Enteric Methane Emission, Rumen Fermentation and Microbial Profiles of Meat-Master Lambs Supplemented with Barley Fodder Sprouts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Ethics Approval
2.2. Hydroponic Barley Fodder Sprout Production
2.3. Study Animals and Diets
2.4. Data Collection
2.5. Rumen Fermentation
2.6. Deoxyribonucleic Acid Extraction and Amplicon Sequencing
2.7. Data Analysis
3. Results
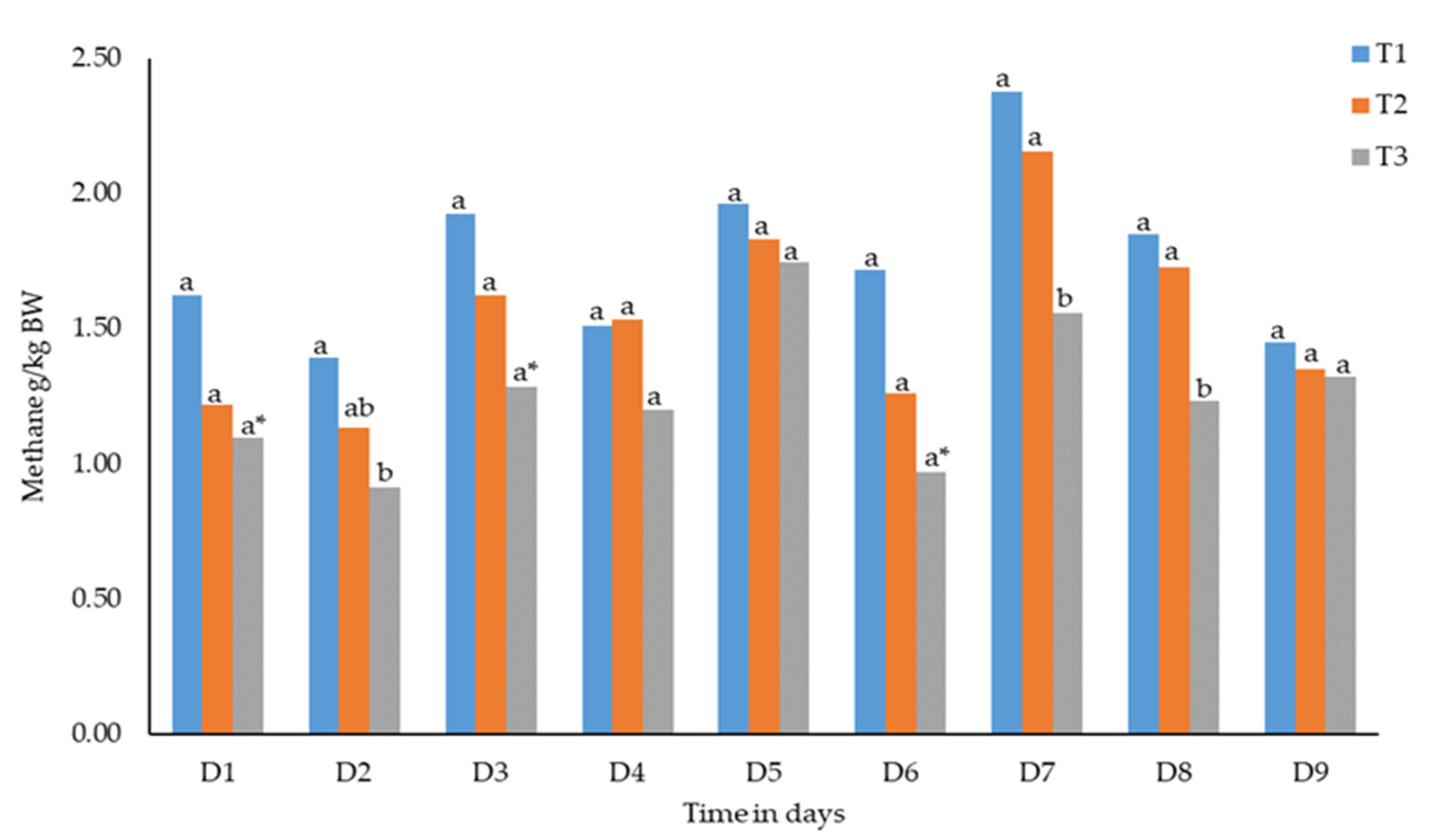
3.1. Methane Emission as Influenced by Barley Sprout Supplementation
3.2. Rumen Fermentation Profile
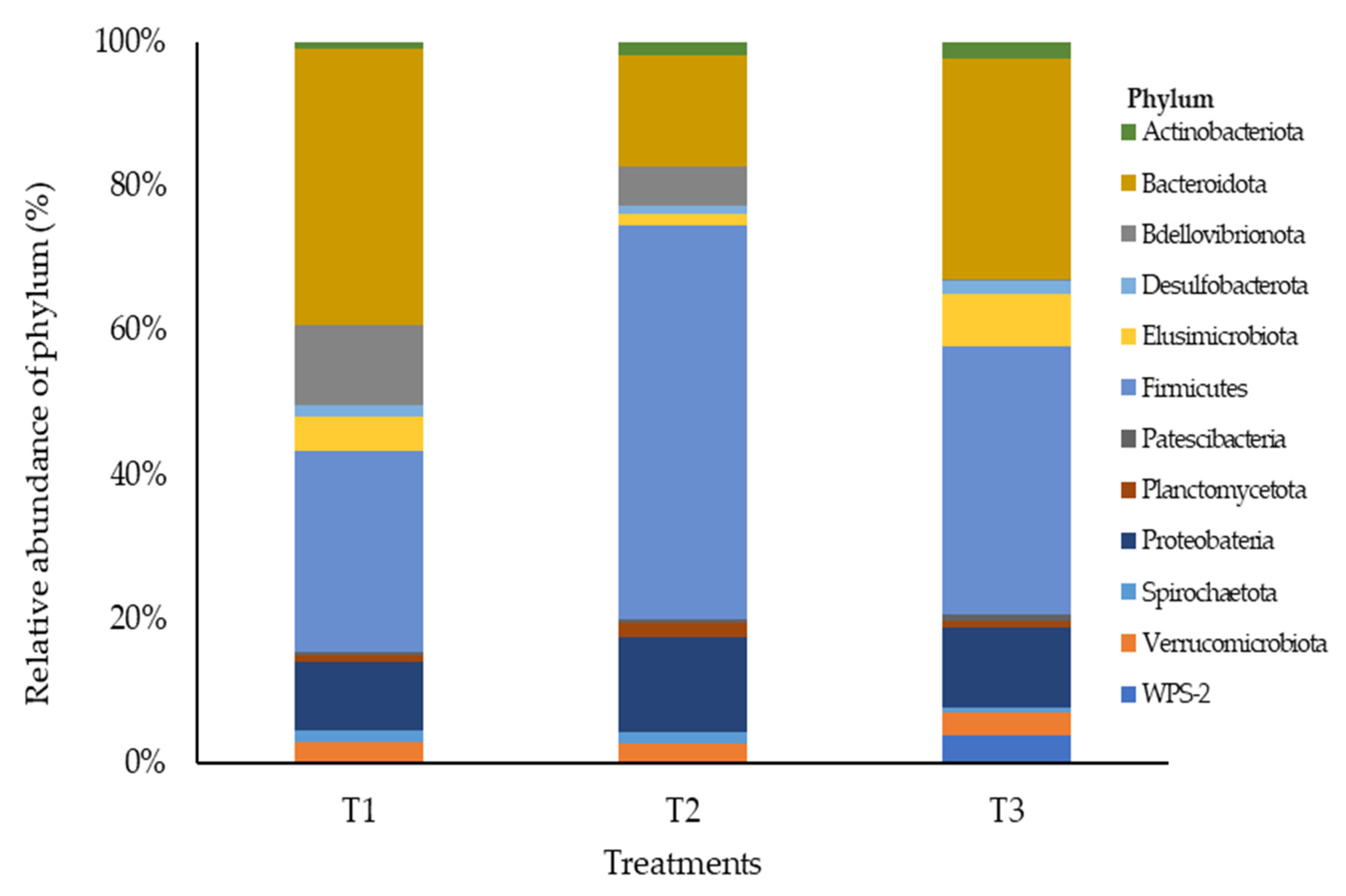
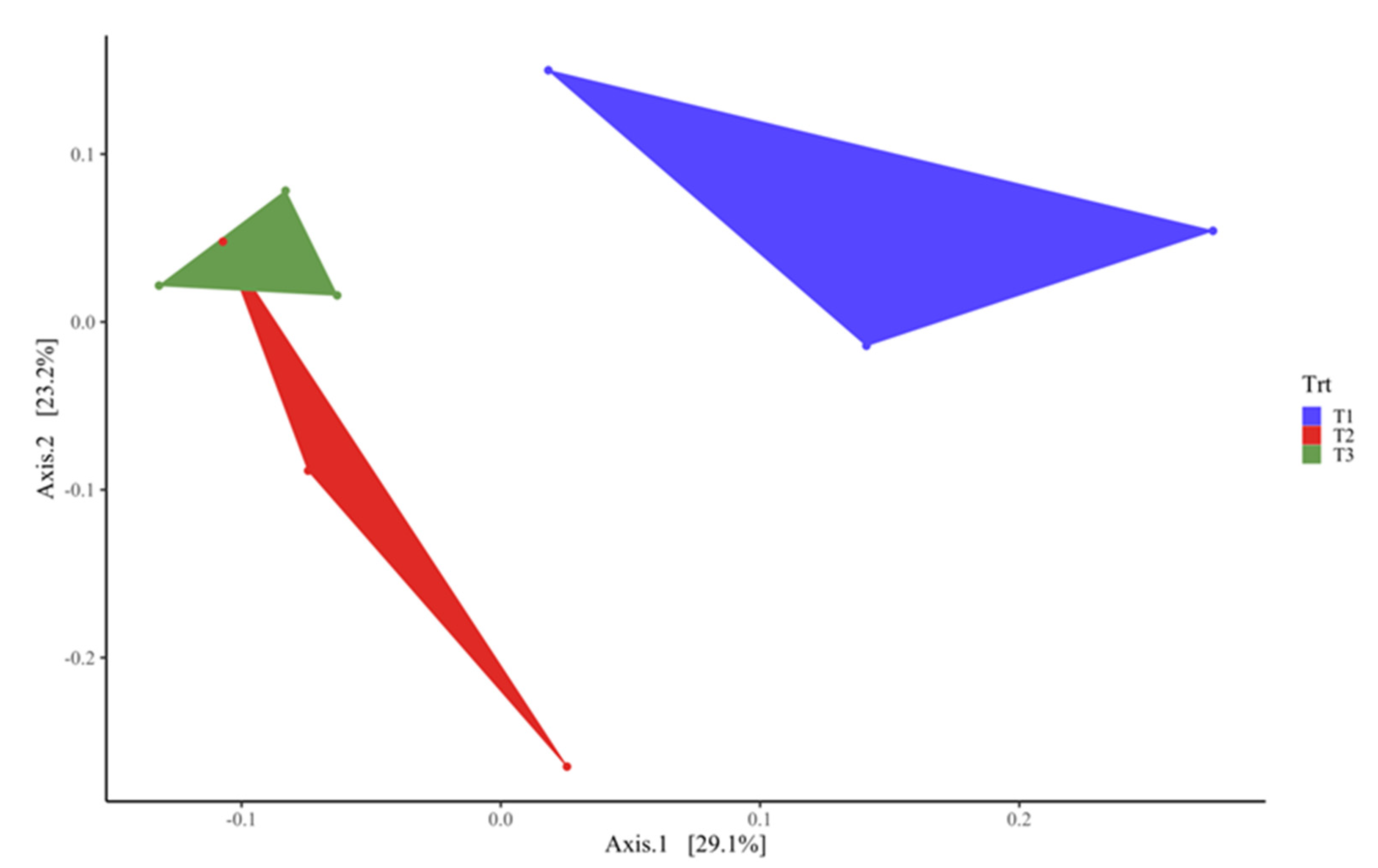
3.3. Rumen Microbial Composition of Meat-Master Lambs as Influenced by Barley Sprouts Supplementation
4. Discussion
4.1. Methane Emission and Ruminal Fermentation as Influenced by Barley Sprouts Supplementation
4.2. Ruminal Microbial Composition as Influenced by Barley Sprouts Supplementation
5. Conclusions and Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Mamuad, L.L.; Lee, S.S.; Lee, S.S. Recent insight and future techniques to enhance rumen fermentation in dairy goats. Asian-Australas J. Anim. Sci. 2019, 32, 1321–1330. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Mei, J.; Huang, L.; Liu, H. Effects of Mulberry branch and leaves silage on microbial community, rumen fermentation characteristics, and milk yield in lactating dairy cows. Fermentation 2022, 8, 86. [Google Scholar] [CrossRef]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Müller, B.; Schnürer, A.; Bertilsson, J. Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, P.; Liu, S.; Miao, B.; Zeng, B.; Jiang, Y.; Li, L.; Wang, L.; Chen, Y.; Zhang, H. Characterization of the rumen microbiota and volatile fatty acid profile of weaned goat kid under shrub-grassland grazing and indoor feeding. Animals 2020, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Mamuad, L.L.; Kim, S.H.; Biswas, A.A.; Yu, Z.; Cho, K.-K.; Kim, S.-B.; Lee, K.; Lee, S.S. Rumen fermentation and microbial community composition influenced by live Enterococcus faecium supplementation. AMB Express. 2019, 9, 123. [Google Scholar] [CrossRef]
- Doi, R.H.; Kosugi, A. Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2004, 2, 541–551. [Google Scholar] [CrossRef]
- Ahmad, A.; Yang, C.; Zhang, J.; Kalwar, Q.; Liang, Z.; Li, C.; Du, M.; Yan, P.; Long, R.; Han, J.; et al. Effect of dietary energy levels on rumen fermentation,, microbial diversity and feed efficiency of yaks (Bos grunniens). Front. Microbiol. 2020, 11, 625. [Google Scholar] [CrossRef]
- Geber, P.J.; Mottet, A.; Opio, C.I.; Falcucci, A.; Teillard, F. Environmental impacts of beef production: Review of challenges and perspectives for durability. Meat Sci. 2015, 109, 2–12. [Google Scholar] [CrossRef]
- Hassan, F.-u.; Arshad, M.A.; Ebeit, H.M.; Rehman, M.S.-u.; Khan, M.S.; Shahid, S.; Yang, C. Phytogenic additive can modulated rumen microbiome to mediate fermentation kinetics and methanogenesis through exploiting Diet –Microbes interaction. Front. Vet. Sci. 2020, 7, 575801. [Google Scholar] [CrossRef]
- Newbold, C.J.; Ramos-Morales, E. Review: Ruminal microbiome and microbial metabolome: Effects of diet and ruminant host. Animal 2020, 14 (Suppl. S1), 78–86. [Google Scholar] [CrossRef] [PubMed]
- Judy, J.V.; Bachman, G.C.; Brown-Brandl, T.M.; Fernando, S.C.; Hales, K.E.; Miller, P.S.; Stowell, R.R.; Kononoff, P.J. Reducing methane production with corn oil and calcium sulfate: Responses on whole-animal energy and nitrogen balance in dairy cattle. J. Dairy Sci. 2019, 102, 2054–2067. [Google Scholar] [CrossRef] [PubMed]
- Theil, A.; Schoenmakers, A.C.M.; Verbaan, I.A.J.; Chenal, E.; Etheve, S.; Beilstein, P. 3- NOP: Mutagenicity and genotoxicity assessment. Food Chem. Toxicol. 2019, 123, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.E.; Andres, S.; López-Ferreras, L.; Snelling, T.J.; Yáñez-Ruíz, D.R.; García-Estrada, C.; López, S. Dietary supplemental plant oils reduce methanogenesis from anaerobic microbial fermentation in the rumen. Sci. Rep. 2020, 10, 1613. [Google Scholar] [CrossRef]
- Roque, B.M.; Venegas, M.; Kinley, R.D.; de Nys, R.; Duarte, T.L.; Yang, X.; Kebreab, E. Red seaweed (Asparagopsis taxiformis) supplementation reduces enteric methane by over 80 percent in beef steers. PLoS ONE 2021, 16, e0247820. [Google Scholar] [CrossRef]
- Sahebi Ala, M.; Pirmohammadi, R.; Khalilvandi-Behroozyar, H. Changes in vitro rumen fermentation, methane production and microbial populations in response to green tea extract. Ital. J. Anim. Sci. 2021, 20, 1114–1125. [Google Scholar] [CrossRef]
- Bowen, J.M.; Cormican, P.; Lister, S.J.; McCabe, M.S.; Duthie, C.-A.; Roehe, R.; Dewhurst, R.J. Links between the rumen microbial, methane emissions and feed efficiency of finishing steers offered dietary lipid and nitrate supplementation. PLoS ONE 2020, 15, e0231759. [Google Scholar] [CrossRef]
- Zhou, Z.; Fang, L.; Meng, Q.; Li, S.; Chai, S.; Liu, S.; Schonewille, J.T. Assessment of ruminal bacterial and archaeal community structure in yak (Bos grunniens). Front. Microboil. 2017, 8, 179. [Google Scholar] [CrossRef]
- Qiu, Q.; Gao, C.; Gao, Z.; Rahman, A.M.; He, Y.; Cao, B.; Su, H. Temporal dynamics in rumen bacterial community composition of finishing steers during an adaptation period of three months. Microorganisms 2019, 7, 410. [Google Scholar] [CrossRef]
- Belanche, A.; Kingston-Smith, A.H.; Griffith, G.W.; Newbold, C.J. A multi-kingdom study reveals the plasticity of the rumen microbiota in response to a shift from non-grazing to grazing diets in sheep. Front. Microbiol. 2019, 10, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekuma, A. Nutritional benefit and economic value of hydroponics fodder production technology in sustainable livestock production against climate change—A mini-review. Adv. Appl. Sci. 2019, 4, 23–25. [Google Scholar] [CrossRef]
- Agius, A.; Pastorelli, G.; Attard, E. Cows fed hydroponic fodder and conventional diet: Effects on milk quality. Arch. Anim. Breed. 2019, 62, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Getachew, A.; Mengistu, U.; Getachew, A.; Getnet, A. Effect of variety and seed rate on hydroponic maize fodder biomass yield, chemical composition, and water use efficiency. Biotechnol. Anim. Husb. 2020, 36, 87–100. [Google Scholar] [CrossRef]
- Salo, S. Effects of hydroponic fodder feeding on milk yield and composition of dairy cow: Review. J. Nat. Sci. Res. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Farghaly, M.M.; Abdullah, M.A.M.; Yuossef, I.M.I.; Abdel-Rahim, I.A.; Abouelezz, K. Effect of feeding hydroponic barley sprouts to sheep on feed intake, nutrient digestibility, nitrogen retention, rumen fermentation and ruminal enzymes. Livest. Sci. 2019, 228, 31–37. [Google Scholar] [CrossRef]
- Badran, E.; Omor, J.O.; Qaisy, A.L.; Amsha, R.A.; Al Jammal, M.; Qadri, M. Milk yield and quality and performance of Awassi ewes fed two levels of hydroponic barley. J. New Sci. Agric. Biotechnol. 2017, 39, 2136–2143. [Google Scholar]
- Chagunda, M.G.G.; Ross, D.; Roberts, D.J. On the use of laser methane detector in dairy cows. Comput. Electron. Agric. 2009, 68, 157–160. [Google Scholar] [CrossRef]
- Shen, J.S.; Chai, Z.; Song, L.J.; Liu, J.X.; Wu, Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, Y.; Zeng, X.; Wang, X.; Wang, Y.; Dai, C.; Li, J.; Huang, P.; Huang, J.; Hussain, T.; et al. Effects of dietary energy levels on rumen fermentation, gastrointestinal tract histology, and bacterial community diversity in fattening male Hu lambs. Front. Microbiol. 2021, 12, 695445. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Edgar, R.C.; Flyvbjerg, H. Error Filtering, Pair Assembly and Error Correction for Next-Generation Sequencing Reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Metwally, A.A.; Yang, J.; Ascoli, C.; Dai, Y.; Finn, P.W.; Perkins, D.L. MetaLonDA: A flexible R package for identifying time intervals of differentially abundant features in metagenomic longitudinal studies. Microbiome 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- SAS. Statistical Analysis Systems User’s Guide: Statistics, Version 9.0; SAS Institute Inc.: Cary, NC, USA, 2002. [Google Scholar]
- R Studio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015; Available online: http://www.rstudio.com/ (accessed on 5 May 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 27 July 2022).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.I.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Package ‘Vegan’. Community Ecology Package, Version 2013. pp. 1–295. Available online: https://cran.r-project.org; https://github.com/vegandevs/vegan (accessed on 27 July 2022).
- Kumar, S.; Dagar, S.S.; Puniya, A.K.; Upadhyay, R.C. Changes in methane emission, rumen fermentation in response to diet and microbial interactions. Res. Vet. Sci. 2013, 94, 263–268. [Google Scholar] [CrossRef]
- Swainson, N.; Muetzel, S.; Clark, H. Updated prediction of enteric methane emissions from sheep suitable for use in the New Zealand national greenhouse gas inventory. Anim. Prod. Sci. 2018, 58, 973–979. [Google Scholar] [CrossRef]
- Tiklebrhan, T.; Wang, R.; Wang, M.; Wen, J.N.; Li, W.T.; Zhang, X.M.; Ma, Z.Y.; Tan, Z.L. Effect of dietary corn gluten inclusion on rumen fermentation, microbial and methane emissions in goats. Anim. Feed Sci. Technol. 2020, 259, 114314. [Google Scholar] [CrossRef]
- Congio, G.F.S.; Bannink, A.; Mogollón, O.L.M.; Latin America Methane Project Collaborators; Hristov, A.N. Enteric methane mitigation strategies for ruminant livestock systems in the Latin America and Caribbean region: A meta-analysis. J. Clean. Prod. 2021, 312, 127693. [Google Scholar] [CrossRef]
- Gaviria-Uribe, X.; Bolivar, D.M.; Rosenstock, T.S.; Molina-Botero, I.C.; Chirinda, N.; Barahona, R.; Arango, J. Nutritional quality, voluntary intake and enteric methane emissions of diets based on novel cayman grass and its associations with two Leucaena shrub legumes. Front. Vet. Sci. 2020, 7, 579189. [Google Scholar] [CrossRef] [PubMed]
- Min, B.R.; Solaiman, S.; Waldrip, H.M.; Parker, D.; Toddy, R.W.; Brauer, D. Dietary mitigation of enteric methane emissions from ruminants: A review of plants tannin mitigation options. Anim. Nutr. 2020, 6, 231–246. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.; Edwards, R.A.; Greenhalgh, J.F.D.; Morgan, C.A.; Sinclair, L.A.; Wilkinson, R.G. Animal Nutrition, 7th ed.; Printice Hall: Harlow, UK, 2010. [Google Scholar]
- Panyawoot, N.; So, S.; Cherdthong, A.; Chanjula, P. Effect of feeding discarded durian peel ensiled with Lactobacillus casei TH14 and additives in total mixed rations on digestibility, ruminal fermentation, methane mitigation, and nitrogen balance of Thai Native-Anglo-Nubian goats. Fermentation 2022, 8, 43. [Google Scholar] [CrossRef]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shah, A.M.; Shoa, Y.; Wang, Z.; Zou, H.; Hu, R.; Peng, Q.; Kang, K.; Wanapat, M. Effects of yeast cell wall on the growth performance, ruminal fermentation and microbial community of weaned calves. Livest. Sci. 2020, 239, 104170. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Martin, C.; Boudra, H. Fungal secondary metabolites from Monascus spp. reduce rumen methane production in vitro and in vivo. J. Anim. Sci. 2013, 91, 848–860. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Zhang, J.B.; Liang, Z.; Yang, C.; Kalwar, Q.; Shan, T.; Du, M.; Muhammad, I.; Zheng, J.; Yan, P.; et al. Dynamics of rumen bacterial composition of yak (Bos grunniens) in response to dietary supplements during the cold season. Peer J. 2021, 9, e11520. [Google Scholar] [CrossRef]
- Thoetkiattikul, H.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Pattarajinda, V.; Eurwilaichitr, L.; Champreda, V. Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Current Microbiol. 2013, 67, 130–137. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Zhang, Y.; Wang, L. The effects of different concentrate-to-forage ration diets on rumen bacterial microbiota and the structures of Holstein cows during the feeding cycle. Animals 2020, 10, 957. [Google Scholar] [CrossRef]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ration condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Liu, H.; Hu, L.; Han, X.; Zhao, N.; Xu, T.; Ma, L.; Wang, X.; Zhang, X.; Kang, S.; Zhoa, X.; et al. Tibetan sheep adapt to plant phenology in alpine meadows by changing rumen microbial community structure and function. Front. Microbiol. 2020, 11, 587558. [Google Scholar] [CrossRef] [PubMed]
- Linde, D.A.; van Marle-Köster, E.; du Toit, C.J.L.; Scholtz, M.M.; Schokker, D. Rumen microbial diversity of Bonsmara cattle using amplicon sequencing during a 120-day growth trial. S. Afr. J. Anim. Sci. 2022, 52, 148–161. [Google Scholar] [CrossRef]
- Mani, S.; Aiyegoro, O.A.; Adeleke, M.A. Characterization of rumen microbiota of two sheep breeds supplemented with direct-fed lactic acid bacteria. Front. Vet. Sci. 2021, 7, 570074. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Chen, H.; Zhao, X.; Xu, S.; Hu, L.; Xu, T.; Jiang, L.; Zhang, W. Rumen prokaryotic communities of ruminants under different feeding paradigms on the Qinghai-Tibetan Plateau. Syst. Appl. Microbiol. 2017, 40, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhoa, R. Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Guo, X. Comparative analysis of rumen fermentation parameters and bacterial profiles during adaptation to different fattening stages in beef cattle fed TMR with various forage silage. Anim. Feed Sci. Technol. 2021, 278, 115006. [Google Scholar] [CrossRef]
- Dassa, B.; Borovok, I.; Ruimy-Israeli, V.; Lamed, R.; Flint, H.J.; Duncan, S.H.; Henrissat, B.; Coutinho, P.; Morrison, M.; Mosoni, P.; et al. Rumen cellulosomics: Divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS ONE 2014, 9, e99221. [Google Scholar] [CrossRef]
- Fu, Z.; Xu, X.; Zhang, J.; Zhang, L. Effect of different feeding methods on rumen microbes in growing Chinese Tan sheep. R. Bras. Zootec. 2020, 49, e20190258. [Google Scholar] [CrossRef]
- Myer, P.R.; Smith, T.P.; Wells, J.E.; Kuehm, L.A.; Freetly, H.C. Rumen microbiome from steers differing in feed efficiency. PLoS ONE 2015, 10, e0129174. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Huo, W.; Zhu, W.; Mao, S. Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World J. Microbiol. Biotechnol. 2014, 30, 669–680. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Treatments | SEM 1 | p-Value | ||
|---|---|---|---|---|---|
| T1 | T2 | T3 | |||
| Intake (kg) | 1.0 c | 1.4 b | 1.6 a | 0.05 | <0.0001 |
| Initial body weight (kg) | 23.2 | 23.3 | 23.5 | 1.16 | 0.9871 |
| Final body weight (kg) | 28.6 b | 31.9 ab | 33.5 a | 1.12 | 0.0186 |
| Methane (ppm) 2 | 23.4 | 22.0 | 19.1 | 1.11 | 0.0608 |
| Methane (g/day) | 51.1 | 48.2 | 41.9 | 2.44 | 0.0608 |
| Methane (g/kg BW) 3 | 1.8 a | 1.5 ab | 1.3 b | 0.11 | 0.0264 |
| Methane (g/DMI) 4 | 53.0 a | 35.1 b | 25.5 c | 2.50 | <0.0001 |
| Parameters | Treatments | SEM 1 | p-Value | ||
|---|---|---|---|---|---|
| T1 | T2 | T3 | |||
| NH3-N (mg/dL) 2 | 19.4 a | 11.1 b | 9.8 b | 0.66 | <0.0001 |
| Total VFA (mmol/L) 3 | 68.9 | 68.3 | 60.1 | 5.90 | 0.5344 |
| Molar proportion of VFA | |||||
| Acetate (A) | 73.3 a | 67.3 b | 67.8 b | 1.23 | 0.0286 |
| Propionate (P) | 16.5 | 20.1 | 20.8 | 1.04 | 0.0536 |
| Butyrate | 7.1 | 8.8 | 9.0 | 0.71 | 0.2068 |
| Iso-butyrate | 1.2 | 1.5 | nd 4 | 0.09 | 0.2671 |
| Valerate | 0.8 | 1.0 | 0.8 | 0.06 | 0.1276 |
| Iso-valerate | 1.2 | 1.3 | 1.0 | 0.18 | 0.4426 |
| A:P ratio | 4.5 a | 3.4 b | 3.3 b | 0.28 | 0.0408 |
| Bacteria | Treatments | SEM 1 | p-Value | ||
|---|---|---|---|---|---|
| T1 | T2 | T3 | |||
| Acholeplasmataceae | 2.6 | 0.5 | 0.5 | 0.67 | 0.1097 |
| Anaerovoracaceae | 1.3 | 2.4 | 2.3 | 0.62 | 0.4303 |
| Butyricicoccaceae | 1.0 a | 0.2 b | 0.1 b | 0.20 | 0.0388 |
| Atopobiaceae | 0.3 | 0.9 | 1.8 | 0.47 | 0.1529 |
| Chitinophagaceae | 0.5 | 1.9 | 1.0 | 0.93 | 0.5826 |
| COB P4-1 termite group | 3.3 | 0.1 | 0.2 | 1.05 | 0.1246 |
| Desulfovibrionaceae | 1.0 | 0.9 | 1.9 | 0.70 | 0.5806 |
| Erwiniaceae | 1.5 | 0.2 | 0.2 | 0.81 | 0.4529 |
| Erysipelatoclostridiaceae | 1.5 | 24.7 | 1.5 | 13.09 | 0.4084 |
| Erysipelotrichaceae | 0.7 | 1.2 | 2.6 | 0.55 | 0.1279 |
| Hungateiclostridiaceae | 4.2 a | 0.8 b | 0.7 b | 0.64 | 0.013 |
| Lachnospiraceae | 7.9 | 11.0 | 12.4 | 2.44 | 0.4656 |
| Moraxellaceae | 1.2 | 1.3 | 0.5 | 0.68 | 0.7049 |
| Oscillospiraceae | 6.0 | 4.6 | 3.6 | 1.93 | 0.6761 |
| Oxalobacteraceae | 0.2 | 1.4 | 0.8 | 0.56 | 0.4259 |
| p-2534-18B5 gut group | 26.5 | 0.2 | 0.2 | 7.98 | 0.0924 |
| Pasteurellaceae | 1.7 | 1.3 | 1.9 | 0.89 | 0.8862 |
| PeH15 | 2.7 | 5.5 | 0.2 | 3.16 | 0.5275 |
| Pirellulaceae | 1.0 | 1.9 | 0.6 | 0.46 | 0.1884 |
| Prevotellaceae | 1.5 | 5.9 | 26.2 | 12.10 | 0.3677 |
| Rikenellaceae | 7.8 | 0.8 | 0.8 | 3.96 | 0.4087 |
| Ruminococcaceae | 1.7 | 1.7 | 1.9 | 0.44 | 0.9587 |
| Selenomonadaceae | 0.9 | 4.6 | 10.5 | 3.90 | 0.2895 |
| Spirochaetaceae | 1.1 | 1.7 | 0.6 | 0.64 | 0.5574 |
| Succinivibrionaceae | 2.1 | 5.7 | 4.9 | 3.25 | 0.7317 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mpanza, T.D.E.; Dhlamini, T.C.; Pierneef, R.E.; Mbatha, K.R. Enteric Methane Emission, Rumen Fermentation and Microbial Profiles of Meat-Master Lambs Supplemented with Barley Fodder Sprouts. Fermentation 2022, 8, 434. https://doi.org/10.3390/fermentation8090434
Mpanza TDE, Dhlamini TC, Pierneef RE, Mbatha KR. Enteric Methane Emission, Rumen Fermentation and Microbial Profiles of Meat-Master Lambs Supplemented with Barley Fodder Sprouts. Fermentation. 2022; 8(9):434. https://doi.org/10.3390/fermentation8090434
Chicago/Turabian StyleMpanza, Thamsanqa Doctor Empire, Thabo Creswell Dhlamini, Rian Ewald Pierneef, and Khanyisile R. Mbatha. 2022. "Enteric Methane Emission, Rumen Fermentation and Microbial Profiles of Meat-Master Lambs Supplemented with Barley Fodder Sprouts" Fermentation 8, no. 9: 434. https://doi.org/10.3390/fermentation8090434
APA StyleMpanza, T. D. E., Dhlamini, T. C., Pierneef, R. E., & Mbatha, K. R. (2022). Enteric Methane Emission, Rumen Fermentation and Microbial Profiles of Meat-Master Lambs Supplemented with Barley Fodder Sprouts. Fermentation, 8(9), 434. https://doi.org/10.3390/fermentation8090434






