Abstract
This study highlights the decomposing role through the hydrolytic activities of fungi isolated from natural environments represented by brackish and hypersaline lakes in Romania. Novel strains belonging to the Penicillium, Aspergillus, and Emericellopsis genera were isolated and screened for the ability to produce extracellular hydrolytic enzymes, i.e., proteases, lipases, amylases, cellulases, xylanases, and pectinases. According to salt requirements, they were classified as moderate halophilic and halotolerant strains. Agar plate-based assays with Tween 80, slide cultures with organic deposits, and quantitative evaluation allowed the selection of Aspergillus sp. BSL 2-2, Penicillium sp. BSL 3-2, and Emericellopsis sp. MM2 as potentially good decomposers of organic matter not only in lakes but also on deposits covering the mural paintings. Experiments performed on painted experimental models revealed that only Penicillium sp. BSL 3-2 decomposed Paraloid B72, transparent dispersion of casein, beeswax, sunflower oil, and soot. Moreover, using microscopic, spectroscopic, and imaging methods, it was proved the efficiency of Penicillium sp. BSL 3-2 for decomposition of organic deposits artificially applied on frescoes fragments.
1. Introduction
By receiving and processing large amounts of leaf litter, wood fragments, and dead animal bodies, terrestrial and aquatic ecosystems work as veritable factories of decay. Debris is either assimilated into biomass or mineralized to CO2. Fungi primarily contribute to terrestrial carbon turnover by providing litter carbon to the microbial loop, while bacteria determine whether the supplied carbon substrate is assimilated into biomass [1,2,3]. There are significant changes in microbial colonization after the collapse of plants, i.e., when plant material accumulates on the ecosystem’s floor [4].
Fungi evolved, increased size and complexity, and metabolic functioning trying to adapt and interact with different environmental conditions or hosts. As decomposers or through their interactions with plants, fungi changed terrestrial ecology and geology and modified the Earth’s atmosphere [5]. Fungi are key organisms of both terrestrial and aquatic habitats, being involved in organic-matter decomposition, cycling of vitally important chemical elements, as well as plant pathogenicity and symbioses [6]. As decomposers, fungi are attached to the substrate by hyphae and produce enzymes, such as cellulases, amylases, pectinases, proteases, and lipases, which hydrolyze plant polysaccharides (cellulose, starch, pectin, xylan), proteins and lipids. In biotechnology, selected fungi serve as veritable enzyme-producing factories. Fungal enzymes are exploited in various industries. Fungal amylases (α-amylases, β-amylases, and glucoamylases) are involved in starch depolymerization and are used mainly in the food, detergent, and textile industries [7,8] as well as in the field of restoration of cultural heritage, namely for removing the starch glue. The enzymes involved in cellulose decomposition, i.e., endoglucanases, exoglucanases, and β-glucosidases, act synergistically and have important roles in the food, feed, detergent, textile, pulp and paper, and biofuels industries [9]. Similarly, due to its heterogeneity and complexity, the complete hydrolysis of xylan requires the synergistic action of a wide variety of enzymes, the most important being endo-1,4-β-xylanases and β-xylosidases. Xylanolytic enzymes are used in various industrial applications beyond biomass saccharification, e.g., in food, animal feed, biofuel, pulp, and paper industries [10]. Proteases hydrolyze the peptide bonds of proteins into peptides and amino acids and have potential application in a wide number of industrial processes such as food, laundry detergent [11], and in the field of cultural heritage for removing bone glue. Fungal pectinases are mixed enzymes that decompose pectic substances from plants and have various applications in the textile, plant fiber processing, and oil extraction industries, as well as in the treatment of industrial wastewater [12].
Lipolytic enzymes, including lipases and esterases, represent a versatile group of enzymes with diverse amino acid sequences but related three-dimensional structures. Kovacic et al. [13] proposed 19 families of lipolytic enzymes based on the conserved features of sequences and structures and correlated the biochemical properties of some enzymes with the nature of the microorganism from which the respective enzyme was isolated. Both lipases and esterases catalyze the hydrolysis of esters. Many different criteria have been proposed to distinguish between lipases and esterases, but all are indicative and not decisive [14]. Esterases are included in the class of hydrolases and catalyze the cleavage and formation of ester bonds [15], being involved in esterification, interesterification, and transesterification reactions. Esterases hydrolyze short-chain carboxylic acids (C ≤ 12), while lipases hydrolyze insoluble long chain (C ≥ 12) triglycerides and secondary alcohols [16]. Fungal feruloyl esterase releases ferulic acids and other phenolic acids from plant debris and facilitates the degradation of polysaccharides by removing the ester bonds between plant polymers [17]. Fungal glucuronyl esterase catalyzes the cleavage of the ester bond in glucuronoxylans, being able to degrade the lignin–carbohydrate complex in nature or for biotechnological applications such as woody biomass utilization [18]. Based on the finding that degradation of poly butylene succinate-co-adipate (PBSA) film takes place in soil [19], esterase-producing fungi were recommended as highly valuable decomposers both in aquatic and terrestrial ecosystems, opening a window for their application in other fields such as the biocleaning of frescoes concomitantly.
Although previously considered hostile to normal life forms, the salty ecosystems represent nowadays a promising source of novel halophilic microorganisms harboring metabolic pathways that function in saline conditions. For example, halophilic and halotolerant enzymes are biologically active in which the structure and functionality of common enzymes are generally critically affected.
The unique properties of halophilic enzymes (high content of acidic residues on the surface, low frequency of basic amino acids, a high number of salt bridges, and low hydrophobicity), the requirement of salt for stability and activity, and high resistance to denaturation methods gained a special interest throughout the scientific community. Several halophilic enzymes, including glycosidases, proteases, and lipases, have been purified and characterized in recent years to be used in biofuel production, for the biodegradation of organic pollutants, or in food processing [20]. Apart from being stable in the presence of significant amounts of salt, the halo-proteases could also display polyextremophilic characteristics such as tolerance to alkaline pH, high temperature, organic solvents. Furthermore, halophilic lipases and esterases represent a useful tool for many syntheses in the production of detergents, biosurfactants, and cosmetics, in the pharmaceutical, food, paper, and oil–chemical industries.
In addition to the beneficial roles of fungi as decomposers in natural environments, their growth on artistic items may cause the deterioration of cultural heritage. The bio deteriorative contribution of fungi involves calcium carbonate solubilization or dissolution, mineralization or crystallization development, and enzymatic action (ligninolytic, lipolytic, proteolytic, cellulolytic abilities) [21]. Apart from the fact that fungi produce irreversible damage to artifacts, they also represent a risk to human health. To avoid this, decontamination is carried out before any type of activity, including manipulation. In this respect, different chemicals (biocides) or green products are used [22,23,24,25,26,27].
Cultural heritage buildings and artifacts require restoration, which brings them as close as possible to the original shape and aspect by consolidation and cleaning. Traditional conservation treatments are gradually replaced by green technologies (less aggressive, more specific, and more sustainable), including bioconsolidation, protection, and biocleaning [28,29,30,31] based on living microorganisms or their metabolites. Bioconsolidation is performed with selected carbonatogenic bacteria or by stimulation of relevant autochthonous microorganisms, including fungi [30,31,32]. The biocleaning of various surfaces is based on applying living microorganisms for removing inorganic deposits (e.g., sulfate-reducing bacteria such as Desulfovibrio desulfuricans, nitrite-reducing bacteria such as Pseudomonas stutzeri), or hydrolytic enzymes for removing of organic materials (delivery system + hydrolytic enzymes such as proteases, lipases) [28,29,33,34]. To our knowledge, there has been no research on the isolation of fungi from brackish and hypersaline environments to be tested in order to highlight their role as decomposers of organic deposits on mural surfaces affected by salts. In this context, the perspective of the current study is to use moderately halophilic and halotolerant fungal isolates in the decomposition of organic deposits (i.e., consolidants such as acrylic resin Paraloid® B72 and Transparent Dispersion of Casein or contaminants such as beeswax, sunflower oil, and soot) from murals covered by efflorescences.
Under limiting conditions such as salt crystallization, biocatalyzts capable of retaining structural stability and catalytic activity over a wide range of salinities could represent the optimal solution to remove unwanted inorganic or organic from the surfaces of wall paintings and historical monuments [28].
The present study aims to: (I) prospect for hydrolase-producing fungi from brackish and hypersaline lakes; (II) perform the molecular identification of the isolates of interest; (III) establish the optimal methodology for selecting lipase-producing strains with possible applications in the field of mural restoration; (IV) test the best lipase-producing strains for the decomposition of deposits on the surface of painted laboratory models and fresco fragments. The research starts from the hypothesis that brackish and hypersaline lakes are reservoirs of fungi-producing salt-tolerant hydrolases with potential applicability in the biocleaning of salt-loaded mural paintings.
2. Materials and Methods
2.1. Salt Requirements of Fungal Isolates and Qualitative Evaluation of Hydrolytic Enzymes
Microbial strains were isolated from natural environments represented by brackish lakes (Lake Amara—AM, Lake Balta Alba—BA) and hypersaline lakes (Movila Miresei Salt Lake—MM and Braila Salt lake—BSL). From these lakes, water, leaves, and small fragments of branches and sediment samples were collected and analyzed. From a total of 244 microbial isolates, 182 (74.6%) were represented by bacteria, 22 (9%) by archaea, and 40 (16.4%) by fungi [20]. Water sample was the source for Aspergillus sp. BA 1-4; leaves and small fragments of branches samples were sources for Emericelopsis sp. AM1, Aspergillus sp. AM 1-1, Aspergillus sp. BA 2-2 and BA 2-3, Aspergillus sp. BSL 1-2; sediment samples have content Emericellopsis sp. MM 2-3, MM2 and MM 1-2, Aspergillus sp. MM 1-1 and MM 3-3, Aspergillus sp. BSL 3-1 and BSL 2-2, Penicillium sp. MM 1-4, MM 3-2 and BSL 3-2. The salinity level measured in situ with a portable multiparameter instrument for water analysis (Hanna HI98194) was as follows: AM = 11.31 0.15; BA = 12.36 0.11; MM = >70; BSL = >70. According to previous classification, brackish lakes have a salinity of 36 g∙L−1 and hypersaline lakes >50 g∙L−1 [20].
Salt requirements were evaluated by inoculation of fungal isolates on HM growth medium containing (g/L): yeast extract (10), 7.0 MgCl2 × 6H2O (7), MgSO4 × 7H2O (9.6), CaCl2 × 2H2O (0.36), KCl (2.0), NaHCO3 (0.06), NaBr (0.026) [34] and different NaCl concentrations (0, 0.5, 1.2, 3.0, 3.5, 4.0 and 4.5 M) followed by incubation at 28 °C for 30 days. Positive results were considered when growth was observed. Halotolerant fungi were considered as a result of their growth on HM nutrients with 0 and 2 M NaCl (optimally at 0–1 M). Moderately halophilic fungi grew slowly (more than 15 days) on HM nutrients with 3.0–3.5 M NaCl (optimally at 3.0 M).
The ability of fungal strains to produce amylase, cellulase, lipase, pectinase, protease, and xylanase was tested by agar plate-based assays. The HM growth media containing, instead of glucose, one of the substrates of interest (i.e., starch, carboxymethyl cellulose, Tween 80, pectin, and casein) were inoculated in triplicate with fungal spores and incubated at 28 °C for 14 days. The production of enzymes was indicated by clear zones around the colonies after flooding the plates with KI solution (amylase and pectinase), Congo red solution (cellulase and xylanase), or 1N HCl (protease). Lipolytic activity was put in evidence by an opaque halo. The levels of enzyme activities (LEA) were evaluated using the formula LEA = diameter of the hydrolysis zone divided by the diameter of the microbial colonies (in millimeters) [20].
2.2. Taxonomic Identification
The fungal strains were identified by PCR amplification and sequencing of the ITS1-5.8S-ITS2 region [35]. Briefly, DNA was extracted from mycelia using the Quick-DNA Fungal/Bacterial Kit (Zymo Research, Irvine, CA, USA), and the ITS sequence was amplified using the primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATTGATATGC) under the conditions described previously [35]. Amplicons were purified using the DNA Clean and Concentrator Kit (Zymo Research, Irvine, CA, USA) and directly sequenced by Macrogen Europe (Amsterdam, The Netherlands). The partial ITS sequences were deposited in GenBank under the accession numbers listed in Table S1. The phylogenetic tree was constructed in MEGA X from multiple sequence alignments (CLUSTALW algorithm), using the Neighbor-joining method and the Tamura–Nei model.
2.3. Establishing the Optimal Methodology for Selecting Lipase-Producing Strains
Lipase-producing strains were selected based on the criteria described in Figure 1.
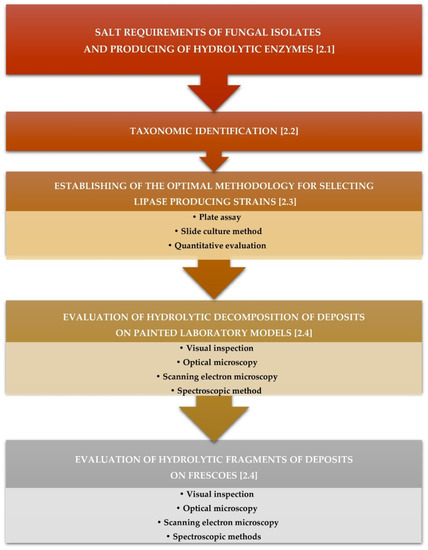
Figure 1.
Synopsis of the experiments.
The qualitative evaluation of lipase activity by agar plate-based assay involved the spot-inoculation of HM media containing Tween-80 instead of glucose with 17 fungal strains, followed by incubation at 28 °C for 14 days as previously described (Section 2.1).
The qualitative evaluation of lipase activity by the slide cultures method involved the inoculation with a 2 mm square fungal colony of cleaned microscope glass slides covered with three layers of acrylic resin Paraloid B-72 (Paraloid B72), transparent dispersion of casein (TDC), sunflower oil (SO) or beeswax (BW) followed by incubation at 28 °C for 21 days. The following strains were inoculated: Aspergillus sp. BA 2-2, Aspergillus sp. BSL 2-2, Emericellopsis sp. MM2, and Penicillium sp. BSL 3-2. Lipase activity was evidenced indirectly by measuring the diameters of the colonies. The ability of the fungal isolates to use the organic deposits as nutrients was interpreted as the capacity to synthesize extracellular enzymes. Photos were taken with the NIKON D200 camera.
Quantitative evaluation of lipase activities was performed as follows: Aspergillus sp. BSL 2-2, Emericellopsis sp. MM2, and Penicillium sp. BSL 3-2 were inoculated in 250 mL Erlenmeyer flasks with 100 mL of modified HM containing (g/L): yeast extract (10), NaCl (10), 7.0 MgCl2 × 6H2O (7), MgSO4 × 7H2O (9.6), CaCl2 × 2H2O (0.36), KCl (2.0), NaHCO3 (0.06), NaBr (0.026) and Tween-80 (10). Incubation was allowed for 7 days at 28 °C, under stirring conditions (150 rpm). At specific times (3, 5, and 7 days), pellets were harvested by centrifugation at 9500 rpm for 20 min at 4 °C, and the supernatant was used for enzymatic analysis. Lipase activity was quantified by using pNPB (p-nitrophenylbutyrate) as substrate [36]. The assay mixture consisted of 250 µL of the suitably diluted enzyme, 730 µL of 50 mM potassium phosphate buffer (pH 7.5), and 20 µL of 50 mM p-NPB, and incubated at 37 °C for 15 min. Optical density was measured at 410 nm against a blank. For the enzyme activity calculation, the molar extinction coefficient of 10,400 M−1 cm−1 for p-NP (p-nitrophenol) was used [37]. One unit of esterase activity was defined as the amount of enzyme needed to release 1 µmol of p-NP per minute under the assay conditions. All experiments were conducted in triplicate.
2.4. Evaluation of the Hydrolytic Decomposition of the Deposits on the Surface of the Painted Laboratory Models and Fragments of Frescoes
Painted laboratory models were created for the purpose of testing the selected strains by using bricks (22.5 × 11 cm), 2–3% of 0.5 mm sand, lime, tow, and pigments. Each laboratory model had four sections, onto which red, ochre, blue, and green pigments were applied. Pigments (Venetian red 0315, Yellow ochre 0324, Pure Ultramarine blue 0516, and Green earth 0264) were acquired from CTSR Company. According to the manufacturer, the chemical composition of the selected pigments is as follows: the red pigment contains Fe2O3, the yellow one contains α-FeO(OH), CaCO3, and CaSO4, the blue pigment contains, approximately, (Na,Ca)4(Al,SiO4)3(SO4,S,Cl), and the green contains ferrous and ferric silicates of potassium, manganese, and aluminum [38]. Following the preparation of painted laboratory models, three layers of Paraloid B72, TDC, BW, and SO were applied to the painted surfaces using a brush [35]. BW was applied by dipping the melted candle in the painted laboratory models. The soot (S) was provided by burning beeswax candle. In the end, each color area was divided into two rows and four columns by mechanically scratching the surface. Two types of inoculums were prepared: one represented by 50 μL of conidia suspension in distilled water (105 spores ml−1), which was spread onto all row-1 squares of each pigment, and another one, represented by 50 μL of conidia suspension in liquid HM nutrient (105 spores ml−1) which was spread onto all row-2 squares of each pigment. The following fungal strains were used: Penicillium sp. BSL 3-2, Aspergillus sp. BSL 2-2, Emericellopsis sp. MM2 and H1 (Supplementary Figure S1). All inoculated painted laboratory models were incubated in special devices at around 20 °C, with relative humidity of 90%, for 10 days. Then, the fungal cultures were removed from the treated surfaces using cotton swabs moistened in distilled water. Cleaned painted experimental models were further dried, visually inspected, and then samples were cut and analyzed using different methods.
Fragments of frescoes coming from the Fresco Library belonging to Department of Conservation and Restoration, Faculty of Art History, Bucharest National University of Arts were covered with the same deposits as those used for painted laboratory models and inoculated with 50 μL of Penicillium sp. BSL 3-2 conidia suspension in liquid HM nutrient.
Production of lipases was evaluated as an expression of fungal growth to use of deposits as nutrients on experimental painted models and frescoes by visual inspection (photo shoot with Nikon D200 camera).
The hydrolytic decomposition of the deposits was evaluated by visual inspection (which revealed well-developed and sporulated fungal colonies or very thin layer of mycelium of 2 mm diameter, nonsporulated) as well as under Nikon AZ 100 microscope without any preparation. It was also evaluated under Scanning Electron Microscope (SEM). The samples were gold sputtered and then observed under a variable pressure scanning electron microscope JEOL JSM6610LV operated at high vacuum (accelerated voltage 10–20 kV) [39].
The effect of the hydrolytic decomposition of the deposits was also evaluated by spectroscopic and imaging methods. The color variation of the samples was evaluated through colorimetry [40,41]. For this, an X-rite Ci64UV colorimeter was used, with aperture 8 mm and 4 mm, standard illuminant D65/10°, CIELAB color space, averaging three measurements in different points for each treated area. Principal component analysis was performed using the Unscrambler 11 software in order to better understand the data. The input dataset was created using the luminosity (L*), red-green (a*), and yellow-blue (b*) values recorded with the colorimeter. Prior to each measurement session, the colorimeter was calibrated using a black trap and a white ceramic plaque (L*a*b* coordinates 96.32/₋₋0.43/1.24).
After the inoculation process, the presence of the surface deposit layers was evaluated through Fourier-transform infrared (FTIR) spectroscopy [40,42] performed in duplicate on samples collected from each inoculated area. FTIR spectra were recorded with Perkin Elmer Spectrum Two FTIR spectrometer equipped with a PIKE GladiATR accessory in the spectral region between 4000 and 380 cm−1, 4 cm−1 resolutions, an average of 32 scans per each analyzed area. Existing literature was used for the identification of the bands related to Paraloid B72, oil and beeswax [43], casein [44,45], pigments [46], and fungus [47].
Additionally, hyperspectral imaging was applied in order to see if it would be suitable for assessing the ability of the method to decompose the surface layers. Hyperspectral data were recorded with a short-wavelength infrared (SWIR) HySpex SWIR 384 scanning hyperspectral camera (NEO). Data were recorded in push-broom mode in 288 spectral bands, between 950–2500 nm, and were afterward processed using the Spectral Angle Mapper supervised classification method in ENVI to better observe the differences between inoculated and control areas. For each sample, an inoculated and a control area were chosen: the first one was defined based on the location where the inoculum was applied, while the second was defined in a location outside this area. The classification images obtained with the SAM method help to visualize how much of the sample resembles what was designated as inoculated area.
3. Results and Discussion
3.1. Taxonomy of the Fungal Strains
According to the phylogenetic affiliation, the isolated fungal strains belong to three different genera: Penicillium, Aspergillus, and Emericellopsis (Figure 2). Table S1 (Supplementary) contains the GenBank accession numbers of the ITS sequences.
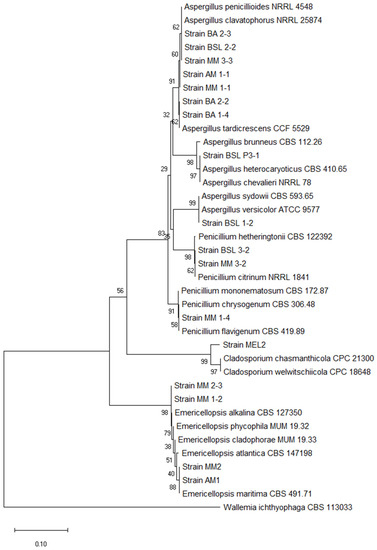
Figure 2.
Neighbor-joining phylogenetic tree based on the analysis of the ITS regions of the fungal isolates.
3.2. Salt Requirements of Fungal Isolates and Qualitative Evaluation of Hydrolytic Enzymes
Aspergillus strains were identified in both types of lakes as moderately halophilic (MM 1-1, MM 3-3, BSL 2-2, BSL 2-3, BA 2-3, AM 1-1, and BA 2-2) and halotolerant representatives (BSL 1-2, BSL 3-1, BA 1-4).
Penicillium sp. MM 1-4, MM 3-2, and BSL 3-2, as well as Emericellopsis sp. MM 2-3, MM 1-2, and MM2 recovered from the hypersaline lakes were halotolerant. Contrary, the Emericelopsis sp. AM1 isolated from a brackish lake was moderately halophilic.
All fungal isolates were able to produce hydrolases (Figure 3). Emericellopsis sp. MM2 and Penicillium sp. BSL 3-2 produced all tested hydrolytic enzymes. Most of the fungal isolates were able to produce proteases, pectinases, cellulases, lipases, and xylanases (Aspergillus sp. MM 3-3 and BA 2-2, Penicillium sp. BSL 3-2, Emericellopsis sp. MM 1-2, 2-3, MM 2 and AM1). Some of the strains produced amylases Emericellopsis sp. MM 1-2, Penicillium sp. MM 1-4, Aspergillus sp. MM 1-1, Penicillium sp. BSL 3-2, Aspergillus sp. BSL 1-2. Aspergillus sp. BA 2-2 and BA 2-3 isolated from a brackish environment had in common the ability to produce cellulase and pectinase. Penicillium sp. BSL 3-2, Aspergillus sp. BSL 2-2 and Aspergillus sp. MM 1-1 showed the ability to produce lipase.
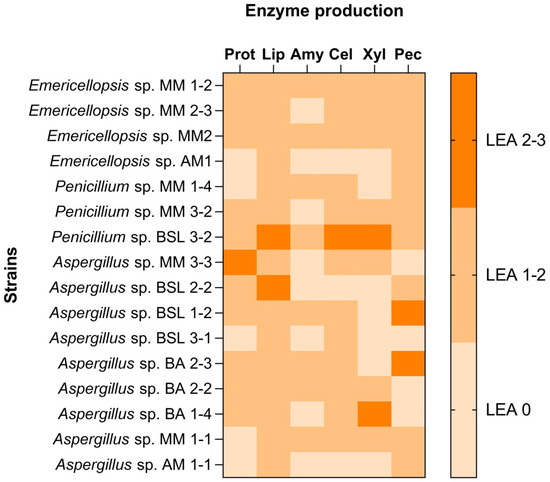
Figure 3.
Production of hydrolytic enzymes by different strains of Aspergillus, Penicillium and Emericellopsis (LEA: Level of Enzyme Activity).
The organic-rich content of the analyzed lakes resulting from the decomposition of fauna, plant debris, and microorganisms had contributed to the development of fungi producing hydrolytic enzymes. The increase in organic debris in the lakes is expected to stimulate their synthesis, mostly in the autumn. Our results suggested the adaptation of fungi to an aquatic environment. In the natural environments, the biodiversity of the cultivable fungal community was noticed, as well as the diversity of the synthesized hydrolases. Our results confirm previous reports, which found that in water samples, all isolates have proteolytic activity, but the amylolytic activity was rarely encountered [48]. They clearly demonstrate that the organic matter composition and other physicochemical conditions of the environment drive the structure of microbial communities and their ability to produce the hydrolases involved in decomposition.
3.3. Screening of Lipase-Producing Strains
Plate assay revealed that all tested isolates showed levels of lipase activities between 0.8 (Aspergillus sp. BA 1-4) and 2.8 (Penicillium sp. BSL 3-2). A remarkable lipase activity (LEA = 2.6) was also shown by Aspergillus sp. BSL 2-2 (Figure 4). Emericellopsis sp. MM 1-2, MM 2-3, MM2, AM1 were found to be also good lipase producers (LEA = 1.3–1.5).
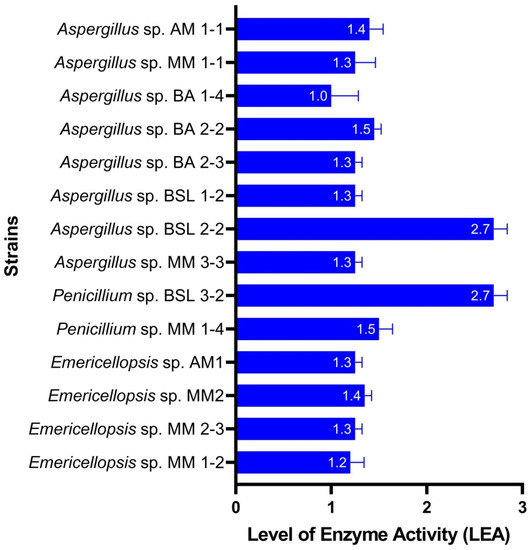
Figure 4.
Level of lipase activity of fungal isolates (plate assay).
We suppose that, in the investigated environments, lipids were in high amounts and that the predominance of lipase was associated with the natural selection of very-well-adapted fungi. It is considered that soil is a very good source for lipase-producers [49], but we extended research to brackish and hypersaline lakes, proving that prospecting of new fungal strains for these types of enzymes is a new direction of application, namely biocleaning of frescoes.
The potential applications of fungal lipase are extended to different fields: degradation of plastic, ester synthesis, preparation of optically active compounds, drinking [15], food, textile and detergent industries, medicine, cosmetics, agriculture, and the environment. Fungal cultures are also used in bioremediation (Geotrichum candidum, P. chrysogenum), biodegradation of oil (Fusarium oxysporum, Aspergillus niger, Candida tropicalis, or Pseudomonas aeruginosa, Penicillium notatum, Escherichia coli and A. niger) [50,51], plastic and polymers (C. rugosa, P. citrinum, F. solani pisi) [50,52].
The slide cultures were used as the second method for evaluating the ability of fungal strains to degrade specific substrates. The lipase activity was interpreted based on the size of colonies and the photographic recording of the mycelium. According to the first method of evaluation, all fungal strains grew on Paraloid B72, except Aspergillus sp. BSL 1-2. On TDC, only Penicillium sp. BSL 3-2 was able to grow. On BW, the following strains had the best growth: Emericellopsis sp. MM2, MM 2-3, and Penicillium sp. BSL 3-2. The other strains had good growth (Penicillium sp. MM 1-4 and Aspergillus sp. BA 2-2). On SO many strains had a very good growth: Aspergillus sp. BA 2-3, BSL 2-2, MM 3-3, Penicillium sp. BSL 3-2, MM 1-4, Emericellopsis MM 2-3, and MM2 (Figure 5 and Supplementary Figure S2). Photographic recordings revealed both non-sporulated and sporulated mycelium.
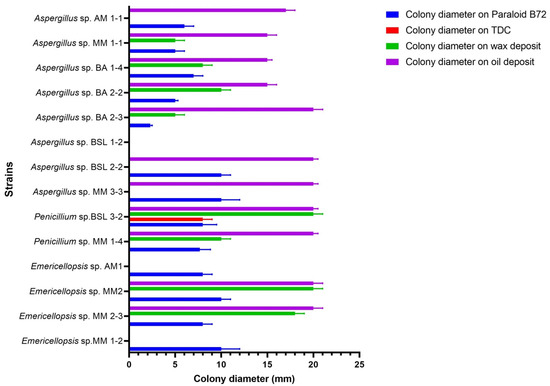
Figure 5.
Qualitative evaluation of lipase activity on Paraloid B72, TDC, BW and SO (slide cultures method).
The qualitative evaluation of lipase activity on slide cultures allowed the selection of the following species for testing the decomposition of deposits from experimental models: Aspergillus sp. BSL 2-2, Penicillium sp. BSL 3-2 for Emericellopsis sp.
The lipase activities of the tested fungal strains are shown in Figure 6. The activity increased after 3 days of growth, with an optimum activity after 5 days for Emericellopsis sp. MM2. After incubation for 5 days, Aspergillus sp. BSL 2-2 lipase retained approximately 30% of the activity (Figure 6). A similar percentage of activity (32%) was obtained for the Penicillium sp. BSL 3-2. After 7 days of growth, about 55% residual activity was detectable for Emericellopsis sp. MM2, while Aspergillus sp. BSL 2-2 produced low levels of lipase activity. No residual activity was observed for Penicillium sp. BSL 3-2. The biosynthesis time for lipase was found at a maximum of 5 days for all fungal species. In the case of Aspergillus niger, Lloyd et al. found maximum of lipase production at the late stage of conidiation [53].
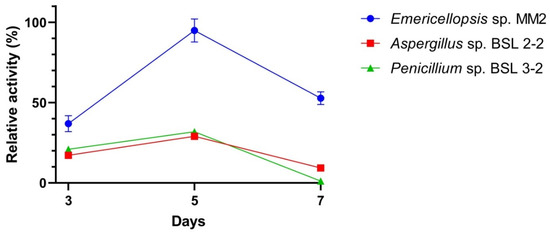
Figure 6.
Lipase activity in the crude extracts of the fungal strains after 3, 5, or 7 days of growth.
3.4. Testing of the Best Lipase-Producing Strains for the Decomposition of Deposits on the Painted Laboratory Models
Visual inspection, as the basic method for evaluating the state of conservation of mural painting, was applied in the case of the inoculated areas of the painted laboratory models (Figure S3).
Controls of painted laboratory models covered with Paraloid B72, TDC, and SO had the same characteristics: thin, slightly shiny, and pigments well fixed. Those covered with BW were thick and yellowish, and those covered with S looked blackish, but the soot layer came off in some areas.
Compared to the controls, the inoculated painted laboratory models had a totally different appearance. Those covered with Paraloid B72 and BW showed very good growth of Penicillium sp. BSL 3-2 and Aspergillus sp. BSL 2-2. SO and S allowed a good development of Penicillium sp. BSL 3-2, Aspergillus sp. BSL 2-2 and Emericellopsis sp. MM2. On TDC, Penicillium sp. BSL 3-2 had good growth. Aspergillus sp. BSL 2-2 and Emericellopsis sp. MM2 showed weak growth, i.e., they were observed under an optical microscope as very thin mycelium layers. After removing the colonies and cleaning with a moistened swab, white halos appeared most probably as a result of esterification, interesterification, and transesterification reactions.
Optical Microscopy (OM) and Scanning Electron Microscopy Analysis (SEM) showed morphological changes in inoculated samples when compared with control samples due to lipase activities. Due to the fact that the morphological changes produced by fungal lipase were similar, below are presented in detail will be only those caused by Penicillium sp. BSL 3-2 inoculated as spore suspension in the HM liquid nutrient. The four parts of each painted laboratory model to which red, yellow, blue, and green pigments were applied will be called squares or areas.
Painted laboratory models covered with the consolidant Paraloid B72 had partially or totally decomposed areas. OM examination showed areas where Paraloid B72 was totally decomposed by lipase, and so, the original pained layer (red, yellow, blue, and green) became visible, still attached to the mortar (yellow area—Figure 7a). SEM images revealed areas uncovered by Paraloid B72, such as true gaps (Figure 7b) where individual particles of mortar or the original paint layer were visible. Some areas were still completely covered by the consolidant, mostly towards the margins of colonies. The control surface was bright and unchanged (Supplementary Figure S4a,b).
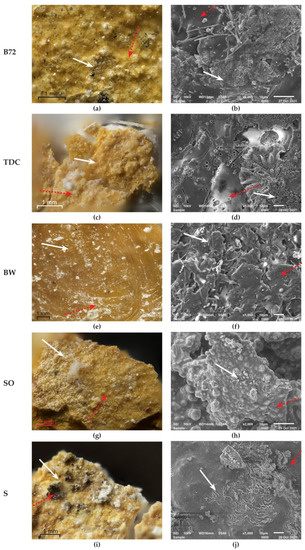
Figure 7.
Organic compounds decomposition by lipases synthesized by Penicillium sp. BSL 3-2 on the yellow areas of the painted laboratory models; white arrows show decomposed areas; red dotted arrows show unchanged areas. (a) OM on Paraloid B72; (b) SEM on Paraloid B72; (c) OM on TDC; (d) SEM on TDC; (e) OM on Beeswax; (f) SEM on Beeswax; (g) OM on sunflower oil; (h) SEM on sunflower oil; (i) OM on Soot; (j) SEM on Soot.
TDC is also a consolidant of mural painting. The layer of TDC had different thicknesses because it was manually applied. The whitish areas situated under the colonies were the result of TDC degradation. OM revealed small but completely clean areas on red and green squares; on yellow and blue squares, large parts of the TDC layer were completely removed (Figure 7c). Yellow pigment particles were noticed on the back of colonies. Powdery TDC layer (red square), folded and decomposed (yellow square—Figure 7d), completely removed (blue square), or irregularly shaped fragments (green square) were observed by SEM. The control surface was bright and unchanged (Supplementary Figures S4c,d).
The OM images showed that BW was entirely decomposed by lipase in areas with a thin layer (red and blue squares) or was fragmented into small particles (yellow square—Figure 7e). SEM images revealed wax layers of different thicknesses in decomposition (red, blue, and green areas) and their fragmentation (yellow area—Figure 7f). The control surface was opaque and unchanged (Supplementary Figure S4e,f).
SO was partially absorbed by the paint layer and what remained on the surface provided it a shiny aspect. Observations under OM showed the emulsification and partial decomposition of the oil on the red square and the partial removal on the yellow squares (Figure 7g). These forms of SO decomposition were confirmed by SEM analysis. Thus, on the red square, the decomposition of SO was almost complete; on the yellow square (Figure 7h), the decomposed areas alternated with the undecomposed ones, and on the blue and green squares, both the intact oil layer and the completely decomposed areas could be clearly observed. The control surface was bright and unchanged (Supplementary Figure S4g,h).
Soot, like the other materials, was unevenly deposited. No soot particles were detected by visual examination under the fungal colony; however, they were sporadically highlighted by OM (all squares, Figure 7i). Under SEM, the unaltered soot showed a network structure, but as a result of the action of lipase, it was either disorganized in the sense of total hydrolysis (red, blue and green squares) or areas of accumulation of decomposition products appeared (yellow square, Figure 7j). The control surface was black and unchanged (Supplementary Figure S4i,j).
Our screening methods revealed the successful identification of lipase-producing fungal strains, becoming of interest for the decomposition of consolidants used in the restoration of mural painting or organic deposits accumulated over time.
3.5. Spectroscopic and Imaging Methods also Revealed Changes on the Surface of Deposits
On Paraloid B72, the statistical analysis performed on the colorimetry data (Figure 8a) showed a clear difference between control and inoculated areas with Penicillium sp. BSL 3-2 and Emericellopsis sp. MM2 only for the blue squares, while a slightly smaller difference could be seen for the red squares and lesser for the yellow and green ones. The hyperspectral data (Figure 8b) showed that it could be inferred from the images that a large area from the red and yellow squares resembles what was defined as inoculated. However, this does not correlate with the areas in which the inoculum was actually applied, meaning that for the Paraloid B72 consolidant, this would not be a very accurate identification method, and further post-processing methods should be tested in the future for better discrimination.
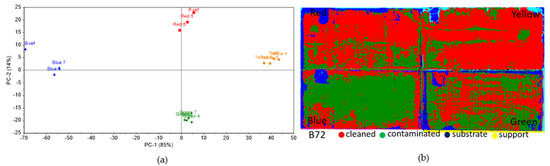
Figure 8.
Colorimetric and hyperspectral evaluation of Paraloid B72 decomposition by fungal lipase. (a) Score plot of the first 2 PCs, based on colorimetry data; (b) SAM images based on hyperspectral data.
The changes observed with the colorimetry and microscopy revealed alterations in the consolidant layer. However, the consolidant had infiltrated the lower layers; this could be inferred from the FTIR analysis (Supplementary Figure S5), which found all bands associated with it in all areas where inoculum had been applied: the C-H stretching vibrations at 2984 and 2954 cm−1, the C=O stretching vibration (ester) at 1722 cm−1, the C-H bending vibration at 1447 and 1386 cm−1, and several bands coming from the C–O stretching, at 1233, 1140, 1024, 968 cm−1. No fungal residues were detected on the treated areas.
On TDC, the colorimetric analysis (Figure 9a) indicated that the blue, red, and yellow areas had significant differences between the areas inoculated with Penicillium sp. BSL 3-2 and the reference data, but a much smaller one was noticed for the green pigment. This means that Penicillium sp. BSL 3-2 produced lesser lipase on the green pigment than for the others. The SAM images (Figure 9b) indicate that most of the surface resembles the control area rather than the inoculated one, except for the red area, where the decomposition of the surface deposits seemed to be successful. Similar to the B72 sample, this does not match the areas where the inoculum had been applied.
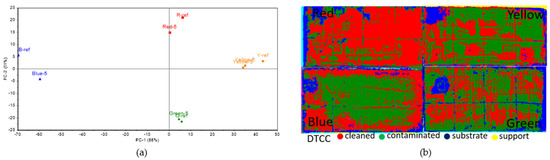
Figure 9.
Colorimetric and hyperspectral evaluation of TDC decomposition by fungal lipase. (a) Score plot of the first 2 PCs, based on colorimetry data; (b) SAM images based on hyperspectral data.
Casein had a very well-defined IF signal, but in real practice, TDC is used at very low concentrations, so the identification of peaks was not always possible. Weak bands associated with the TCD were found in the FTIR spectra collected from the blue pigment (Supplementary Figure S6c) at ~3300 (broad band coming from the O-H and N-H stretching of the amine groups), 1644 (protein amide I), 1576 (amide II), and 1232 cm−1 (N-H deformation vibration). After the treatment, these bands are no longer visible, and the bands of the pigments remain unaltered. TDC was barely visible for the yellow pigment (Supplementary Figure S6b) spectra, as the main protein amide bands are overlapped with the gypsum sharp bands at 1682 cm−1 and 1618 cm−1 in that region. Similarly, for the red pigment (Supplementary Figure S6a), the sharp gypsum bands overlapped the TCD bands, which were not visible. No TDC bands were noticed for the green pigment (Supplementary Figure S6d); furthermore, because of the sampling, some of the spectra contain bands related to the support (the presence of hydrated lime −3643 cm−1).
For the TDC samples, well-resolved bands coming from the fungus were identified on the surface of the pictorial layer at 3271 cm−1 (N–H and the O–H stretching vibrations), 1645 cm−1 (protein amide I), 1153 cm−1 (C–O stretching vibrations). The best results appear in the area inoculated with Penicillium sp. BSL 3-2 on the yellow squares, where there are no visible bands of TDC or fungi.
On both red and yellow squares, there is an altering of the pigment bands, as the main silicate band (Si–O stretching vibration) between 900–1200 cm−1 is deformed, and the bands in the region 490–650 cm−1 are broadened (marked with dashed circles), and the gypsum bands have disappeared. At the opposite end, the bands of the blue pigment (Supplementary Figure S6c) were not altered, and the green pigment-related bands seemed partially unaltered, as only the bands of the gypsum were not present in the treated spectrum.
On BW, the colorimetry data (Figure 10a) varied greatly, as some of the inoculated areas were far from the reference. For the blue square, the area inoculated with Aspergillus sp. BSL 2-2 was close to the reference, while a clear difference was seen for the area inoculated with Penicillium sp. BSL 3-2 and a smaller one for the area inoculated with Emericellopsis sp. MM2. For the red square, the area inoculated with Aspergillus sp. BSL 2-2 was close to the reference, but the area inoculated with Penicillium sp. BSL 3-2 was not. The same situation was seen for the yellow square. This could suggest that Penicillium sp. BSL 3-2 was much more efficient for lipase production than others. For the green square, no significant difference has been determined between the inoculated areas and the reference. The processed hyperspectral data (Figure 10b) showed that several small areas suggested a partially successful decomposition of the surface layer.
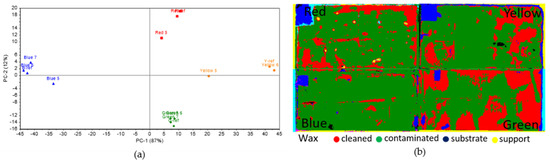
Figure 10.
Colorimetric and hyperspectral evaluation of BW decomposition by fungal lipase. (a) Score plot of the first 2 PCs, based on colorimetry data; (b) SAM images based on hyperspectral data.
Regarding the FTIR results, due to the thickness of the layer, the wax bands identified at 2917 (CH2 asymmetric stretching band), 2850 (CH2 symmetric stretching band), 1738 (C=O stretching band), 1463 (CH2 scissor deformation), 1173 (C–O stretching and C-H bending mode), 956 (C–O stretching mode), 729, 719 cm−1 (CH2 rocking mode) were still visible after the treatment on all of the samples. For the blue pigment (Supplementary Figure S7c), the areas inoculated with Penicillium sp. BSL 3-2 and Aspergillus sp. BSL 2-2 had fungal residues detected by the band at 1648 cm−1 assigned to the protein amide I. The red (Supplementary Figure S7a) and yellow (Supplementary Figure S7b) areas also showed fungal residues detected by the band at 3278 cm−1 assigned to the N–H and the O–H stretching vibrations, the band at 1648 cm−1 assigned to the protein amide I and, the broad bands between 1200–900 cm−1 and 650–450 cm−1. No fungal residues have been detected on the surface of the green pigment (Supplementary Figure S7d) after the treatment. Bands coming from the pigments and the calcite layer (at 2516, 1795, 1394, 871 cm−1) were also present in some of the spectra.
On SO, from the statistical analysis of the colorimetry data (Figure 11a), clear differences could be seen for the blue, red, and yellow inoculated squares, whereas the data for the green square were very close to the reference. Results were similar to those obtained on Paraloid B72, where Penicillium sp. BSL 3-2 produced lesser lipase on the green square. The processed hyperspectral data (Figure 11b) indicated variable results, with higher efficiency in removing the surface deposits for the green, yellow and blue squares.
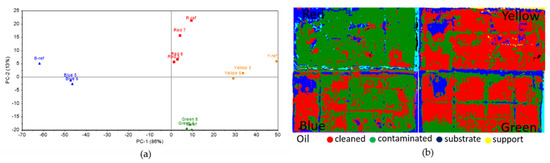
Figure 11.
Colorimetric and hyperspectral evaluation of SO decomposition by fungal lipase (a) Score plot of the first 2 PCs, based on colorimetry data; (b) SAM images based on hyperspectral data.
The infrared spectroscopy showed that all the bands related to the SO (the stretching vibrations of the methylene bands (-CH2) at 2924 cm−1 and 2855 cm−1, and the sharp carbonyl band at 1742 cm−1 are missing from all blue (Supplementary Figure S8c), and green (Supplementary Figure S8d) analyzed areas after the treatment. Similarly, no oil traces were detected for the red areas inoculated with Aspergillus sp. BSL 2-2 (Supplementary Figure S8a). For the yellow (Supplementary Figure S8b) and red squares, in the areas inoculated with Penicillium sp. BSL 3-2, the stretching vibrations of the methylene bands (-CH2) at 2924 cm−1 and 2855 cm−1 of the sunflower oil are still present, whilst the carbonyl band at 1742 cm−1 is missing, meaning that oil was partially removed. A plausible interpretation could be that positive results have been obtained, but not for all inoculated fungal strains.
The greatest difference from the point of view of the colorimetric analysis was observed for the S samples (Figure 12a). All the data from the inoculated areas were distanced from the reference, which means significant changes had occurred; thus, the inoculation was efficient. In the case of S samples, SAM images (Figure 12b) show relevant differences between the inoculated and the control samples, especially for the blue, yellow, and red areas, thus identifying very precisely the areas where the inoculum had been applied and the decomposition of the soot layer was obtained.
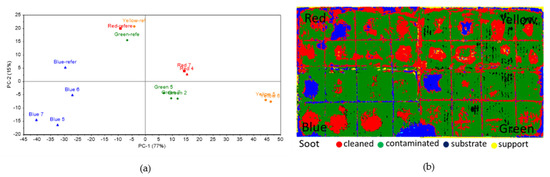
Figure 12.
Colorimetric and hyperspectral evaluation of soot decomposition by Penicillium sp. BSL 3-2 esterases. (a) Score plot of the first 2 PCs, based on colorimetry data; (b) SAM images based on hyperspectral data. As soot cannot be identified via FTIR, the efficiency of the inoculation procedures could not be assessed with this method. The FTIR spectra (Supplementary Figure S9) only showed that some pigments appeared unaltered. No specific fungal traces have been detected for any of the examined areas.
3.6. Testing of the Best Lipase-Producing Strains for the Decomposition of Deposits on the Fresco Fragments
By visual inspection, all the deposits on the surface of the frescoes fragments placed under the Penicillium sp. BSL 3-2 colonies showed morphological changes compared to the uninoculated control samples (Supplementary Figure S10). After removing the mycelium and cleaning it with a damp swab, white areas under it became visible.
Optical Microscopy and Scanning Electron Microscopy Analysis showed morphological changes in inoculated fragments of frescoes due to fungal growth and lipase activities.
OM showed that Paraloid B72, TDC, SO, and S deposits on frescoes fragments were almost totally decomposed by lipase, the original pictorial layer becoming visible (Figure 13a,c,g,i). In Figure 13e was observed that in the thinner layer areas of BW, hydrolysis took place. These areas correspond to the decomposition areas highlighted by SEM (Figure 13f).
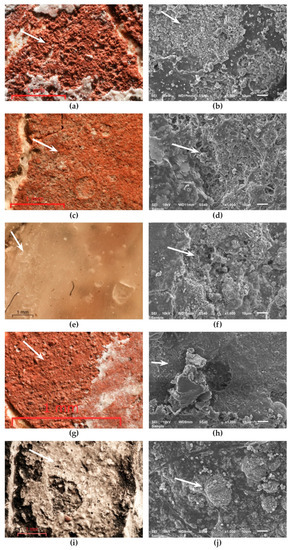
Figure 13.
Degradation of deposits on the surface of fresco fragments. (a) OM on Paraloid B72; (b) SEM on Paraloid B72; (c) OM on TDC; (d) SEM on TDC; (e) OM on BW; (f) SEM on BW; (g) OM on SO; (h) SEM on SO; (i) OM on S; (j) SEM on S.
SEM images revealed large areas of decomposed Paraloid B72 and SO (Figure 13b,h), large areas of decomposed TDC and S, with still present consolidant strips (Figure 13d,j). No changes were noticed in control samples (Supplementary Figure S10).
The colorimetric analysis of the fresco samples was performed by comparison between two areas, with and without surface deposits. The ΔE values were calculated for all samples and are plotted in Figure 14a. Similar to the data obtained for the laboratory samples, the highest variation was obtained for the S samples. In this case, a high ΔE value indicates that the surface S layer was decomposed by the lipases synthesized by the fungal colonies. At the opposite end, lower ΔE values were calculated for the BW samples, but this is also in accordance with the previously-obtained data for the laboratory samples and can be interpreted by the fact that the BW layer was thick, so no major changes had been induced.
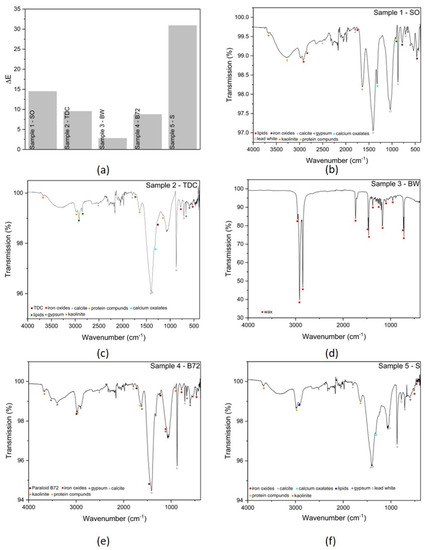
Figure 14.
Colorimetry and FTIR spectroscopy performed on the fresco samples. (a) ΔE values calculated from the colorimetric data; (b) FTIR spectrum of the sample with SO; (c) FTIR spectrum of the sample with TDC; (d) FTIR spectrum of the sample with BW; (e) FTIR spectrum of the samples with Paraloid B72; (f) FTIR spectrum of the sample with S.
As the analyzed samples came from real frescoes, their structure was slightly more complex as compared to the laboratory samples. Thus, several bands could be seen in the infrared spectra (Figure 14b–f), belonging to the nature of the frescoes (structure, pigments), the surface deposits, or the decomposition experiment. For example, in Figure 14b, corresponding to the fragment on which SO had been applied, the bands associated with lipids were clearly visible at 2920, 2851, and 1747 cm−1. Bands from the calcite (~712, 870, 1396, 1410, 1793, 2510 cm−1) and kaolinite (~ 3668, 909 cm−1) were seen in samples covered with SO (Figure 14b), S (Figure 14f), Paraloid B72 (Figure 14e) and TDC (Figure 14c), along with some calcium degradation products—calcium oxalates (~ 1320 cm−1). Pigment bands, coming from iron oxides, were found at 781 and 466 cm−1 for the sample covered with SO, at 466, 781, 1100 cm−1 for the sample with Paraloid B72, at 531 and 781 cm−1 for the TDC fresco sample, and at 462 and 520 cm−1 for the soot sample (Figure 14f). The identification of pigment bands suggested that there were areas where the deposits were completely decomposed by the lipase produced by Penicillium sp. BSL 3-2. The latter also showed bands coming from a lead white pigment at ~ 787, 1044, 1397 cm−1. It has been observed the presence of some protein compounds of fungal origin (3267, 1462 cm−1) could be correlated with the fungal activity present in all samples except the wax-covered one. For this sample, the presence of BW was highlighted through several wax-correlated bands, as can be seen in Figure 14d.
The spectroscopic and imaging analysis pointed out that the fungal inoculation decomposed more S, TDC, and SO and less BW and Paraloid B72. The interpretation of the results obtained by spectroscopic methods must be correlated both with each other and with the microbiology and microscopy data because they complemented each other.
Organic materials used in restoration, as well as organic deposits, are susceptible to degradation by heterotrophic microorganisms. Linen textiles consolidated by different polymers, including Paraloid B72, were sensitive to fungal deterioration due to the ability of Aspergillus and Penicillium species to decompose acrylic resin [54]. Milanesi et al. [55] proved that Bacillus sp. and Brevibacillus ruber decomposed the painted surface in the Chapel of the Holy Nail in the medieval ex-hospital of Siena (Italy) restored with Paraloid B72. Furthermore, Elhagrassy [56] showed that by using Pseudomonas stutzeri, any organic matter, including acrylic polymers and animal glue, was removed from mural paintings.
Microorganisms and their metabolites can be used in conservation and restoration and generally for the safeguarding of artworks [57]. Cleaning the murals involves the use of toxic chemicals, metabolic products of microbial origin, or cultures of decomposing microorganisms. Thus, aged consolidants and organic deposits are decomposed and chemically (cleaning) or biologically (biocleaning) removed. Bacterial cultures such as Pseudomonas stutzeri [58], Desulfovibrio desulfuricans and Desulfovibrio vulgaris have been used so far [33]. They were applied directly to the work of art together with the nutrient medium or were included in gels [59,60]. D. vulgaris subsp. vulgaris ATCC 29579 was also used coupled with a non-ionic detergent pre-treatment [61].
Over time, some materials used in restoration (consolidants) and organic residues age represent a risk from an aesthetic and physical point of view, and so removal is recommended. A new type of protocol is proposed in this paper to remove Paraloid B72, TDC, BW, SO, and S by fungal lipase produced during the development of the colonies. This approach has an ecological role and potential economic advantages, and it is healthy for restorers. There is no danger of contaminating the mural painting since the inoculation is performed only in the areas covered by organic deposits, and those areas are isolated. Although the fungal removal proved in this paper was not totally performed for the entire surface, it was safe for mural painting. It has a high level of selectivity towards the aged materials and organic deposits, which encourages further research in the context of the European Green Deal and white technologies with low environmental impact.
Future investigation is needed to develop and monitor the safety and effectiveness of this approach. Rapid analyses (such as viability, ATP content, and dehydrogenase activity) must be applied to monitor the biological activity and avoid undesirable effects of biocleaning with fungal cultures. Furthermore, a few repeated inoculations should be tried for the total removal of the deposits without aesthetically and structurally affecting the mural.
The identified connections prove the complex relationship between fungi and the decomposition process in different environments, yet a deep analysis of the cleaning process taking place on the surface of murals requires more investigation.
4. Conclusions
In the present study, different species of fungi isolated from brackish and hypersaline lakes were screened for their ability to produce hydrolytic enzymes involved in the decomposition of organic matter. At the same time, they were great resources for biocleaning of mural paintings and other biotechnological applications such as: the production of biofuel, detergents, biosurfactants, and cosmetics, in the pharmaceutical, paper, and oil–chemical industries, for the biodegradation of organic pollutants, or in the processing of food. According to the phylogenetic affiliation, the fungal isolates belong ed to the Penicillium, Aspergillus, and Emericellopsis genera. It was proved that the fungal strains isolated from brackish and hypersaline lakes in Romania produced proteases, lipases, amylases, cellulases, xylanases, and pectinases, which involved them in the decomposition of the organic matter in the original environment. Applying selective testing methods, Aspergillus sp. BSL 2-2, Penicillium sp. BSL 3-2 and Emericellopsis sp. MM2 were selected for the decomposition of organic deposits (Paraloid B72, TDC, BW, SO, and S) covering painted experimental models. The most efficient strain, Penicillium sp. BSL 3-2 was inoculated successfully on frescoes fragments covered with the same deposits, becoming very promising for applications not only in biocleaning but also in bioremediation treatments.
We demonstrated that by cultivating fungi directly on the target polymeric compounds, it induced the production and secretion of hydrolytic enzymes, including lipase, and so the decomposition process took place.
Supplementary Materials
Supplementary Materials: The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8090462/s1, Table S1: List of fungal strains used in the present study and accession numbers of their partial ITS1-5.8S-ITS2 sequence; Figure S1. Pained laboratory models divided in squares for inoculation of fungi. Row 1.1 and Row 2.5—inoculation with Penicillium sp. BSL 3-2; Row 1.2 and Row 2.6—inoculation with Aspergillus sp. BSL 2-2; Row 1.3 and Row 2.7—inoculation with Emericellopsis sp. MM2; Row 1.4 and Row 2.8—inoculation with strain H1; Figure S2. Qualitative evaluation of lipase activity by slide cultures. (a) Decomposition of beeswax by Aspergillus sp. BSL 1-2 and Emericellopsis sp. MM2; (b) Decomposition of sunflower oil by Emericellopsis sp. MM2 and Penicillium sp. BSL 3-2; Figure S3. Images of morphological aspects of organic deposits on painted laboratory models (control). (a) OM on B72; (b) SEM on B72; (c) OM on TDC; (d) SEM on TDC; (e) OM on Beeswax; (f) SEM on Beeswax; (g) OM on sunflower oil; (h) SEM on sunflower oil; (i) OM on Soot; (j) SEM on Soot; Figure S4. FTIR spectra acquired for the B72 sample. (a) red square; (b) yellow square; (c) blue square; (d) green square; Figure S5. FTIR spectra acquired for the TDC sample. (a) red square; (b) yellow square; (c) blue square; (d) green square; Figure S6. FTIR spectra acquired for BW sample. (a) red square; (b) yellow square; (c) blue square; (d) green square; Figure S7. FTIR spectra acquired for the SO sample. (a) red square; (b) yellow square; (c) blue square; (d) green square; Figure S8. FTIR spectra acquired for the S sample. (a) red square; (b) yellow square; (c) blue square; (d) green square; Figure S9. Colonization of different deposits applied on the surface of frescoes fragments by Penicillum sp. BSL 2-3. (a) Fresco covered with B72—control; (b) Fresco covered with B72—colonized; (c) Fresco covered with TDC—control; (d) Fresco with TDC—colonized; (e) Fresco covered with BW—control; (f) Fresco covered with BW—colonized; (g) Fresco covered with SO—control; (h) Fresco covered with SO—colonized; (i) Fresco covered with S—control; (j) Fresco covered with S—colonized; Figure S10. Morphological aspects of frescoes uninoculated—control. (a) OM on B72; (b) SEM on B72; (c) OM on TDC; (d) SEM on TDC; (e) OM on BW; (f) SEM on BW; (g) OM on SO; (h) SEM on SO; (i) OM on S; (j) SEM on S.
Author Contributions
Conceptualization, I.G., R.C., R.R. (Robert Ruginescu) and L.G.; methodology, I.G., R.R. (Robert Ruginescu), S.N., G.M., R.R. (Roxana Rădvan) and L.G; formal analysis, M.E. and M.D.; investigation, R.C, L.-C.R. and V.A.; writing—original draft preparation, I.G., R.C. and L.G; writing—review and editing, I.O., R.C. and R.R. (Roxana Rădvan); visualization, I.G and R.C.; funding acquisition, I.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Executive Agency for Higher Education, Research, Development and Innovation Funding (UEFISCDI), grant number 570PED/2020 and by The Romanian Academy, grant number RO1567-IBB05/2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funder had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Fabian, J.; Zlatanovic, S.; Mutz, M.; Premke, K. Fungal–Bacterial Dynamics and Their Contribution to Terrigenous Carbon Turnover in Relation to Organic Matter Quality. ISME J. 2017, 11, 415–425. [Google Scholar] [CrossRef]
- Zanne, A.E.; Abarenkov, K.; Afkhami, M.E.; Aguilar-Trigueros, C.A.; Bates, S.; Bhatnagar, J.M.; Busby, P.E.; Christian, N.; Cornwell, W.K.; Crowther, T.W.; et al. Fungal Functional Ecology: Bringing a Trait-based Approach to Plant-associated Fungi. Biol. Rev. 2020, 95, 409–433. [Google Scholar] [CrossRef]
- Geethanjali, P.A.; Jayashankar, M. A Review on Litter Decomposition by Soil Fungal Community. IOSR 2016, 11, 1–3. [Google Scholar] [CrossRef]
- Likar, M.; Grašič, M.; Stres, B.; Regvar, M.; Gaberščik, A. Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats. JoF 2022, 8, 284. [Google Scholar] [CrossRef]
- Berbee, M.L.; James, T.Y.; Strullu-Derrien, C. Early Diverging Fungi: Diversity and Impact at the Dawn of Terrestrial Life. Annu. Rev. Microbiol. 2017, 71, 41–60. [Google Scholar] [CrossRef]
- Gadd, G.M. Fungal Biomineralization. Curr. Biol. 2021, 31, R1557–R1563. [Google Scholar] [CrossRef]
- Saranraj, P.; Stella, D. Fungal Amylase—A Review. Int. J. Microbiol. Res. 2013, 4, 203–211. [Google Scholar] [CrossRef]
- Ayansina, A.D.V.; Adelaja, A.O.; Mohammed, S.S.D. Characterization of Amylase from Some Aspergillus and Bacillus Species Associated with Cassava Waste Peels. AiM 2017, 7, 280–292. [Google Scholar] [CrossRef]
- Li, J.-X.; Zhang, F.; Jiang, D.-D.; Li, J.; Wang, F.-L.; Zhang, Z.; Wang, W.; Zhao, X.-Q. Diversity of Cellulase-Producing Filamentous Fungi from Tibet and Transcriptomic Analysis of a Superior Cellulase Producer Trichoderma harzianum LZ117. Front. Microbiol. 2020, 11, 1617. [Google Scholar] [CrossRef]
- Li, X.; Dilokpimol, A.; Kabel, M.A.; de Vries, R.P. Fungal Xylanolytic Enzymes: Diversity and Applications. Bioresour. Technol. 2022, 344, 126290. [Google Scholar] [CrossRef]
- Souza, P.M.; Bittencourt, M.L.A.; Caprara, C.C.; Freitas, M.; Almeida, R.P.C.; Silveira, D.; Fonseca, Y.M.; Ferreira Filho, E.X.; Pessoa Junior, A.; Magalhães, P.O. A Biotechnology Perspective of Fungal Proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef]
- Haile, S.; Ayele, A. Pectinase from Microorganisms and Its Industrial Applications. Sci. World J. 2022, 2022, 1881305. [Google Scholar] [CrossRef]
- Kovacic, F.; Babic, N.; Krauss, U.; Jaeger, K.E. Classification of Lipolytic Enzymes from Bacteria. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids; Rojo, F., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 255–289. ISBN 978-3-319-50417-9. [Google Scholar]
- Bracco, P.; van Midden, N.; Arango, E.; Torrelo, G.; Ferrario, V.; Gardossi, L.; Hanefeld, U. Bacillus subtilis Lipase A—Lipase or Esterase? Catalysts 2020, 10, 308. [Google Scholar] [CrossRef]
- Sayali, K.; Sadichha, P.; Surekha, S. Microbial Esterases: An Overview. Int. J. Curr. Microbiol. App. Sci. 2013, 2, 135–146. [Google Scholar]
- Faiz, O.; Colak, A.; Saglam, N.; Canakçi, S.; Beldüz, A.O. Determination and Characterization of Thermostable Esterolytic Activity from a Novel Thermophilic Bacterium Anoxybacillus gonensis A4. J. Biochem. Mol. Biol. 2007, 40, 588–594. [Google Scholar] [CrossRef]
- Dilokpimol, A.; Mäkelä, M.R.; Varriale, S.; Zhou, M.; Cerullo, G.; Gidijala, L.; Hinkka, H.; Brás, J.L.A.; Jütten, P.; Piechot, A.; et al. Fungal Feruloyl Esterases: Functional Validation of Genome Mining Based Enzyme Discovery Including Uncharacterized Subfamilies. New Biotechnol. 2018, 41, 9–14. [Google Scholar] [CrossRef]
- Lin, M.I.; Hiyama, A.; Kondo, K.; Nagata, T.; Katahira, M. Classification of Fungal Glucuronoyl Esterases (FGEs) and Characterization of Two New FGEs from Ceriporiopsis subvermispora and Pleurotus eryngii. Appl. Microbiol. Biotechnol. 2018, 102, 9635–9645. [Google Scholar] [CrossRef]
- Yamamoto-Tamura, K.; Hiradate, S.; Watanabe, T.; Koitabashi, M.; Sameshima-Yamashita, Y.; Yarimizu, T.; Kitamoto, H. Contribution of Soil Esterase to Biodegradation of Aliphatic Polyester Agricultural Mulch Film in Cultivated Soils. AMB Expr. 2015, 5, 10. [Google Scholar] [CrossRef]
- Ruginescu, R.; Gomoiu, I.; Popescu, O.; Cojoc, R.; Neagu, S.; Lucaci, I.; Batrinescu-Moteau, C.; Enache, M. Bioprospecting for Novel Halophilic and Halotolerant Sources of Hydrolytic Enzymes in Brackish, Saline and Hypersaline Lakes of Romania. Microorganisms 2020, 8, 1903. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A. Current Knowledge on the Fungal Degradation Abilities Profiled through Biodeteriorative Plate Essays. Appl. Sci. 2021, 11, 4196. [Google Scholar] [CrossRef]
- Minotti, D.; Vergari, L.; Proto, M.R.; Barbanti, L.; Garzoli, S.; Bugli, F.; Sanguinetti, M.; Sabatini, L.; Peduzzi, A.; Rosato, R.; et al. Il Silenzio: The First Renaissance Oil Painting on Canvas from the Uffizi Museum Restored with a Safe, Green Antimicrobial Emulsion Based on Citrus aurantium var. amara Hydrolate and Cinnamomum zeylanicum Essential Oil. JoF 2022, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Paolo Cremonesi L’ambiente Acquoso per Il Trattamento Di Opere Policrome. In Metodologie, Tecniche e Formazione Nel Mondo Del Restauro; Il Prato Publishing House: Villatora, Italy, 2012; ISBN 978-88-6336-156-8.
- Borgioli, L.; de Comelli, A.; Pressi, G. Indagini Microbiologiche per La Verifica Dell’efficacia Di Alcuni Biocidi Esenti Da Metalli Pesanti. Progett. Restauro 2006, 11, 24–29. [Google Scholar]
- Ortega-Morales, B.O.; Gaylarde, C.C. Bioconservation of Historic Stone Buildings—An Updated Review. Appl. Sci. 2021, 11, 5695. [Google Scholar] [CrossRef]
- da Silva, M.T.C. Caldeira Novel Biocides for Cultural Heritage; Universidade de Évora: Évora, Portugal, 2017. [Google Scholar]
- Masi, M.; Petraretti, M.; De Natale, A.; Pollio, A.; Evidente, A. Fungal Metabolites with Antagonistic Activity against Fungi of Lithic Substrata. Biomolecules 2021, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Gomoiu, I.; Radvan, R.; Ghervase, L.; Mohanu, I.; Enache, M.; Neagu, S.; Ruginescu, R.; Cojoc, R. Cleaning of Mural Paintings and Mortars: Review. Rev. Rom. Mater. 2020, 50, 485–492. [Google Scholar]
- Gomoiu, I.; Enache, M.; Neagu, S.; Ruginescu, R.; Dumbrăvician, M.; Radvan, R.; Ghervase, L.; Mohanu, I.; Cojoc, R. Green Biotechnologies Used in the Restoration of Mural Painting and Lithic Support: Review. Rev. Rom. Mater. 2021, 51, 495–504. [Google Scholar]
- Marvasi, M.; Mastromei, G.; Perito, B. Bacterial Calcium Carbonate Mineralization in Situ Strategies for Conservation of Stone Artworks: From Cell Components to Microbial Community. Front. Microbiol. 2020, 11, 1386. [Google Scholar] [CrossRef]
- Castro-Alonso, M.J.; Montañez-Hernandez, L.E.; Sanchez-Muñoz, M.A.; Macias Franco, M.R.; Narayanasamy, R.; Balagurusamy, N. Microbially Induced Calcium Carbonate Precipitation (MICP) and Its Potential in Bioconcrete: Microbiological and Molecular Concepts. Front. Mater. 2019, 6, 126. [Google Scholar] [CrossRef]
- Nazel, T. Bioconsolidation of Stone Monuments. An Overview. Restor. Build. Monum. 2016, 22, 37–45. [Google Scholar] [CrossRef]
- Balloi, A.; Palla, F. Biocleaning. In Biotechnology and Conservation of Cultural Heritage; Palla, F., Barresi, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 67–84. ISBN 978-3-319-46166-3. [Google Scholar]
- Bosch-Roig, P.; Ranalli, G. The Safety of Biocleaning Technologies for Cultural Heritage. Front. Microbiol. 2014, 5, 155. [Google Scholar] [CrossRef][Green Version]
- Ruginescu, R.; Enache, M.; Popescu, O.; Gomoiu, I.; Cojoc, R.; Batrinescu-Moteau, C.; Maria, G.; Dumbravician, M.; Neagu, S. Characterization of Some Salt-Tolerant Bacterial Hydrolases with Potential Utility in Cultural Heritage Bio-Cleaning. Microorganisms 2022, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Shirai, K.; Jackson, R.L. Lipoprotein Lipase-Catalyzed Hydrolysis of p-Nitrophenyl Butyrate. Interfacial Activation by Phospholipid Vesicles. J. Biol. Chem. 1982, 257, 1253–1258. [Google Scholar] [CrossRef]
- de Lourdes Moreno, M.; García, M.T.; Ventosa, A.; Mellado, E. Characterization of Salicola Sp. IC10, a Lipase- and Protease-Producing Extreme Halophile. FEMS Microbiol. Ecol. 2009, 68, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, N. CTS Pure Artis’s Pigments. 2010. Available online: file:///D:/Downloads/CTS%20PURE%20ARTISTS%E2%80%99%20PIGMENTS.pdf (accessed on 31 July 2022).
- Cojoc, L.R.; Enache, M.I.; Neagu, S.E.; Lungulescu, M.; Setnescu, R.; Ruginescu, R.; Gomoiu, I. Carotenoids Produced by Halophilic Bacterial Strains on Mural Paintings and Laboratory Conditions. FEMS Microbiol. Lett. 2019, 366, fnz243. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.E.; Kolisis, F.N. An Investigation into the Removal of Starch Paste Adhesives from Historical Textiles by Using the Enzyme α-Amylase. J. Cult. Herit. 2011, 12, 169–179. [Google Scholar] [CrossRef]
- Gioventù, E.; Lorenzi, P. Bio-Removal of Black Crust from Marble Surface: Comparison with Traditional Methodologies and Application on a Sculpture from the Florence’s English Cemetery. Procedia Chem. 2013, 8, 123–129. [Google Scholar] [CrossRef]
- Bosch-Roig, P.; Pozo-Antonio, J.S.; Sanmartín, P. Identification of the Best-Performing Novel Microbial Strains from Naturally-Aged Graffiti for Biocleaning Research. Int. Biodeterior. Biodegrad. 2021, 159, 105206. [Google Scholar] [CrossRef]
- Derrick, M.R.; Stulik, D.; Landry, J.M. Infrared Spectroscopy in Conservation Science; Scientific Tools for Conservation; Getty Conservation Institute: Los Angeles, CA, USA, 1999; ISBN 978-0-89236-469-5. [Google Scholar]
- Minaei, F.; Ravandi, S.A.H.; Hejazi, S.M.; Alihosseini, F. The Fabrication and Characterization of Casein/PEO Nanofibrous Yarn via Electrospinning. e-Polymers 2019, 19, 154–167. [Google Scholar] [CrossRef]
- Cortea, I.M.; Ratoiu, L.; Ghervase, L.; Țentea, O.; Dinu, M. Investigation of Ancient Wall Painting Fragments Discovered in the Roman Baths from Alburnus maior by Complementary Non-Destructive Techniques. Appl. Sci. 2021, 11, 10049. [Google Scholar] [CrossRef]
- Genestar, C.; Pons, C. Earth Pigments in Painting: Characterisation and Differentiation by Means of FTIR Spectroscopy and SEM-EDS Microanalysis. Anal. Bioanal. Chem. 2005, 382, 269–274. [Google Scholar] [CrossRef]
- Naumann, A. Fourier Transform Infrared (FTIR) Microscopy and Imaging of Fungi. In Advanced Microscopy in Mycology; Dahms, T.E.S., Czymmek, K.J., Eds.; Fungal Biology; Springer International Publishing: Cham, Switzerland, 2015; pp. 61–88. ISBN 978-3-319-22436-7. [Google Scholar]
- Alves, P.D.D.; Siqueira, F.d.F.; Facchin, S.; Horta, C.C.R.; Victória, J.M.N.; Kalapothakis, E. Survey of Microbial Enzymes in Soil, Water, and Plant Microenvironments. TOMICROJ 2014, 8, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.C.; Cantão, M.E.; Nogueira, M.A.; Vasconcelos, A.T.R.; Hungria, M. Outstanding Impact of Soil Tillage on the Abundance of Soil Hydrolases Revealed by a Metagenomic Approach. Braz. J. Microbiol. 2018, 49, 723–730. [Google Scholar] [CrossRef] [PubMed]
- El-Gendi, H.; Saleh, A.K.; Badierah, R.; Redwan, E.M.; El-Maradny, Y.A.; El-Fakharany, E.M. A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges. J. Fungi 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Obahiagbon, K.O.; Owabor, C.N. Bio-Treatment of Crude Oil Polluted Water Using Mixed Microbial Populations of P. aureginosa, Penicillium notatum, E. coli and Aspergillus niger. AMR 2009, 62–64, 802–807. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial Lipases and Their Industrial Applications: A Comprehensive Review. Microb. Cell Fact. 2020, 19, 169. [Google Scholar] [CrossRef]
- Lloyd, G.I.; Morris, E.O.; Smith, J.E. A Study of the Esterases and Their Function in Candida lipolytica, Aspergillus niger and a Yeast-like Fungus. J. Gen. Microbiol. 1970, 63, 141–150. [Google Scholar] [CrossRef]
- Abdel-Kareem, O. Microbiological Testing of Polymers and Resins Used in Conservation of Linen Textiles; Associazione Italiana Prove non Distruttive: Roma, Italy, 2000. [Google Scholar]
- Milanesi, C.; Baldi, F.; Borin, S.; Brusetti, L.; Ciampolini, F.; Iacopini, F.; Cresti, M. Deterioration of Medieval Painting in the Chapel of the Holy Nail, Siena (Italy) Partially Treated with Paraloid B72. Int. Biodeterior. Biodegrad. 2009, 63, 844–850. [Google Scholar] [CrossRef]
- Elhagrassy, A.F. Modification of EAPC-XYL by Pseudomonas Lipases Bacteria to Remove Acrylic from the Mural Oil Paintings. Shedet 2019, 6, 189–202. [Google Scholar] [CrossRef]
- Ranalli, G.; Zanardini, E. Biocleaning on Cultural Heritage: New Frontiers of Microbial Biotechnologies. J. Appl. Microbiol. 2021, 131, 583–603. [Google Scholar] [CrossRef]
- Bosch-Roig, P.; Regidor Ros, J.L.; Estellés, R.M. Biocleaning of Nitrate Alterations on Wall Paintings by Pseudomonas stutzeri. Int. Biodeterior. Biodegrad. 2013, 84, 266–274. [Google Scholar] [CrossRef]
- Ranalli, G.; Zanardini, E.; Rampazzi, L.; Corti, C.; Andreotti, A.; Colombini, M.P.; Bosch-Roig, P.; Lustrato, G.; Giantomassi, C.; Zari, D.; et al. Onsite Advanced Biocleaning System for Historical Wall Paintings Using New Agar-gauze Bacteria Gel. J. Appl. Microbiol. 2019, 126, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Soffritti, I.; D’Accolti, M.; Lanzoni, L.; Volta, A.; Bisi, M.; Mazzacane, S.; Caselli, E. The Potential Use of Microorganisms as Restorative Agents: An Update. Sustainability 2019, 11, 3853. [Google Scholar] [CrossRef]
- Troiano, F.; Gulotta, D.; Balloi, A.; Polo, A.; Toniolo, L.; Lombardi, E.; Daffonchio, D.; Sorlini, C.; Cappitelli, F. Successful Combination of Chemical and Biological Treatments for the Cleaning of Stone Artworks. Int. Biodeterior. Biodegrad. 2013, 85, 294–304. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).