Statistical Optimization for Cost-Effective Production of Yeast-Bacterium Cell-Bound Lipases Using Blended Oily Wastes and Their Potential Applications in Biodiesel Synthesis and Wastewater Bioremediation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Chemicals
2.2. Palm Oil Mill Effluent (POME)-Based Media Preparation and Their Characteristics
2.3. Microbial Strains and Inoculum Preparation
2.4. Microbial Growth and Cell Biomass (CBM) Determination
2.5. The Lipases Hydrolytic Activity Assays
2.6. Experimental Design and Statistical Analysis
2.6.1. Selection of Medium Composition for Valorization Process of Agro-Industrial Wastes into CBLs by One Variable at a Time (OVAT)
2.6.2. Experimental Design by Plackett–Burman Design (PBD) and Optimization of Key Parameters by Response Surface Methodology-Central Composite Design (RSM-CCD)
2.7. Time Course Study of CBLs Production under Optimal Conditions
2.8. Application of CBLs in Biodiesel Production
2.9. Palm Oil Mill Effluent (POME) Bioremediation by CBLs
2.10. Data Analysis
3. Results and Discussion
3.1. Selection of Medium Composition for Valorization Process of Agro-Industrial Wastes into CBLs by One Variable at A Time (OVAT)
3.2. Screening of Significant Parameters Using Plackett-Burman Design (PBD)
+ 35.31252 * D + 314.07504 * E
+ 0.11833 * D + 2.06481 * E
+ 0.51667 * D + 4.38889 * E
3.3. Statistical Optimization Procedure Using Response Surface Methodology-Central Composite Design (RSM-CCD)
+ 289.442 * BC − 83.0406 * A2 − 492.227 * B2 − 230.336 * C2
+ 1.8455 * BC + 0.0609913 * A2 − 3.73308 * B2 − 1.28688 * C2
+ 1.49348 * BC − 0.15020 * A2 − 2.43320 * B2 − 0.71604 * C2
3.4. Time Course Study of Yeast–Bacterium CBLs Production under Optimal Conditions
3.5. Application of CBLs in Biodiesel Synthesis
3.6. Application of CBLs in Palm Oil Mill Effluent (POME) Bioremediation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Amini, Z.; Ilham, Z.; Ong, H.C.; Mazaheri, H.; Chen, W.-H. State of the Art and Prospective of Lipase-Catalyzed Transesterification Reaction for Biodiesel Production. Energy Convers. Manag. 2017, 141, 339–353. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Enzymes in Lipid Modification. Annu. Rev. Food Sci. Technol. 2018, 9, 85–103. [Google Scholar] [CrossRef]
- Joshi, R.; Kuila, A. Lipase and Their Different Industrial Applications: A Review. Brazilian J. Biol. Sci. 2018, 5, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From Production to Applications. Sep. Purif. Rev. 2019, 49, 143–158. [Google Scholar] [CrossRef]
- Fibriana, F.; Upaichit, A.; Cheirsilp, B. Low-Cost Production of Cell-Bound Lipases by Pure and Co-Culture of Yeast and Bacteria in Palm Oil Mill Effluent and the Applications in Bioremediation and Biodiesel Synthesis. Biomass Convers. Biorefinery 2021, 1–22. [Google Scholar] [CrossRef]
- Louhasakul, Y.; Cheirsilp, B.; Prasertsan, P. Valorization of Palm Oil Mill Effluent into Lipid and Cell-Bound Lipase by Marine Yeast Yarrowia lipolytica and Their Application in Biodiesel Production. Waste Biomass Valori 2016, 7, 417–426. [Google Scholar] [CrossRef]
- Miranda, S.M.; Pereira, A.S.; Belo, I.; Lopes, M. Waste Cooking Oils: Low-Cost Substrate for Co-Production of Lipase and Microbial Lipids. In WASTES–Solutions, Treatments and Opportunities II; CRC Press: Boca Raton, FL, USA, 2017; pp. 99–104. [Google Scholar]
- Silveira, E.A.; Tardioli, P.W.; Farinas, C.S. Valorization of Palm Oil Industrial Waste as Feedstock for Lipase Production. Appl. Biochem. Biotechnol. 2016, 179, 558–571. [Google Scholar] [CrossRef]
- Komesli, S.; Akbulut, S.; Arslan, N.P.; Adiguzel, A.; Taskin, M. Waste Frying Oil Hydrolysis and Lipase Production by Cold-Adapted Pseudomonas yamanorum LP2 under Non-Sterile Culture Conditions. Environ. Technol. 2021, 42, 3245–3253. [Google Scholar] [CrossRef]
- Baloch, K.A.; Upaichit, A.; Cheirsilp, B. Use of Low-Cost Substrates for Cost-Effective Production of Extracellular and Cell-Bound Lipases by a Newly Isolated Yeast Dipodascus capitatus A4C. Biocatal. Agric. Biotechnol. 2019, 19, 101012. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z.; AbdulKarim, M.I.; Salleh, H.M. Optimization of Lipase Production by Candida cylindracea in Palm Oil Mill Effluent Based Medium Using Statistical Experimental Design. J. Mol. Catal. B Enzym. 2011, 69, 66–73. [Google Scholar] [CrossRef]
- Hermansyah, H.; Maresya, A.; Putri, D.N.; Sahlan, M.; Meyer, M. Production of Dry Extract Lipase from Pseudomonas Aeruginosa by the Submerged Fermentation Method in Palm Oil Mill Effluent. Int. J. Technol. 2018, 9, 325–334. [Google Scholar] [CrossRef] [Green Version]
- Rachmadona, N.; Quayson, E.; Amoah, J.; Alfaro-Sayes, D.A.; Hama, S.; Aznury, M.; Kondo, A.; Ogino, C. Utilizing Palm Oil Mill Effluent (POME) for the Immobilization of Aspergillus Oryzae Whole-cell Lipase Strains for Biodiesel Synthesis. Biofuels Bioprod. Biorefining 2021, 15, 804–814. [Google Scholar] [CrossRef]
- Prasertsan, P.; Binmaeil, H. Treatment of Oil Mill Effluent by Thermotolerant Polymer-Producing Fungi. J. Water Environ. Technol. 2018, 16, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.A.; Yousuf, A.; Karim, A.; Pirossi, D.; Khan, M.R.; Wahid, Z.A. Bioremediation of Palm Oil Mill Effluent and Lipid Production by Lipomyces starkeyi: A Combined Approach. J. Clean. Prod. 2018, 172, 1779–1787. [Google Scholar] [CrossRef]
- Karim, A.; Islam, M.A.; Mishra, P.; Muzahid, A.J.M.; Yousuf, A.; Khan, M.M.R.; Faizal, C.K.M. Yeast and Bacteria Co-Culture-Based Lipid Production through Bioremediation of Palm Oil Mill Effluent: A Statistical Optimization. Biomass Convers. Biorefinery 2021, 1–12. [Google Scholar] [CrossRef]
- Baloch, K.; Upaichit, A.; Cheirsilp, B.; Fibriana, F. The Occurrence of Triple Catalytic Characteristics of Yeast Lipases and Their Application Prospects in Biodiesel Production from Non-Edible Jatropha Curcas Oil in a Solvent-Free System. Curr. Microbiol. 2021, 78, 1914–1925. [Google Scholar] [CrossRef]
- Rosenstein, R.; Götz, F. Staphylococcal Lipases: Biochemical and Molecular Characterization. Biochimie 2000, 82, 1005–1014. [Google Scholar] [CrossRef]
- Venkatesagowda, B.; Ponugupaty, E.; Barbosa, A.M.; Dekker, R.F.H. Solid-State Fermentation of Coconut Kernel-Cake as Substrate for the Production of Lipases by the Coconut Kernel-Associated Fungus Lasiodiplodia theobromae VBE-1. Ann. Microbiol. 2015, 65, 129–142. [Google Scholar] [CrossRef]
- Kyeong, J.S.; Yeom, S.H. Preparation of Immobilized Whole Cell Biocatalyst and Biodiesel Production Using a Packed-Bed Bioreactor. Bioprocess Biosyst. Eng. 2014, 37, 2189–2198. [Google Scholar] [CrossRef]
- Adachi, D.; Koh, F.; Hama, S.; Ogino, C.; Kondo, A. A Robust Whole-Cell Biocatalyst That Introduces a Thermo- and Solvent-Tolerant Lipase into Aspergillus oryzae Cells: Characterization and Application to Enzymatic Biodiesel Production. Enzym. Microb. Technol. 2013, 52, 331–335. [Google Scholar] [CrossRef]
- Babaki, M.; Yousefi, M.; Habibi, Z.; Mohammadi, M. Process Optimization for Biodiesel Production from Waste Cooking Oil Using Multi-Enzyme Systems through Response Surface Methodology. Renew. Energy 2017, 105, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Sandoval, G.; Casas-Godoy, L.; Bonet-Ragel, K.; Rodrigues, J.; Ferreira-Dias, S.; Valero, F. Enzyme-Catalyzed Production of Biodiesel as Alternative to Chemical-Catalyzed Processes: Advantages and Constraints. Curr. Biochem. Eng. 2017, 4, 109–141. [Google Scholar] [CrossRef]
- Toro, E.C.; Rodríguez, D.F.; Morales, N.; García, L.M.; Godoy, C.A. Novel Combi-Lipase Systems for Fatty Acid Ethyl Esters Production. Catalysts 2019, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- Tongboriboon, K.; Cheirsilp, B.; H-Kittikun, A. Mixed Lipases for Efficient Enzymatic Synthesis of Biodiesel from Used Palm Oil and Ethanol in a Solvent-Free System. J. Mol. Catal. B Enzym. 2010, 67, 52–59. [Google Scholar] [CrossRef]
- Rodriguez-Mateus, Z.; Agualimpia, B.; Zafra, G. Isolation and Molecular Characterization of Microorganisms with Potential for the Degradation of Oil and Grease from Palm Oil Refinery Wastes. Chem. Eng. Trans. 2016, 49, 517–522. [Google Scholar] [CrossRef]
- Fibriana, F.; Upaichit, A.; Hongpattarakere, T. Selection, Identification, Optimization and Characterization of Lipase Producing Fungal Strain Isolated from Palm Oil Contaminated Waste; Prince of Songkla University: Songkla, Thailand, 2014. [Google Scholar]
- Lee, S.Y.; Rhee, J.S. Hydrolysis of Triglyceride by the Whole Cell of Pseudomonas putida 3SK in Two-Phase Batch and Continuous Reactor Systems. Biotechnol. Bioeng. 1994, 44, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Miranda, S.M.; Alves, J.M.; Pereira, A.S.; Belo, I. Waste Cooking Oils as Feedstock for Lipase and Lipid-rich Biomass Production. Eur. J. Lipid Sci. Technol. 2019, 121, 1800188. [Google Scholar] [CrossRef] [Green Version]
- Rywińska, A.; Witkowska, D.; Juszczyk, P.; Rymowicz, W.; Kita, A. Waste Frying Oil as Substrate for Lipase Production by Geotrichum candidum Strains. Polish J. Environ. Stud 2008, 17, 925–931. [Google Scholar]
- Nurul-Adela, B.; Nasrin, A.-B.; Loh, S.-K. Palm Oil Mill Effluent as a Low-Cost Substrate for Bioflocculant Production by Bacillus Marisflavi NA8. Bioresour. Bioprocess. 2016, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, S.; Zahri, K.N.M.; Convey, P.; Khalil, K.A.; Gomez-Fuentes, C.; Zulkarnain, A.; Alias, S.A.; González-Rocha, G.; Ahmad, S.A. Optimisation of Biodegradation Conditions for Waste Canola Oil by Cold-Adapted Rhodococcus Sp. AQ5-07 from Antarctica. Electron. J. Biotechnol. 2020, 48, 1–12. [Google Scholar] [CrossRef]
- Saenge, C.; Cheirsilp, B.; Suksaroge, T.T.; Bourtoom, T. Efficient Concomitant Production of Lipids and Carotenoids by Oleaginous Red Yeast Rhodotorula Glutinis Cultured in Palm Oil Mill Effluent and Application of Lipids for Biodiesel Production. Biotechnol. Bioprocess Eng. 2011, 16, 23–33. [Google Scholar] [CrossRef]
- Çağatay, Ş.; Aksu, Z. Use of Different Kinds of Wastes for Lipase Production: Inductive Effect of Waste Cooking Oil on Activity. J. Biosci. Bioeng. 2021, 132, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Nunes, P.M.B.; Fraga, J.L.; Ratier, R.B.; Rocha-Leão, M.H.M.; Brígida, A.I.S.; Fickers, P.; Amaral, P.F.F. Waste Soybean Frying Oil for the Production, Extraction, and Characterization of Cell-Wall-Associated Lipases from Yarrowia lipolytica. Bioprocess Biosyst. Eng. 2021, 44, 809–818. [Google Scholar] [CrossRef]
- Potumarthi, R.; Subhakar, C.; Vanajakshi, J.; Jetty, A. Effect of Aeration and Agitation Regimes on Lipase Production by Newly Isolated Rhodotorula mucilaginosa–MTCC 8737 in Stirred Tank Reactor Using Molasses as Sole Production Medium. Appl. Biochem. Biotechnol. 2008, 151, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Casas-Godoy, L.; Gasteazoro, F.; Duquesne, S.; Bordes, F.; Marty, A.; Sandoval, G. Lipases: An Overview. In Lipases and Phospholipases; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–38. [Google Scholar]
- Awad, G.E.A.; Mostafa, H.; Danial, E.N.; Abdelwahed, N.A.M.; Awad, H.M. Enhanced Production of Thermostable Lipase from Bacillus cereus ASSCRC-P1 in Waste Frying Oil Based Medium Using Statistical Experimental Design. J. Appl. Pharm. Sci. 2015, 5, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Theerachat, M.; Tanapong, P.; Chulalaksananukul, W. The Culture or Co-Culture of Candida rugosa and Yarrowia lipolytica Strain RM-4A, or Incubation with Their Crude Extracellular Lipase and Laccase Preparations, for the Biodegradation of Palm Oil Mill Wastewater. Int. Biodeterior. Biodegrad. 2017, 121, 11–18. [Google Scholar] [CrossRef]
- Lima, V.M.G.; Krieger, N.; Sarquis, M.I.M.; Mitchell, D.A.; Ramos, L.P.; Fontana, J.D. Effect of Nitrogen and Carbon Sources on Lipase Production by Penicillium Aurantiogriseum. Food Technol. Biotechnol. 2003, 41, 105–110. [Google Scholar]
- Jamilu, H.; Ibrahim, A.H.; Abdullahi, S.Z. Isolation, Optimization and Characterization of Lipase Producing Bacteria from Abbatoir Soil. Int. J. Sci. Adv. 2021, 3, 75–82. [Google Scholar] [CrossRef]
- Nuylert, A.; Hongpattarakere, T. Improvement of Cell-Bound Lipase from Rhodotorula mucilaginosa P11I89 for Use as a Methanol-Tolerant, Whole-Cell Biocatalyst for Production of Palm-Oil Biodiesel. Ann. Microbiol. 2012, 63, 929–939. [Google Scholar] [CrossRef]
- Salehizadeh, H.; Yan, N. Recent Advances in Extracellular Biopolymer Flocculants. Biotechnol. Adv. 2014, 32, 1506–1522. [Google Scholar] [CrossRef]
- Fibriana, F.; Upaichit, A.; Cheirsilp, B. Turning Waste into Valuable Products: Utilization of Agroindustrial Oily Wastes as the Low-Cost Media for Microbial Lipase Production. J. Phys. Conf. Ser. 2021, 1918, 52028. [Google Scholar] [CrossRef]
- Suci, M.; Arbianti, R.; Hermansyah, H. Lipase Production from Bacillus subtilis with Submerged Fermentation Using Waste Cooking Oil. IOP Conf. Ser. Earth Environ. Sci. 2018, 105, 12126. [Google Scholar] [CrossRef]
- Rajendran, A.; Thirugnanam, M.; Thangavelu, V. Statistical Evaluation of Medium Components by Plackett-Burman Experimental Design and Kinetic Modeling of Lipase Production by Pseudomonas fluorescens. Indian J. Biotechnol. 2007, 6, 469–478. [Google Scholar]
- Ganapathy, B.; Yahya, A.; Ibrahim, N. Bioremediation of Palm Oil Mill Effluent (POME) Using Indigenous Meyerozyma guilliermondii. Environ. Sci. Pollut. Res. 2019, 26, 11113–11125. [Google Scholar] [CrossRef]
- Wrenn, B.A.; Haines, J.R.; Venosa, A.D.; Kadkhodayan, M.; Suidan, M.T. Effects of Nitrogen Source on Crude Oil Biodegradation. J. Ind. Microbiol. 1994, 13, 279–286. [Google Scholar] [CrossRef]
- Thompson, D.S.; Carlisle, P.L.; Kadosh, D. Coevolution of Morphology and Virulence in Candida Species. Eukaryot. Cell 2011, 10, 1173–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshimizu, S.; Otake, I.; Yoshioka, H.; Saito, Y.; Taki, H. Development of Kitchen Wastewater-Treatment System Using Fats and Oils Degrading Microorganisms; Biseibutsu Seizai Wo Shiyoshita Chubo Haisui Shori System No Kaihatsu. Kuki Chowa Eisei Kogaku (J. Soc. Heat. Air-Cond. Sanit. Eng. Jpn.) 1997, 71, 999–1009. [Google Scholar]
- Ibrahim, N.A.; Guo, Z.; Xu, X. Enzymatic Interesterification of Palm Stearin and Coconut Oil by a Dual Lipase System. J. Am. Oil Chem. Soc. 2008, 85, 37–45. [Google Scholar] [CrossRef]
- Binhayeeding, N.; Klomklao, S.; Prasertsan, P.; Sangkharak, K. Improvement of Biodiesel Production Using Waste Cooking Oil and Applying Single and Mixed Immobilised Lipases on Polyhydroxyalkanoate. Renew. Energy 2020, 162, 1819–1827. [Google Scholar] [CrossRef]
- Marsidi, N. Biological Treatment of POME (High Acidic Content): Factorial Analysis. Doctoral Dissertation, Universiti Malaysia Pahang, Pahang, Malaysia, 2013. [Google Scholar]
- Yousuf, A.; Sannino, F.; Addorisio, V.; Pirozzi, D. Microbial Conversion of Olive Oil Mill Wastewaters into Lipids Suitable for Biodiesel Production. J. Agric. Food Chem. 2010, 58, 8630–8635. [Google Scholar] [CrossRef]
- Soleimaninanadegani, M.; Manshad, S. Enhancement of Biodegradation of Palm Oil Mill Effluents by Local Isolated Microorganisms. Int. Sch. Res. Not. 2014, 727049. [Google Scholar] [CrossRef]
- Bala, J.D.; Lalung, J.; Al-Gheethi, A.A.S.; Kaizar, H.; Ismail, N. Reduction of Organic Load and Biodegradation of Palm Oil Mill Effluent by Aerobic Indigenous Mixed Microbial Consortium Isolated from Palm Oil Mill Effluent (POME). Water Conserv. Sci. Eng. 2018, 3, 139–156. [Google Scholar] [CrossRef]
- Nwuche, C.O.; Aoyagi, H.; Ogbonna, J.C. Treatment of Palm Oil Mill Effluent by a Microbial Consortium Developed from Compost Soils. Int. Sch. Res. Not. 2014, 762070. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calviño, D.; Bååth, E. Growth Response of the Bacterial Community to PH in Soils Differing in PH. FEMS Microbiol. Ecol. 2010, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]


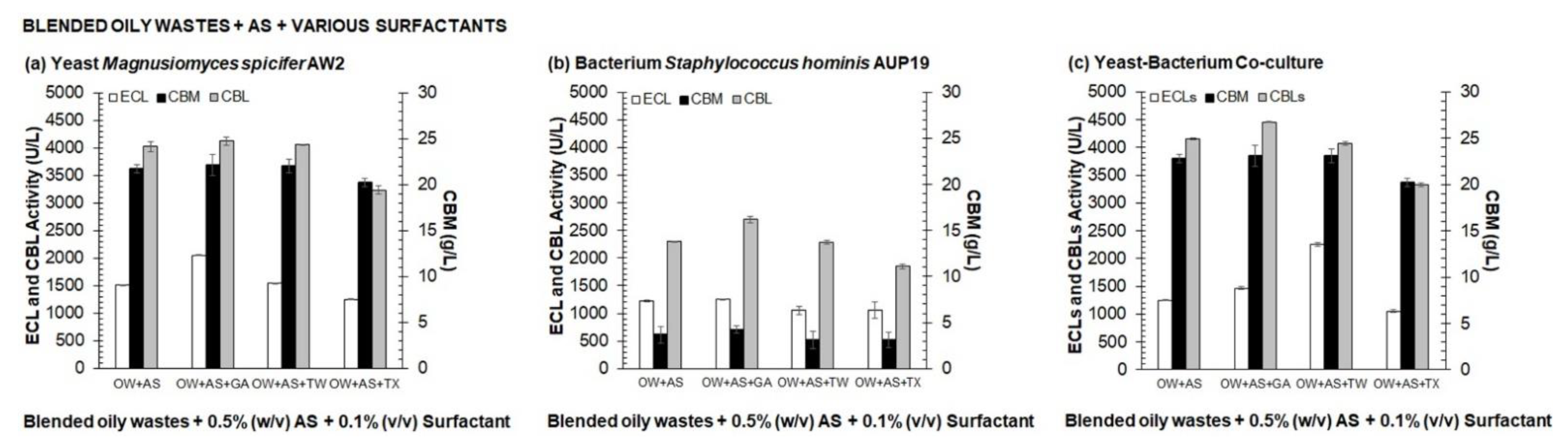


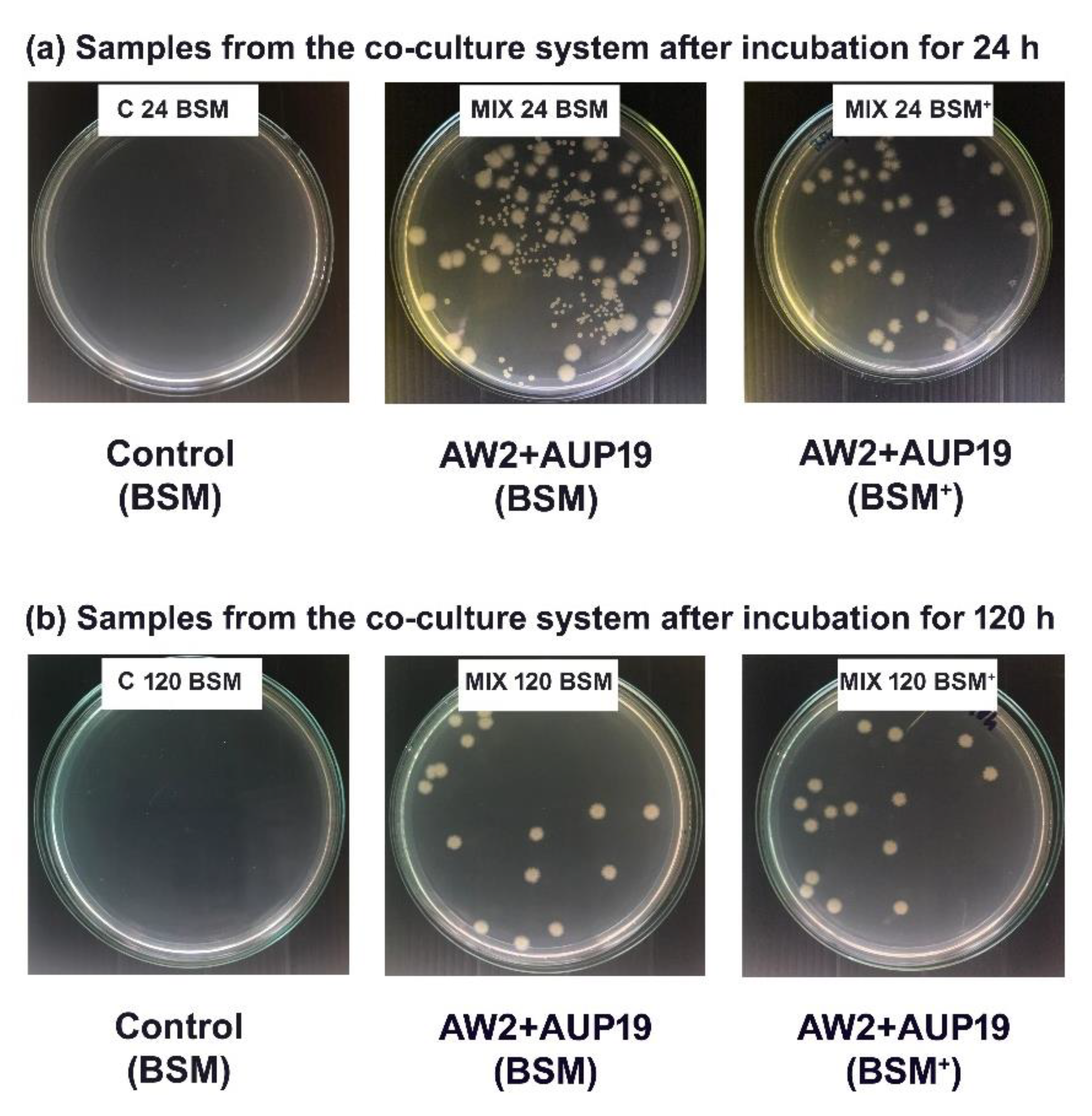
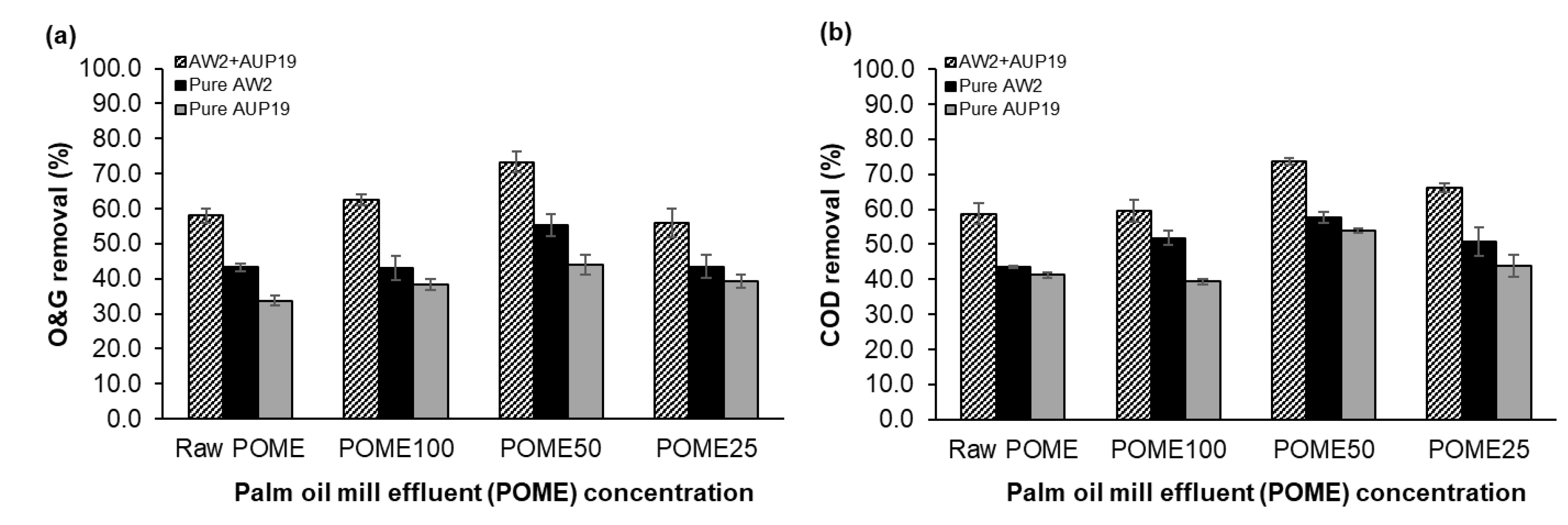
| Strain | POME Medium | Nutrient Supplementation | Product and Applications | Ref. |
|---|---|---|---|---|
| Magnusiomyces spicifer AW2 Staphylococcus hominis AUP19 Magnusiomyces spicifer AW2 + Staphylococcus hominis AUP19 (Co-culture) | 25%, 50%, 100% (v/v) POME | - | Cell-bound lipases through the bioremediation approach for biodiesel synthesis | [5] |
| Yarrowia lipolytica TISTR5151 | Raw POME 50% (v/v) POME | 1 g-N/L (NH4)2SO4 | Cell-bound lipase and lipid for biodiesel synthesis | [6] |
| Candida cylindracea ATCC14830 | POME | 0.45% (w/v) Peptone 0.65% (v/v) Tween-80 | Lipase | [11] |
| Pseudomonas aeruginosa | POME | 0.4% (v/v) Olive oil 0.9% (w/v) Peptone 0.9% (v/v) Tween-80 | Dry extract lipase | [12] |
| Candida antartica lipase B Fusarium heterosporum Bacillus thermocatenulatus | 2.0% (w/v) POME | 2.0% (w/v) Polypeptone 0.5% (w/v) KH2PO4 0.1% (w/v) NaNO3 0.05% (w/v) MgSO4.7H2O | Immobilized whole-cell lipases for biodiesel production | [13] |
| Magnusiomyces spicifer AW2 + Staphylococcus hominis AUP19 (Co-culture) | 50% (v/v) POME | 2.08% (v/v) Waste frying oil 1.72.0% (w/v) (NH4)2SO4 0.1% (w/v) Gum Arabic | Cell-bound lipases for biodiesel synthesis and bioremediation | This study |
| Run | Factor: A Waste Frying Oil % (v/v) | Factor: B Ammonium Sulfate % (w/v) | Factor: C Gum Arabic % (w/v) | Factor: D Inoculum Size % (v/v) | Factor: E Initial pH | CBLs Activity (U/L) | CBM (g/L) | O&G Removal (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Act. * | Pred. ** | Act. * | Pred. ** | Act. * | Pred. ** | ||||||
| 1 | 4.0 | 0.5 | 0.5 | 20.0 | 4.00 | 3413 | 2879 | 16.6 | 13.7 | 49.5 | 43.3 |
| 2 | 4.0 | 2.0 | 0.1 | 20.0 | 7.00 | 3132 | 3027 | 17.9 | 16.1 | 45.4 | 45.4 |
| 3 | 4.0 | 2.0 | 0.1 | 10.0 | 4.00 | 1558 | 1732 | 6.9 | 8.7 | 27.2 | 27.1 |
| 4 | 2.0 | 0.5 | 0.5 | 10.0 | 7.00 | 4342 | 4098 | 17.8 | 18.2 | 53.9 | 56.2 |
| 5 | 4.0 | 0.5 | 0.1 | 10.0 | 7.00 | 3451 | 3349 | 17.1 | 15.9 | 48.2 | 51.5 |
| 6 | 4.0 | 0.5 | 0.5 | 20.0 | 7.00 | 3299 | 3822 | 16.9 | 19.9 | 54.4 | 56.5 |
| 7 | 2.0 | 0.5 | 0.1 | 20.0 | 4.00 | 2908 | 3389 | 7.9 | 10.4 | 49.7 | 48.4 |
| 8 | 2.0 | 2.0 | 0.5 | 10.0 | 7.00 | 3172 | 3424 | 17.9 | 17.2 | 51.1 | 45.0 |
| 9 | 4.0 | 2.0 | 0.5 | 10.0 | 4.00 | 1807 | 1852 | 10.3 | 11.5 | 26.0 | 26.9 |
| 10 | 2.0 | 2.0 | 0.1 | 20.0 | 7.00 | 3981 | 3657 | 15.3 | 15.6 | 51.9 | 50.3 |
| 11 | 2.0 | 0.5 | 0.1 | 10.0 | 4.00 | 3160 | 3036 | 10.8 | 9.2 | 43.5 | 43.2 |
| 12 | 2.0 | 2.0 | 0.5 | 20.0 | 4.00 | 2876 | 2835 | 13.2 | 12.2 | 30.0 | 37.0 |
| Variable | Unit | Level Code | ||||
|---|---|---|---|---|---|---|
| −1.68 | −1 | 0 | 1 | +1.68 | ||
| Waste Frying Oil | % (v/v) | 1.32 | 2.00 | 3.00 | 4.00 | 4.68 |
| (NH4)2SO4 | % (w/v) | 0 | 0.5 | 1.25 | 2.0 | 2.51 |
| Initial pH | - | 2.98 | 4.00 | 5.50 | 7.00 | 8.02 |
| Run | Factor: A Waste Frying Oil % (v/v) | Factor: B Ammonium Sulfate % (w/v) | Factor: C Initial pH | CBLs Activity (U/L) | CBM (g/L) | O&G removal (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Act. * | Pred. ** | Act. * | Pred. ** | Act. * | Pred. ** | ||||
| 1 | 4.00 | 2.00 | 7.00 | 4293 | 4476 | 21.6 | 22.2 | 57.7 | 57.8 |
| 2 | 3.00 | 1.25 | 5.50 | 4375 | 4399 | 18.3 | 20.4 | 52.1 | 54.7 |
| 3 | 4.00 | 2.00 | 4.00 | 2885 | 2951 | 9.9 | 11.8 | 47.6 | 49.2 |
| 4 | 3.00 | 1.25 | 2.98 | 1705 | 1716 | 5.2 | 3.7 | 43.9 | 42.4 |
| 5 | 2.00 | 0.50 | 4.00 | 2618 | 2554 | 7.2 | 7.9 | 42.6 | 44.3 |
| 6 | 3.00 | 1.25 | 5.50 | 4245 | 4399 | 22.5 | 20.4 | 55.3 | 54.7 |
| 7 | 4.00 | 0.50 | 4.00 | 3504 | 3269 | 15.0 | 13.6 | 50.8 | 50.0 |
| 8 | 3.00 | 1.25 | 5.50 | 4088 | 4399 | 19.9 | 20.4 | 55.6 | 54.7 |
| 9 | 3.00 | 1.25 | 5.50 | 4474 | 4399 | 22.1 | 20.4 | 56.1 | 54.7 |
| 10 | 3.00 | 1.25 | 5.50 | 4314 | 4399 | 18.7 | 20.4 | 54.7 | 54.7 |
| 11 | 4.00 | 0.50 | 7.00 | 3682 | 3492 | 16.8 | 15.7 | 51.6 | 51.8 |
| 12 | 1.32 | 1.25 | 5.50 | 4404 | 4074 | 22.3 | 19.9 | 56.0 | 53.2 |
| 13 | 3.00 | 1.25 | 5.50 | 4766 | 4399 | 20.4 | 20.4 | 53.9 | 54.1 |
| 14 | 3.00 | 2.51 | 5.50 | 4426 | 3941 | 21.3 | 17.3 | 56.0 | 53.2 |
| 15 | 2.00 | 2.00 | 7.00 | 4663 | 5016 | 23.6 | 26.4 | 58.2 | 60.9 |
| 16 | 3.00 | 1.25 | 8.02 | 4296 | 4117 | 21.0 | 20.7 | 58.9 | 57.9 |
| 17 | 4.68 | 1.25 | 5.50 | 4059 | 4221 | 20.6 | 21.2 | 55.1 | 55.3 |
| 18 | 3.00 | 0.00 | 5.50 | 2941 | 3257 | 9.4 | 11.5 | 48.2 | 48.3 |
| 19 | 2.00 | 2.00 | 4.00 | 2074 | 2383 | 5.9 | 8.3 | 42.7 | 44.3 |
| 20 | 2.00 | 0.50 | 7.00 | 3832 | 3885 | 18.3 | 17.6 | 53.9 | 54.7 |
| Factor: A Waste Frying Oil % (v/v) | Factor: B Ammonium Sulfate % (w/v) | Factor: C Initial pH | Responses a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CBLs Activity (U/L) | CBM (g/L) | O&G Removal (%) | |||||||||
| Act. * | Pred. ** | Error (%) b | Act. * | Pred. ** | Error (%) b | Act. * | Pred. ** | Error (%) b | |||
| 2.08 | 1.72 | 7.00 | 4708.9 | 4954.9 | 4.9 | 23.4 | 25.6 | 4.7 | 58.3 | 59.8 | 2.5 |
| Reaction Time (h) | FAME (%) | |||
|---|---|---|---|---|
| Esterification | Transesterification | |||
| CBLs from Optimized Medium | CBLs from Non-Optimized Medium * | CBLs from Optimized Medium | CBLs from Non-Optimized Medium * | |
| 24 | 35.5 ± 0.5 A,a | 35.6 ± 0.3 A,a | 58.4 ± 0.5 A,a | 57.0 ± 2.4 A,a |
| 48 | 76.1 ± 0.2 B,b | 58.2 ± 1.5 A,b | 65.5 ± 1.5 A,b | 64.9 ± 2.3 A,b |
| 72 | 76.0 ± 0.1 B,b | 73.5 ± 3.3 A,c | 85.1 ± 0.9 B,c | 82.5 ± 0.3 A,c |
| 96 | 76.2 ± 0.1 B,b | 73.4 ± 1.2 A,c | 87.7 ± 0.5 B,d | 84.1 ± 0.1 A,d |
| Microorganisms | Wastewater | Modes of Interactions | % Removal | Ref. |
|---|---|---|---|---|
| Magnusiomyces spicifer AW2 + Staphylococcus hominis AUP19 (Starter cultures added into POME) | POME and diluted POME with pH adjustment to 7.0 | Yeast–bacterium A synergic effect of the microbial interactions | 80.1% O&G removal, 75.9% COD removal after 72 h | [5] |
| Candida palmioleophila Bacillus sp. | POME | Yeast–bacterium A synergic effect of the microbial interactions | 84% O&G removal after 48 h | [26] |
| Micrococcus species Bacillus species Pseudomonas species Staphylococcus aureus Aspergillus niger Aspergillus fumigatus Candida species Fusarium species Mucor species Penicillium species | Raw POME | Bacteria-Fungi Mutual interactions for POME degradation | 40% COD removal after 180 h | [55] |
| Micrococcus luteus 101PB Stenotrophomonas maltophilia 102PB Bacillus cereus 103PB Providencia vermicola 104PB Klebsiella pneumonia 105PB Bacillus subtilis 106PB Aspergillus fumigatus 107PF Aspergillus nomius 108PF Aspergillus niger 109PF Meyerozyma guilliermondii 110PF | POME | Bacteria-Fungi Mutual interactions in biodegradation | 90.2.0% BOD removal, 91.1% COD removal, and 92.2.0% TSS removal | [56] |
| Pseudomonas fluorescens Flavobacterium sp. Micrococcus sp. Bacillus subtilis Aspergillus niger Aspergillus tamari Aspergillus sp. Penicillium sp. Trichoderma sp. Mucor sp. | Diluted and enriched POME | Bacteria-Fungi Mutual interactions for POME degradation | 90.3% COD removal after 7 days | [57] |
| Magnusiomyces spicifer AW2 + Staphylococcus hominis AUP19 (Cell-bound lipases added into POME) | POME and diluted POME without pH adjustment | Yeast–bacterium A synergic effect of the microbial interactions | 73.3% O&G removal, 73.6% COD removal after 72 h | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fibriana, F.; Upaichit, A.; Cheirsilp, B. Statistical Optimization for Cost-Effective Production of Yeast-Bacterium Cell-Bound Lipases Using Blended Oily Wastes and Their Potential Applications in Biodiesel Synthesis and Wastewater Bioremediation. Fermentation 2022, 8, 411. https://doi.org/10.3390/fermentation8080411
Fibriana F, Upaichit A, Cheirsilp B. Statistical Optimization for Cost-Effective Production of Yeast-Bacterium Cell-Bound Lipases Using Blended Oily Wastes and Their Potential Applications in Biodiesel Synthesis and Wastewater Bioremediation. Fermentation. 2022; 8(8):411. https://doi.org/10.3390/fermentation8080411
Chicago/Turabian StyleFibriana, Fidia, Apichat Upaichit, and Benjamas Cheirsilp. 2022. "Statistical Optimization for Cost-Effective Production of Yeast-Bacterium Cell-Bound Lipases Using Blended Oily Wastes and Their Potential Applications in Biodiesel Synthesis and Wastewater Bioremediation" Fermentation 8, no. 8: 411. https://doi.org/10.3390/fermentation8080411
APA StyleFibriana, F., Upaichit, A., & Cheirsilp, B. (2022). Statistical Optimization for Cost-Effective Production of Yeast-Bacterium Cell-Bound Lipases Using Blended Oily Wastes and Their Potential Applications in Biodiesel Synthesis and Wastewater Bioremediation. Fermentation, 8(8), 411. https://doi.org/10.3390/fermentation8080411






