maier, T.S.; Meiners, T.; Müller, C.; Obermaier, E.; Prati, D.; et al. Choosing and Using Diversity Indices: Insights for Ecological Applications from the German Biodiversity Exploratories. Ecol. Evol. 2014, 4, 3514–3524. [Google Scholar] [CrossRef]
- Barrantes, G.; Sandoval, L. Conceptual and Statistical Problems Associated with the Use of Diversity Indices in Ecology. Rev. De Biol. Trop. 2009, 57, 451–460. [Google Scholar] [CrossRef]
- Smith, A.M.; Sharma, D.; Lappin-Scott, H.; Burton, S.; Huber, D.H. Microbial Community Structure of a Pilot-Scale Thermophilic Anaerobic Digester Treating Poultry Litter. Appl. Microbiol. Biotechnol. 2014, 98, 2321–2334. [Google Scholar] [CrossRef]
- Rademacher, A.; Zakrzewski, M.; Schlueter, A.; Schönberg, M.; Szczepanowski, R.; Goesmann, A.; Pühler, A.; Klocke, M. Characterization of Microbial Biofilms in a Thermophilic Biogas System by High-Throughput Metagenome Sequencing. FEMS Microbiol. Ecol. 2011, 79, 785–799. [Google Scholar] [CrossRef]
- Güllert, S.; Fischer, M.; Turaev, D.; Noebauer, B.; Ilmberger, N.; Wemheuer, B.; Alawi, M.; Rattei, T.; Daniel, R.; Schmitz, R.; et al. Deep Metagenome and Metatranscriptome Analyses of Microbial Communities Affiliated with an Industrial Biogas Fermenter, a Cow Rumen, and Elephant Feces Reveal Major Differences in Carbohydrate Hydrolysis Strategies. Biotechnol. Biofuels 2016, 9, 1–20. [Google Scholar] [CrossRef]
- Li, A.; Chu, Y.; Wang, X.; Ren, L.; Yu, J.; Liu, X.; Yan, J.; Zhang, L.; Wu, S.; Li, S.Z. A Pyrosequencing-Based Metagenomic Study of Methane-Producing Microbial Community in Solid-State Biogas Reactor. Biotechnol. Biofuels 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.; Tiedje, J.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5264–5267. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wang, Z.; Wang, Q.; Zhu, C.; Wu, Z. An Anaerobic Dynamic Membrane Bioreactor (AnDMBR) for Landfill Leachate Treatment: Performance and Microbial Community Identification. Bioresour. Technol. 2014, 161, 29–39. [Google Scholar] [CrossRef]
- Xu, Z.; Hansen, M.A.; Hansen, L.H.; Jacquiod, S.; Sørensen, S.J. Bioinformatic Approaches Reveal Metagenomic Characterization of Soil Microbial Community. PLoS ONE 2014, 9, e93445. [Google Scholar] [CrossRef]
- Zhang, J.; Loh, K.-C.; Li, W.; Lim, J.W.; Dai, Y.; Tong, Y.W. Three-Stage Anaerobic Digester for Food Waste. Appl. Energy 2017, 194, 287–295. [Google Scholar] [CrossRef]
- Zhang, J.; Loh, K.-C.; Lee, J.; Wang, C.-H.; Dai, Y.; Tong, Y. Three-Stage Anaerobic Co-Digestion of Food Waste and Horse Manure. Sci. Rep. 2017, 7, 1269. [Google Scholar] [CrossRef]
- Wong, M.T.; Zhang, D.; Li, J.; Hui, R.; Tun, H.; Brar, M.; Park, T.; Chen, Y.; Leung, F. Towards a Metagenomic Understanding on Enhanced Biomethane Production from Waste Activated Sludge after PH 10 Pretreatment. Biotechnol. Biofuels 2013, 6, 38. [Google Scholar] [CrossRef]
- Pervin, H.M.; Dennis, P.G.; Lim, H.J.; Tyson, G.W.; Batstone, D.J.; Bond, P.L. Drivers of Microbial Community Composition in Mesophilic and Thermophilic Temperature-Phased Anaerobic Digestion Pre-Treatment Reactors. Water Res. 2013, 47, 7098–7108. [Google Scholar] [CrossRef]
- Fernando Herrera Adarme, O.; Eduardo Lobo Baêta, B.; Cardoso Torres, M.; Camilo Otalora Tapiero, F.; Vinicius Alves Gurgel, L.; de Queiroz Silva, S.; Francisco de Aquino, S. Biogas Production by Anaerobic Co-Digestion of Sugarcane Biorefinery Byproducts: Comparative Analyses of Performance and Microbial Community in Novel Single-and Two-Stage Systems. Bioresour. Technol. 2022, 354, 127185. [Google Scholar] [CrossRef]
- Martínez, E.J.; Sotres, A.; Arenas, C.B.; Blanco, D.; Martínez, O.; Gómez, X. Improving Anaerobic Digestion of Sewage Sludge by Hydrogen Addition: Analysis of Microbial Populations and Process Performance. Energies 2019, 12, 1228. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Yu, H.; Wang, Z.; Wu, Z.; Zhu, C. Enhanced Waste Activated Sludge Digestion Using a Submerged Anaerobic Dynamic Membrane Bioreactor: Performance, Sludge Characteristics and Microbial Community. Sci. Rep. 2016, 6, 20111. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing Structure and Assembly Cues for Arabidopsis Root-Inhabiting Bacterial Microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, N. Ggtern: An Extension to “Ggplot2”, for the Creation of Ternary Diagrams. R Package Version 2.1.4. 2016. Available online: http://CRAN.R-project.org/package=ggtern (accessed on 13 November 2022).
- Zhuravleva, E.A.; Shekhurdina, S.V.; Kotova, I.B.; Loiko, N.G.; Popova, N.M.; Kryukov, E.; Kovalev, A.A.; Kovalev, D.A.; Litti, Y.V. Effects of Various Materials Used to Promote the Direct Interspecies Electron Transfer on Anaerobic Digestion of Low-Concentration Swine Manure. Sci. Total Environ. 2022, 839, 156073. [Google Scholar] [CrossRef]
- Lu, Y. Microbial Ecology of Fermentative Microbes in Anaerobic Granules. Ph.D. Thesis, School of Chemical Engineering, the University of Queensland, Brisbane, Australia, 2014. [Google Scholar] [CrossRef]
- Breton-Deval, L.; Salinas-Peralta, I.; Alarcón Aguirre, J.S.; Sulbarán-Rangel, B.; Gurubel Tun, K.J. Taxonomic Binning Approaches and Functional Characteristics of the Microbial Community during the Anaerobic Digestion of Hydrolyzed Corncob. Energies 2021, 14, 66. [Google Scholar] [CrossRef]
- Bibby, K.; Viau, E.; Peccia, J. Pyrosequencing of the 16S RRNA Gene to Reveal Bacterial Pathogen Diversity in Biosolids. Water Res. 2010, 44, 4252–4260. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xue, Y.; Chen, S.; Takahashi, J.; Dai, L.; Dai, X. Methanogenic Population Dynamics Regulated by Bacterial Community Responses to Protein-Rich Organic Wastes in a High Solid Anaerobic Digester. Chem. Eng. J. 2017, 317, 444–453. [Google Scholar] [CrossRef]
- Westerholm, M.; Crauwels, S.; Houtmeyers, S.; Meerbergen, K.; Van Geel, M.; Lievens, B.; Appels, L. Microbial Community Dynamics Linked to Enhanced Substrate Availability and Biogas Production of Electrokinetically Pre-Treated Waste Activated Sludge. Bioresour. Technol. 2016, 218, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A. Multivariate Analyses in Microbial Ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef]
- Cole’s, J.R.; Wang, Q.; Fish, J.; Chai, B.; Mcgarrell, D.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.; Tiedje, J. Ribosomal DATABASE PROject: Data and Tools for High Throughput RRNA Analysis. Nucleic Acids Res. 2013, 42, D633–D642. [Google Scholar] [CrossRef]
- Lee, J.; Hwang, B.; Koo, T.; Shin, S.G.; Kim, W.; Hwang, S. Temporal Variation in Methanogen Communities of Four Different Full-Scale Anaerobic Digesters Treating Food Waste-Recycling Wastewater. Bioresour. Technol. 2014, 168, 59–63. [Google Scholar] [CrossRef]
- Ziganshina, E.E.; Ibragimov, E.M.; Vankov, P.Y.; Miluykov, V.A.; Ziganshin, A.M. Comparison of Anaerobic Digestion Strategies of Nitrogen-Rich Substrates: Performance of Anaerobic Reactors and Microbial Community Diversity. Waste Manag. 2017, 59, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Paliy, O.; Shankar, V. Application of Multivariate Statistical Techniques in Microbial Ecology. Mol. Ecol. 2016, 25, 1032–1057. [Google Scholar] [CrossRef]
- Lepš, J.; Šmilauer, P. Multivariate Analysis of Ecological Data Using CANOCO; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar] [CrossRef]
- Tong, J.; Liu, J.; Zheng, X.; Zhang, J.; Ni, X.; Chen, M.; Wei, Y. Fate of Antibiotic Resistance Bacteria and Genes during Enhanced Anaerobic Digestion of Sewage Sludge by Microwave Pretreatment. Bioresour. Technol. 2016, 217, 37–43. [Google Scholar] [CrossRef]
- Alcántara-Hernández, R.; Taş, N.; Carlos, S.; Durán-Moreno, A.; Falcón, L. Microbial Dynamics in Anaerobic Digestion Reactors for Treating Organic Urban Residues during the Start-up Process. Lett. Appl. Microbiol. 2017, 64, 438–445. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.-C.; Estreicher, A.; Gasteiger, E.; Martin, M.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The Swiss-Prot Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Schlueter, A.; Bekel, T.; Diaz, N.; Dondrup, M.; Eichenlaub, R.; Gartemann, K.-H.; Krahn, I.; Krause, L.; Krömeke, H.; Kruse, O.; et al. The Metagenome of a Biogas-Producing Microbial Community of a Production-Scale Biogas Plant Fermenter Analyzed by the 454-Pyrosequencing Technology. J. Biotechnol. 2008, 136, 77–90. [Google Scholar] [CrossRef]
- Meyer, F.; Goesmann, A.; McHardy, A.C.; Bartels, D.; Bekel, T.; Clausen, J.; Kalinowski, J.; Linke, B.; Rupp, O.; Giegerich, R.; et al. GenDB—An Open Source Genome Annotation System for Prokaryote Genomes. Nucleic Acids Res. 2003, 31, 2187–2195. [Google Scholar] [CrossRef]
- Markowitz, V.; Chen, I.-M.; Chu, K.; Szeto, E.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Pagani, I.; Tringe, S.; et al. IMG/M 4 Version of the Integrated Metagenome Comparative Analysis System. Nucleic Acids Res. 2013, 42, D568–D573. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The Integrative Protein Signature Database. Nucleic Acids Res. 2009, 37 (Suppl. 1), D211–D215. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The Gene Ontology (GO) Database and Informatics Resource. Nucleic Acids Res. 2004, 32 (Suppl. 1), D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J. Microarrays for Bacterial Detection and Microbial Community Analysis. Curr. Opin. Microbiol. 2003, 6, 288–294. [Google Scholar] [CrossRef]
- Hoff, K.J. The Effect of Sequencing Errors on Metagenomic Gene Prediction. BMC Genom. 2009, 10, 520. [Google Scholar] [CrossRef]
- Jünemann, S.; Kleinboelting, N.; Jaenicke, S.; Henke, C.; Hassa, J.; Nelkner, J.; Stolze, Y.; Albaum, S.; Schlueter, A.; Goesmann, A.; et al. Bioinformatics for NGS-Based Metagenomics and the Application to Biogas Research. J. Biotechnol. 2017, 261, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Cardona, C.; Weisenhorn, P.; Henry, C.; Jack, G. Network-Based Metabolic Analysis and Microbial Community Modeling. Curr. Opin. Microbiol. 2016, 31, 124–131. [Google Scholar] [CrossRef]
- Faust, K.; Lahti, L.; Gonze, D.; de Vos, W.M.; Raes, J. Metagenomics Meets Time Series Analysis: Unraveling Microbial Community Dynamics. Curr. Opin. Microbiol. 2015, 25, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Bassani, I.; Kougias, P.G.; Treu, L.; Angelidaki, I. Biogas Upgrading via Hydrogenotrophic Methanogenesis in Two-Stage Continuous Stirred Tank Reactors at Mesophilic and Thermophilic Conditions. Environ. Sci. Technol. 2015, 49, 12585–12593. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Li, B.; Li, L.; Zhang, T.; Angelidaki, I. Antibiotic Resistance Genes and Correlations with Microbial Community and Metal Resistance Genes in Full-Scale Biogas Reactors as Revealed by Metagenomic Analysis. Environ. Sci. Technol. 2017, 51, 4069–4080. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.; Shin, S.G.; Lee, J.; Han, G.; Kim, W.; Cho, K.; Hwang, S. Identifying Methanogen Community Structures and Their Correlations with Performance Parameters in Four Full-Scale Anaerobic Sludge Digesters. Bioresour. Technol. 2017, 228, 368–373. [Google Scholar] [CrossRef]
- Hidaka, T.; Tsushima, I.; Tsumori, J. Comparative Analyses of Microbial Structures and Gene Copy Numbers in the Anaerobic Digestion of Various Types of Sewage Sludge. Bioresour. Technol. 2018, 253, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Lee, J.; Loh, K.-C.; Dai, Y.; Tong, Y.W. Enhancement of Biogas Production in Anaerobic Co-Digestion of Food Waste and Waste Activated Sludge by Biological Co-Pretreatment. Energy 2017, 137, 479–486. [Google Scholar] [CrossRef]
- Suksong, W.; Kongjan, P.; Prasertsan, P.; Imai, T.; O-Thong, S. Optimization and Microbial Community Analysis for Production of Biogas from Solid Waste Residues of Palm Oil Mill Industry by Solid-State Anaerobic Digestion. Bioresour. Technol. 2016, 214, 166–174. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.; Loh, K.-C. Activated Carbon Enhanced Anaerobic Digestion of Food Waste–Laboratory-Scale and Pilot-Scale Operation. Waste Manag. 2018, 75, 270–279. [Google Scholar] [CrossRef]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse Hydrogen Production and Consumption Pathways Influence Methane Production in Ruminants. ISME J. 2019, 13, 2617–2632. [Google Scholar] [CrossRef]
- Zhang, L.; Gong, X.; Chen, Z.; Zhou, Y. Genome-Centric Metagenomics Analysis Revealed the Metabolic Function of Abundant Microbial Communities in Thermal Hydrolysis-Assisted Thermophilic Anaerobic Digesters under Propionate Stress. Bioresour. Technol. 2022, 360, 127574. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Pilarski, K.; Adamski, M.; Grzyb, A.; Grządziel, J.; Gałązka, A. Silica/Lignin Carrier as a Factor Increasing the Process Performance and Genetic Diversity of Microbial Communities in Laboratory-Scale Anaerobic Digesters. Energies 2021, 14, 4429. [Google Scholar] [CrossRef]
- Kim, N.-K.; Lee, S.-H.; Kim, Y.; Park, H.-D. Current understanding and perspectives in anaerobic digestion based on genome-resolved metagenomic approaches. Bioresour. Technol. 2022, 344, 126350. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Banks, C.; Zhang, Y.; Heaven, S.; Longhurst, P. Quantifying the Percentage of Methane Formation via Acetoclastic and Syntrophic Acetate Oxidation Pathways in Anaerobic Digesters. Waste Manag. 2018, 71, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Fotidis, I.A.; Mancini, E.; Treu, L.; Mahdy, A.; Ballesteros, M.; González-Fernández, C.; Angelidaki, I. Acclimation to Extremely High Ammonia Levels in Continuous Biomethanation Process and the Associated Microbial Community Dynamics. Bioresour. Technol. 2018, 247, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, W.; He, Y.; Zhang, R.; Liu, G. Effect of Ammonia on Methane Production, Methanogenesis Pathway, Microbial Community and Reactor Performance under Mesophilic and Thermophilic Conditions. Renew. Energy 2018, 125, 915–925. [Google Scholar] [CrossRef]
- Zhang, S.; Xiao, M.; Liang, C.; Chui, C.; Wang, N.; Shi, J.; Liu, L. Multivariate Insights into Enhanced Biogas Production in Thermophilic Dry Anaerobic Co-Digestion of Food Waste with Kitchen Waste or Garden Waste: Process Properties, Microbial Communities and Metagenomic Analyses. Bioresour. Technol. 2022, 361, 127684. [Google Scholar] [CrossRef]
- Fan, Y.; Niu, X.; Zhang, D.-Q.; Lin, Z.; Fu, M.; Zhou, S. Analysis of the Characteristics of Phosphine Production by Anaerobic Digestion Based on Microbial Community Dynamics, Metabolic Pathways, and Isolation of the Phosphate-Reducing Strain. Chemosphere 2021, 262, 128213. [Google Scholar] [CrossRef]
- Cai, G.; Zhao, L.; Wang, T.; Lv, N.; Li, J.; Ning, J.; Pan, X.; Zhu, G. Variation of Volatile Fatty Acid Oxidation and Methane Production during the Bioaugmentation of Anaerobic Digestion System: Microbial Community Analysis Revealing the Influence of Microbial Interactions on Metabolic Pathways. Sci. Total Environ. 2021, 754, 142425. [Google Scholar] [CrossRef]
- Jin, W.; Xu, X.; Yang, F. Application of Rumen Microorganisms for Enhancing Biogas Production of Corn Straw and Livestock Manure in a Pilot-Scale Anaerobic Digestion System: Performance and Microbial Community Analysis. Energies 2018, 11, 920. [Google Scholar] [CrossRef]
- Murillo-Roos, M.; Uribe-Lorío, L.; Fuentes-Schweizer, P.; Vidaurre-Barahona, D.; Brenes-Guillén, L.; Jiménez, I.; Arguedas, T.; Liao, W.; Uribe, L. Biogas Production and Microbial Communities of Mesophilic and Thermophilic Anaerobic Co-Digestion of Animal Manures and Food Wastes in Costa Rica. Energies 2022, 15, 3252. [Google Scholar] [CrossRef]
- Lee, J.; Shin, S.G.; Jang, H.M.; Kim, Y.; Lee, J.; Kim, Y. Characterization of Antibiotic Resistance Genes in Representative Organic Solid Wastes: Food Waste-Recycling Wastewater, Manure, and Sewage Sludge. Sci. Total Environ. 2016, 579, 1692–1698. [Google Scholar] [CrossRef]
- Jang, H.M.; Shin, J.; Choi, S.; Shin, S.G.; Park, K.Y.; Cho, J.; Kim, Y.M. Fate of Antibiotic Resistance Genes in Mesophilic and Thermophilic Anaerobic Digestion of Chemically Enhanced Primary Treatment (CEPT) Sludge. Bioresour. Technol. 2017, 244, 433–444. [Google Scholar] [CrossRef]
- Oulas, A.; Pavloudi, C.; Polymenakou, P.; Pavlopoulos, G.; Papanikolaou, N.; Kotoulas, G.; Arvanitidis, C.; Iliopoulos, I. Metagenomics: Tools and Insights for Analyzing Next-Generation Sequencing Data Derived from Biodiversity Studies. Bioinform. Biol. Insights 2015, 9, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Pu, C.; Liu, H.; Ding, G.; Sun, Y.; Yu, X.; Chen, J.; Ren, J.; Gong, X. Impact of Direct Application of Biogas Slurry and Residue in Fields: In Situ Analysis of Antibiotic Resistance Genes from Pig Manure to Fields. J. Hazard. Mater. 2017, 344, 441–449. [Google Scholar] [CrossRef]
- Yin, X.; Jiang, X.-T.; Chai, B.; Li, L.; Yang, Y.; Cole’s, J.R.; Tiedje, J.; Zhang, T. ARGs-OAP v2.0 with an Expanded SARG Database and Hidden Markov Models for Enhancement Characterization and Quantification of Antibiotic Resistance Genes in Environmental Metagenomes. Bioinformatics 2018, 34, 2263–2270. [Google Scholar] [CrossRef]
- Hanemaaijer, M.; Röling, W.; Olivier, B.; Khandelwal, R.; Teusink, B.; Bruggeman, F. Systems Modeling Approaches for Microbial Community Studies: From Metagenomics to Inference of the Community Structure. Front. Microbiol. 2015, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Antonella, S.; Wanjiku, M.F.; Oliver, E. A Diverse Community to Study Communities: Integration of Experiments and Mathematical Models to Study Microbial Consortia. J. Bacteriol. 2017, 199, e00865-16. [Google Scholar] [CrossRef]
- Shaw, G.; Pao, Y.-Y.; Wang, D. MetaMIS: A Metagenomic Microbial Interaction Simulator Based on Microbial Community Profiles. BMC Bioinform. 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chuai, G.; Qi, T.; Shao, F.; Zhou, C.; Zhu, C.; Yang, J.; Yu, Y.; Shi, C.; Kang, N.; et al. MetaTopics: An Integration Tool to Analyze Microbial Community Profile by Topic Model. BMC Genom. 2017, 18, 962. [Google Scholar] [CrossRef]
- Hanemaaijer, M.; Olivier, B.G.; Röling, W.F.M.; Bruggeman, F.J.; Teusink, B. Model-Based Quantification of Metabolic Interactions from Dynamic Microbial-Community Data. PLoS ONE 2017, 12, e0173183. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.; Gibbons, S.; Jack, G. Modeling Microbial Community Structure and Functional Diversity Across Time and Space. FEMS Microbiol. Lett. 2012, 332, 91–98. [Google Scholar] [CrossRef]
- Succurro, A.; Ebenhöh, O. Review and Perspective on Mathematical Modeling of Microbial Ecosystems. Biochem. Soc. Trans. 2018, 46, BST20170265. [Google Scholar] [CrossRef]
- Chen, Y.; Lan, S.; Wang, L.; Dong, S.; Zhou, H.; Tan, Z.; Li, X. A Review: Driving Factors and Regulation Strategies of Microbial Community Structure and Dynamics in Wastewater Treatment Systems. Chemosphere 2017, 174, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, R.; Liu, F.; Yong, X.; Wu, X.; Zheng, T.; Jiang, M.; Jia, H. Biogas Production and Microbial Community Shift through Neutral PH Control during the Anaerobic Digestion of Pig Manure. Bioresour. Technol. 2016, 217, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulos, A. Soft Sensor Development and Process Control of Anaerobic Digestion. Ph.D. Thesis, University of Exeter, Devon, UK, 2013; p. 281. Available online: http://hdl.handle.net/10871/15068 (accessed on 13 November 2022).
- Xue, B.; Li, Z.; Wang, X.; Xi, H.; Cheng, S.; Xiaofeng, B.; Gao, R. Online Measurement of Alkalinity in Anaerobic Co-Digestion Using Linear Regression Method. Int. J. Agric. Biol. Eng. 2017, 10, 176–183. [Google Scholar] [CrossRef]
- Amha, Y.M.; Anwar, M.Z.; Brower, A.; Jacobsen, C.S.; Stadler, L.B.; Webster, T.M.; Smith, A.L. Inhibition of Anaerobic Digestion Processes: Applications of Molecular Tools. Bioresour. Technol. 2018, 247, 999–1014. [Google Scholar] [CrossRef]
- Dröge, J.; McHardy, A. Taxonomic Binning of Metagenome Samples Generated by Next-Generation Sequencing Technologies. Brief. Bioinform. 2012, 13, 646–655. [Google Scholar] [CrossRef]
- Tabish, M.; Azim, S.; Husain, M.A.; Rehman, S.; Sarwar, T.; Ishqi, H. Bioinformatics Approaches in Studying Microbial Diversity. In Management of Microbial Resources in the Environment; Springer: Berlin/Heidelberg, Germany, 2013; pp. 119–140. [Google Scholar] [CrossRef]
- Campanaro, S.; Treu, L.; Kougias, P.; Zhu, X.; Angelidaki, I. Taxonomy of Anaerobic Digestion Microbiome Reveals Biases Associated with the Applied High Throughput Sequencing Strategies. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Widder, S.; Allen, R.; Pfeiffer, T.; Curtis, T.; Wiuf, C.; Sloan, W.; Cordero, O.X.; Brown, S.; Momeni, B.; Shou, W.; et al. Challenges in Microbial Ecology: Building Predictive Understanding of Community Function and Dynamics. ISME J. 2016, 0, 2557–2568. [Google Scholar] [CrossRef]
- Hiraoka, S.; Yang, C.; Iwasaki, W. Metagenomics and Bioinformatics in Microbial Ecology: Current Status and Beyond. Microbes Environ. 2016, 31, 204–212. [Google Scholar] [CrossRef]
- Tolvanen, K.E.S.; Karp, M.T. Molecular Methods for Characterizing Mixed Microbial Communities in Hydrogen-Fermenting Systems. Int. J. Hydrogen Energy 2011, 36, 5280–5288. [Google Scholar] [CrossRef]
- Palaniswamy, D.; Ramesh, G.; Sivasankaran, S.; Kathiravan, N. Optimising Biogas from Food Waste Using a Neural Network Model. Proc. Inst. Civ. Eng.-Munic. Eng. 2017, 170, 221–229. [Google Scholar] [CrossRef]
- Nair, V.V.; Dhar, H.; Kumar, S.; Thalla, A.K.; Mukherjee, S.; Wong, J.W.C. Artificial Neural Network Based Modeling to Evaluate Methane Yield from Biogas in a Laboratory-Scale Anaerobic Bioreactor. Bioresour. Technol. 2016, 217, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Antwi, P.; Li, J.; Opoku Boadi, P.; Meng, J.; Shi, E.; Deng, K.; Bondinuba, F. Estimation of Biogas and Methane Yields in an UASB Treating Potato Starch Processing Wastewater with Backpropagation Artificial Neural Network. Bioresour. Technol. 2016, 228, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Eng, A.; Borenstein, E. An Algorithm for Designing Minimal Microbial Communities with Desired Metabolic Capacities. Bioinformatics 2016, 32, 2008–2016. [Google Scholar] [CrossRef] [PubMed]


| Brand Name | Country Name | DNA Isolation Kit | Usage Percentage | Reference |
|---|---|---|---|---|
| Clontech | USA | PCR reaction mix | 2% | [40] |
| Zymo Research | USA | ZR soil microbe DNA kit | 2% | [44] |
| Felix bio-tech | USA | DNA extraction kit | 2% | [45] |
| Intron biotechnology | Korea | I-genomic BYF DNA extraction kit | 2% | [46] |
| Magtration System 6GC, Precision System Science | Japan | Automated nucleic acid kit | 6% | [38,47,48] |
| Macherey-Nagel | Germany | NucleoSpin Tissue kit+NucleoSpin soil kit | 6% | [49] |
| OMEGA | USA | E.Z.N.A Soil DNA kit | 6% | [10,50] |
| Q-Bio gene | Australia, Carlsbad, CA, USA | Fast DNA SPIN kit for soil | 25% | [51] |
| MP Biomedicals | Illkirch, France, Australia, Germany, USA | [52] | ||
| MoBio Laboratories | USA | MoBio PowerSoil DNA extraction kit | 43% | [53,54] |
| - | - | CTAB (cetyltrimethylammonium bromide) based DNA extraction method | 6% | [37,55,56,57,58,59] |
| Analysis Type | Name of Software | Reference |
|---|---|---|
| Measurement and analysis of performance, systemic metabolic processes, annotation of genomes, study of principal coordinates, statistics based on canonical correspondence, data from filtered pyrosequencing runs. | MG-RAST | [65] |
| Removing chimaera sequences, ensuring high quality, the assessment of variety, richness, and abundance of base coverage, study of principal coordinates, analysing Good’s scope of coverage, Alignment and quality control of sequences, sample size normalisation | MOTHUR | [52] |
| Shortening of reads | Trimmomatic software | [62] |
| Shortening and aligning reads | HMMER | [67] |
| Identified sequence reads | ARB rRNA database | [67] |
| Assembling of genome | CLC Genomics workbench | [57,62,63] |
| Combination of end-pair reads | FLASH | [17,68] |
| Analysis of microbial population relation | MetaMIS | [17,65,68] |
| Interaction network topology analysis | Gephi | [17] |
| CLUSTAL_X | Iterative sequence alignments; gap editing | [17] |
| PAST | Illustration of beta-diversity metrics | [57,62] |
| mPUMA and Trinity | Assembly and processing sequences | [65] |
| Chimera Slayer | Chromosome complexes elimination. | [69] |
| ClustalW | Alignment of sequences | [49] |
| INFERNAL aligner | Alignment of various clean sequences | [10,49,70] |
| SAMS | Evaluation of the quality of sequences | [70] |
| GenDB genome annotation system | Long-read assembly and functional annotation | [10] |
| Regano | Code-sequence prediction | [60,71] |
| Pipeline | Aligning, trimming, and sorting sequences; analysing biodiversity; naming sequences by taxonomy | [60] |
| RDP (Ribosomal Database Project) | Differences in community architecture | [69] |
| Fast UniFrac | Illustrate co-relations between microbial structure and attributes | [69] |
| CANOCO | Identify causes and effects of microbial communities on reactor efficiency | [69] |
| XLSTAT | Comparison of taxonomies between two samples using pairwise statistics | [72] |
| STAMP | MG-RAST | [65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Upadhyay, A.; Kovalev, A.A.; Zhuravleva, E.A.; Kovalev, D.A.; Litti, Y.V.; Masakapalli, S.K.; Pareek, N.; Vivekanand, V. A Review of Basic Bioinformatic Techniques for Microbial Community Analysis in an Anaerobic Digester. Fermentation 2023, 9, 62. https://doi.org/10.3390/fermentation9010062
Upadhyay A, Kovalev AA, Zhuravleva EA, Kovalev DA, Litti YV, Masakapalli SK, Pareek N, Vivekanand V. A Review of Basic Bioinformatic Techniques for Microbial Community Analysis in an Anaerobic Digester. Fermentation. 2023; 9(1):62. https://doi.org/10.3390/fermentation9010062
Chicago/Turabian StyleUpadhyay, Apoorva, Andrey A. Kovalev, Elena A. Zhuravleva, Dmitriy A. Kovalev, Yuriy V. Litti, Shyam Kumar Masakapalli, Nidhi Pareek, and Vivekanand Vivekanand. 2023. "A Review of Basic Bioinformatic Techniques for Microbial Community Analysis in an Anaerobic Digester" Fermentation 9, no. 1: 62. https://doi.org/10.3390/fermentation9010062
APA StyleUpadhyay, A., Kovalev, A. A., Zhuravleva, E. A., Kovalev, D. A., Litti, Y. V., Masakapalli, S. K., Pareek, N., & Vivekanand, V. (2023). A Review of Basic Bioinformatic Techniques for Microbial Community Analysis in an Anaerobic Digester. Fermentation, 9(1), 62. https://doi.org/10.3390/fermentation9010062










