Mixed Fermentation of Lactiplantibacillus plantarum and Bacillus licheniformis Changed the Chemical Composition, Bacterial Community, and Rumen Degradation Rate of Tea Residue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Tea Residue and Fermentation
2.2. Chemical Composition Analysis
2.3. Bacterial Community Analysis
2.3.1. DNA Extraction and Sequencing
2.3.2. Sequence Analysis
2.4. Animals and Experimental Design
2.4.1. Ethics Statement
2.4.2. Animal Management
2.4.3. In Situ Nutrient Degradability
2.4.4. Rumen Degradation Rate Analysis
2.5. Statistical Analysis
3. Results
3.1. Chemical Composition of Tea Residue Mixture during Fermentation
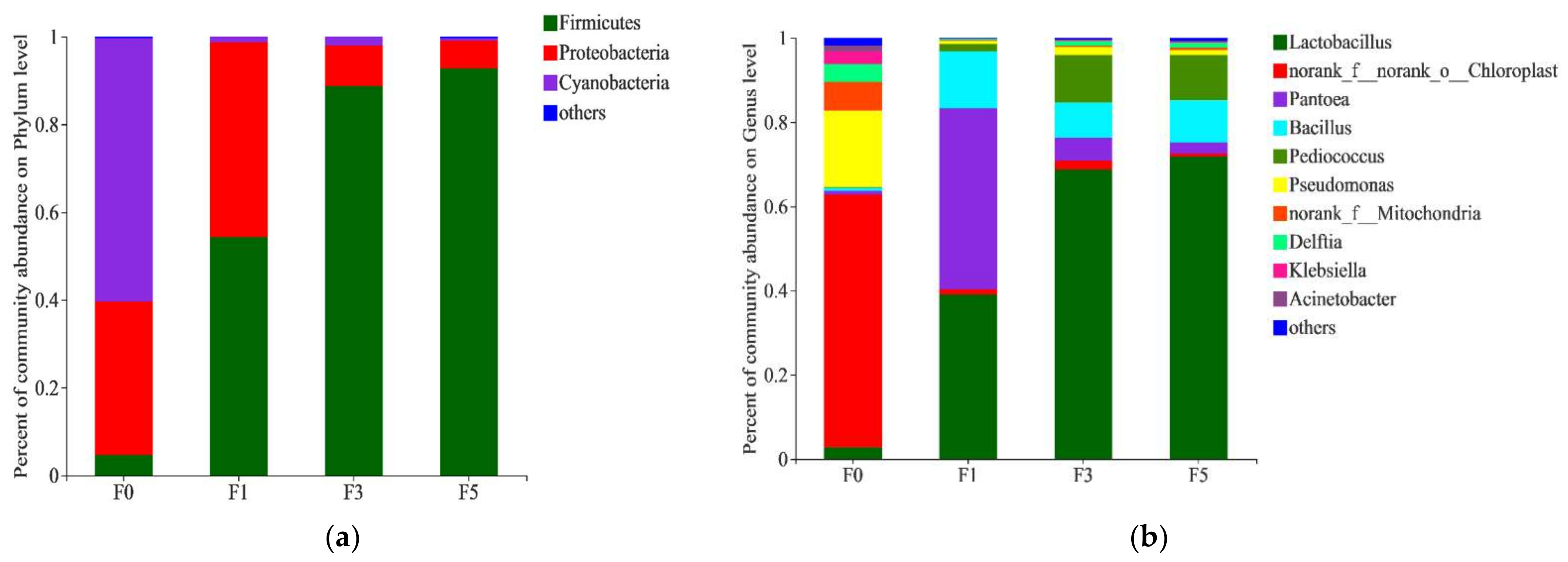
3.2. Bacterial Community of Tea Residue Mixture during Fermentation
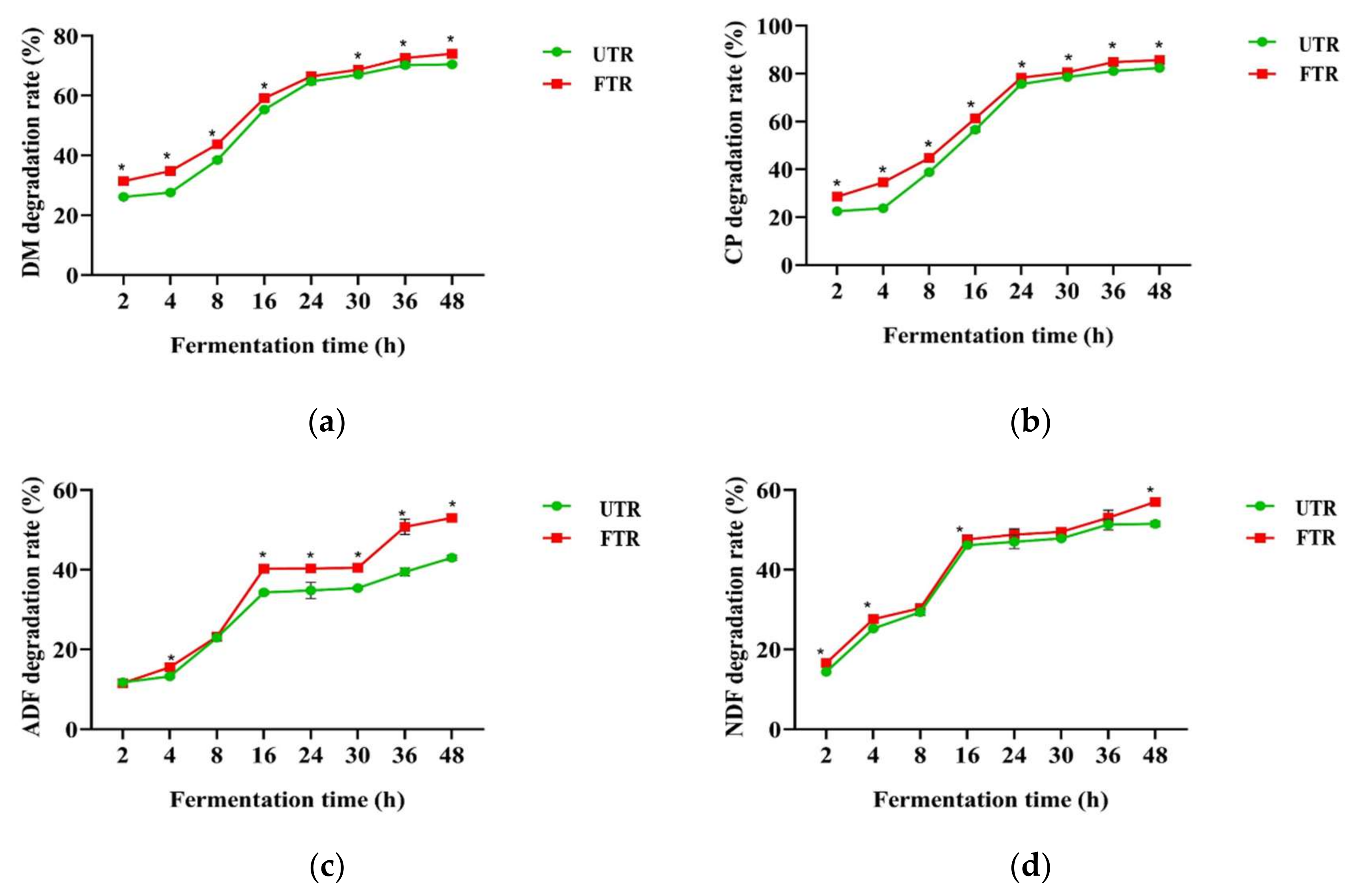
3.3. Rumen Degradation Rates of Unfermented and Fermented Tea Residue Mixtures
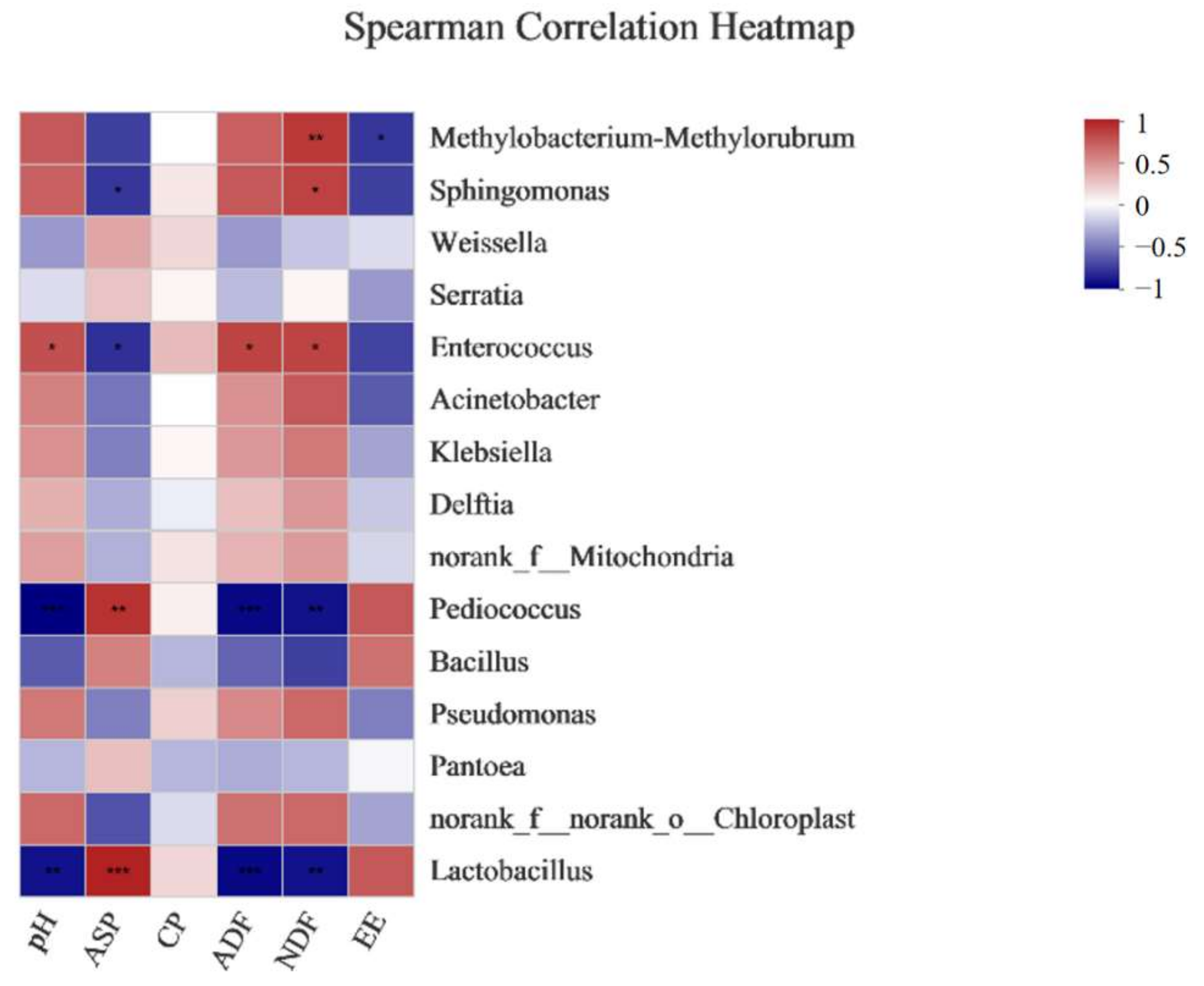
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations, Statistics Division. World Tea Production in 2020. Available online: http://www.fao.org/faostat/en/#data/QCL (accessed on 30 January 2022).
- Statistical Bureau of the People’s Republic of China. China Statistical Yearbook; China Statistics Press: Beijing, China, 2021.
- Ding, X.; Li, H.; Wen, Z.; Hou, Y.; Wang, G.; Fan, J.; Qian, L. Effects of fermented tea residue on fattening performance, meat quality, digestive performance, serum antioxidant capacity, and intestinal morphology in fatteners. Animals 2020, 10, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Guo, X.; Zhao, T.; Li, T. Green waste composting with bean dregs, tea residue, and biochar: Effects on organic matter degradation, humification and compost maturity. Environ. Technol. 2021, 24, 101887. [Google Scholar] [CrossRef]
- Kondo, M.; Kita, K.; Yokota, H.O. Effects of tea leaf waste of green tea, oolong tea, and black tea addition on sudangrass silage quality and in vitro gas production. J. Sci. Food. Agric. 2004, 84, 721–727. [Google Scholar] [CrossRef]
- Ramdani, D.; Chaudhry, A.S.; Seal, C.J. Chemical composition, plant secondary metabolites, and minerals of green and black teas and the effect of different tea-to-water ratios during their extraction on the composition of their spent leaves as potential additives for ruminants. J. Agric. Food Chem. 2013, 61, 4961–4967. [Google Scholar] [CrossRef]
- Basumatary, V.; Saikia, R.; Narzari, R.; Bordoloi, N.; Gogoi, L.; Sut, D.; Bhuyan, N.; Kataki, R. Tea factory waste as a feedstock for thermo-chemical conversion to biofuel and biomaterial. Mater. Today 2018, 5, 23413–23422. [Google Scholar] [CrossRef]
- Zheng, X.; Xie, X.; Yu, C.; Zhang, Q.; Wang, Y.; Cong, J.; Liu, J. Unveiling the activating mechanism of tea residue for boosting the biological decolorization performance of refractory dye. Chemosphere 2019, 233, 110–119. [Google Scholar] [CrossRef]
- Li, S.; Cao, X. Extraction of tea polysaccharides (TPS) using anionic reverse micellar system. Sep. Purif. Technol. 2014, 122, 306–314. [Google Scholar] [CrossRef]
- Kondo, M.; Kita, K.; Yokota, H.O. Ensiled or oven-dried green tea by-product as protein feedstuffs: Effects of tannin on nutritive value in goats. Asian Austral. J Anim. 2007, 20, 880–886. [Google Scholar] [CrossRef]
- Nasehi, M.; Torbatinejad, N.M.; Rezaie, M.; Ghoorchi, T. Effects of partial substitution of alfalfa hay with green tea waste on growth performance and in vitro methane emission of fat-tailed lambs. Small Rumin. Res. 2018, 168, 52–59. [Google Scholar] [CrossRef]
- Sakarika, M.; Delmoitié, B.; Ntagia, E.; Chatzigiannidou, I.; Gabet, X.; Ganigué, R.; Rabaey, K. Production of microbial protein from fermented grass. Chem. Eng. J. 2021, 433, 133631. [Google Scholar] [CrossRef]
- Saeed, A.H.; Salam, A.I. Current limitations and challenges with lactic acid bacteria: A review. Pol. J. Food. Nutr. Sci. 2013, 4, 73–87. [Google Scholar]
- Hoover, G.; Steenson, L.R. Bacteriocins of Lactic Acid Bacteria; Academic Press Inc.: New York, NY, USA, 1993. [Google Scholar]
- Gopal, N.; Hill, C.; Ross, P.R.; Beresford, T.P.; Fenelon, M.A.; Cotter, P.D. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 2015, 6, 1418. [Google Scholar] [CrossRef]
- Contesini, F.J.; Melo, R.R.D.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Factories 2020, 19, 1–12. [Google Scholar] [CrossRef]
- Guo, X.S.; Ke, W.C.; Ding, W.R.; Ding, L.M.; Xu, D.M.; Wang, W.W.; Zhang, P.; Yang, F.Y. Profiling of metabolome and bacterial community dynamics in ensiled Medicago sativa inoculated without or with Lactobacillus plantarum or Lactobacillus buchneri. Sci. Rep. 2018, 8, 357. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; He, R.C.; Zhou, X.K.; Gu, Q.C.; Xia, Z.S.; Liang, M.Z.; Zhou, J.H.; Lin, B.; Zou, C.X. Dynamic changes in fermentation profiles and bacterial community composition during sugarcane top silage fermentation: A preliminary study. Bioresour. Technol. 2019, 285, 121315. [Google Scholar] [CrossRef]
- Zhang, L.Y. Feed Analysis and Quality Test Technology, 4th ed.; China Agricultural University: Beijing, China, 2002; pp. 46–47. [Google Scholar]
- Van Soest, P.V.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- GB/T 28715-2012; Determination of Acidic and Neutral Protease Activity of Feed Additives Spectrophotometric Method. China Agriculture Press: Beijing, China, 2012.
- NY/T 912-2020; Determination of Cellulase Activity of Feed Additives Spectrophotometric Method. China Agriculture Press: Beijing, China, 2020.
- NY/T 3801-2020; Determination of Acid-Soluble Protein in Feed Materials. China Agriculture Press: Beijing, China, 2020.
- GB/T 27985-2011; Determination of Tannin in Feeds-Spectrophotometric. China Agriculture Press: Beijing, China, 2011.
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Wang, A.; Wang, Q.; Lee, D.J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. AMBA 2016, 100, 1421–1426. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. AEM 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Sun, G.; Shah, A.M.; Fan, X.; Li, S.; Yu, X. Effects of different growth stages of amaranth silage on the rumen degradation of dairy cows. Animals 2019, 9, 793. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Khan, M.Z.; Liu, Y.; Xiao, J.; Chen, X.; Ji, S.; Cao, Z.; Li, S. Analysis of nutrient composition, rumen degradation characteristics, and feeding value of Chinese rye grass, barley grass, and naked oat straw. Animals 2021, 11, 2486. [Google Scholar] [CrossRef]
- Nasehi, M.; Torbatinejad, N.M.; Rezaie, M.; Ghoorchi, T. The effect of green tea waste extract on ruminal degradability and intestinal digestibility of barley grain. Turk. J. Vet. Anim. Sci. 2018, 42, 624–632. [Google Scholar] [CrossRef]
- Ørskov, E.R.; McDonald, I. The estimation of protein degradability in the rumen from incubation measurements weighted according to rate of passage. J. Agric. Sci. 1979, 92, 499–503. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.L.; Wei, Y.H.; Zhao, G.Q. Comparative study on rumen degradation rates and small intestinal digestibilities of nutrients of different residue feeds of dairy cows. J. Nutr. 2022, 34, 1592–1603. [Google Scholar]
- Borreani, G.; Tabacco, E.; Schmidt, R.J.; Holmes, B.J.; Muck, R.E. Silage review: Factors affecting dry matter and quality losses in silages. J. Dairy Sci. 2018, 101, 3952–3979. [Google Scholar] [CrossRef] [Green Version]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with functional properties: An approach to increase safety and shelf-life of fermented foods. BioMed Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef] [Green Version]
- Inca-Torres, A.R.; Urbina-Salazar, A.D.R.; Falcón-García, G.; Carbonero-Aguilar, P.; Morgado, B.R.; Parrado, J.; Bautista, J. Hydrolytic enzymes production by Bacillus licheniformis growth on fermentation media formulated with sewage sludge. J. Biotech Res. 2018, 9, 14–26. [Google Scholar]
- Patra, A.K.; Aschenbach, J.R. Ureases in the gastrointestinal tracts of ruminant and monogastric animals and their implication in urea-N/ammonia metabolism: A review. J. Adv. Res. 2018, 13, 39–50. [Google Scholar] [CrossRef]
- Hao, S.; Yu, Y.; Chen, W.; Wu, J.; Xiao, G.; Xu, Y.; Zou, Y. Effect of anaerobic fermentation of Bacillus licheniformis on physicochemical properties of litchi pomace. Feed. Res. 2019, 42, 5. [Google Scholar]
- Van Soest, P.J. Fibre and physicochemical properties of feeds. In Nutritional Ecology of the Ruminant, 2nd ed.; Thomas, L.P., Ed.; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Shi, B.; He, W. Increasing Fat and Acid-Soluble Protein Content in Wheat Bran, by Inoculating Bacteria Liquid Containing Bacillus Subtilis with Sterile Water and Inorganic Salt Nutrient Solution into Wheat Bran, Mixing, and Performing Sealed Fermentation: China, CN112021451-A. 2020. Available online: http://epub.cnipa.gov.cn/ (accessed on 30 January 2022).
- Xu, D.; Ding, Z.; Wang, M.; Bai, J.; Ke, W.; Zhang, Y.; Guo, X. Characterization of the microbial community, metabolome and biotransformation of phenolic compounds of sainfoin (Onobrychis viciifolia) silage ensiled with or without inoculation of Lactobacillus plantarum. Bioresour. Technol. 2020, 316, 123910. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Bolsen, K.K.; Brent, B.E.; Hart, R.A.; Dickerson, J.T.; Feyerherm, A.M.; Aimutis, W.R. Epiphytic microflora on alfalfa and whole-plant corn. J. Dairy Sci. 1992, 75, 2484–2493. [Google Scholar] [CrossRef]
- Li, J.; Wu, J.; Xu, N.; Yu, Y.; Brake, J.; Xu, R.; Wu, X. Dynamic evolution and correlation between microorganisms and metabolites during manufacturing process and storage of Pu-erh tea. LWT 2022, 158, 113128. [Google Scholar] [CrossRef]
- Sun, W.T.; Huang, Y.; Wu, C.R.; Peng, C.; Zheng, Y.L.; Chen, C.; Hao, J. Addition of lactic acid bacteria can promote the quality and feeding value of Broussonetia papyrifera (Paper Mulberry) Silage. Fermentation 2022, 8, 25. [Google Scholar] [CrossRef]
- Xu, Y.; Aung, M.; Sun, Z.; Zhou, Y.; Cheng, Y.; Hao, L.; Padmakumar, V.; Zhu, W. Bio-fermentation improved rumen fermentation and decreased methane concentration of rice straw by altering the particle-attached microbial community. Fermentation 2022, 8, 72. [Google Scholar] [CrossRef]
- Pitta, D.W.; Pinchak, W.E.; Indugu, N.; Vecchiarelli, B.; Sinha, R.; Fulford, J.D. Metagenomic analysis of the rumen microbiome of steers with wheat-induced frothy bloat. Front. Microbiol. 2016, 7, 689. [Google Scholar] [CrossRef] [Green Version]
- Tong, W.; Yu, J.; Wu, Q.; Hu, L.; Tabys, D.; Wang, Y.; Wei, C.; Ling, T.; Inayat, M.; Bennetzen, J.L. Black tea quality is highly affected during processing by its leaf surface microbiome. J. Agric. Food Chem. 2021, 69, 7115–7126. [Google Scholar] [CrossRef]
- Luo, C.; Wang, D.; Lu, N.; Li, H.; Liu, G.; Cao, Z.; Yang, H.; Li, S.; Yu, X.; Shao, W.; et al. Analysis of chemical composition, amino acid content, and rumen degradation characteristics of six organic feeds. Animals 2022, 12, 682. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Q.; Li, W.; Wang, Y.; Zhang, F.; Lv, L.; Li, S.; Yang, H. High-gossypol whole cottonseed exhibited mediocre rumen degradability and less microbial fermentation efficiency than cottonseed hull and cottonseed meal with an in vitro gas production technique. Fermentation 2022, 8, 103. [Google Scholar] [CrossRef]
- Yu, P.; Christensen, D.A.; McKinnon, J.J. In situ rumen degradation kinetics of timothy and alfalfa as affected by culti-var and stage of maturity. Can. J. Anim. Sci. 2004, 84, 255–263. [Google Scholar] [CrossRef]
- Han, L.Y.; Li, J.; Na, R.S.; Yu, Z.; Zhou, H. Effect of two additives on the fermentation, in vitro digestibility and aerobic security of Sorghum-sudan grass hybrid silages. Grass Forage Sci. 2015, 70, 185–194. [Google Scholar] [CrossRef]
- Li, M.; Zi, X.; Zhou, H.; Lv, R.; Tang, J.; Cai, Y. Silage fermentation and ruminal degradation of cassava foliage prepared with microbial additive. AMB Express 2019, 9, 180. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, L.; Ma, G.; Jiang, X.; Yang, J.; Lv, J.; Zhang, Y. Cellulase interacts with lactic acid bacteria to affect fermentation quality, microbial community, and ruminal degradability in mixed silage of soybean residue and corn stover. Animals 2021, 11, 334. [Google Scholar] [CrossRef]
- Li, M.; Zhou, H.; Zi, X.; Cai, Y. Silage fermentation and ruminal degradation of stylo prepared with lactic acid bacteria and cellulase. Anim. Sci. J. 2017, 88, 1531–1537. [Google Scholar] [CrossRef]

| Items | Content (%) |
|---|---|
| Dry matter, DM | 23.12 |
| Crude protein, CP | 22.81 |
| Ether extract, EE | 3.01 |
| Acid detergent fiber, ADF | 25.74 |
| Neutral detergent fiber, NDF | 39.12 |
| Ash | 3.44 |
| Calcium, Ca | 0.62 |
| Phosphorous, P | 0.21 |
| Acid-soluble protein, ASP | 2.35 |
| Tea tannin, TTN | 6.63 |
| Ingredients | Content (% of DM) | Nutrient Levels | Content (% of DM) |
|---|---|---|---|
| Alfalfa hay | 21.0 | NEL (Mcal/kg) 1 | 1.73 |
| Barley silage | 17.5 | Crude protein, CP | 17.60 |
| Corn | 33.6 | Neutral detergent fiber, NDF | 29.17 |
| Soybean meal | 12.8 | Acid detergent fiber, ADF | 17.40 |
| Cottonseed meal | 4.3 | Calcium, Ca | 1.02 |
| Rapeseed meal | 2.5 | Phosphorous, P | 0.45 |
| Cottonseed | 4.5 | ||
| Beet pulp | 2.1 | ||
| Dicalcium phosphate | 0.52 | ||
| Salt | 0.42 | ||
| Sodium bicarbonate | 0.41 | ||
| Vitamin–mineral premix1 | 0.35 |
| Items | F0 (0 day) | F1 (1 day) | F3 (3 day) | F5 (5 day) | p-Value |
|---|---|---|---|---|---|
| Dry matter loss, DM Loss (%) | - | 0.98 ± 0.03 b | 1.08 ± 0.05 b | 1.86 ± 0.01 a | <0.01 |
| pH | 5.91 ± 0.04 a | 4.40 ± 0.04 b | 3.97 ± 0.04 c | 3.93 ± 0.04 c | <0.01 |
| Protease activity, PA (U/g) | 0.15 ± 0.02 d | 328.96 ± 0.98 b | 523.18 ± 2.79 a | 143.41 ± 13.10 c | <0.01 |
| Cellulase activity, CA (U/g) | 0.11 ± 0.01 d | 30.85 ± 0.24 a | 11.78 ± 0.43 b | 6.48 ± 0.23 c | <0.01 |
| Acid-soluble protein, ASP (% DM) | 1.92 ± 0.04 d | 2.48 ± 0.13 c | 3.11 ± 0.03 b | 4.00 ± 0.15 a | <0.01 |
| Crude protein, CP (% DM) | 18.09 ± 0.50 | 18.32 ± 0.15 | 18.32 ± 0.15 | 18.52 ± 0.40 | 0.682 |
| Ether extract, EE (% DM) | 3.59 ± 0.03 c | 3.63 ± 0.13 c | 3.93 ± 0.03 b | 4.38 ± 0.12 a | <0.01 |
| Acid detergent fiber, ADF (% DM) | 23.16 ± 0.54 a | 22.18 ± 0.17 ab | 20.46 ± 0.01 bc | 19.94 ± 1.29 c | 0.030 |
| Neutral detergent fiber, NDF (% DM) | 49.94 ± 1.11 a | 46.53 ± 1.00 b | 45.57 ± 0.79 b | 43.90 ± 0.30 c | <0.01 |
| Items | F0 (0 day) | F1 (1 day) | F3 (3 day) | F5 (5 day) | p-Value |
|---|---|---|---|---|---|
| Chao1 | 153.6 ± 10.58 a | 90.24 ± 22.52 b | 92.73 ± 10.16 b | 107.68 ± 18.32 b | <0.01 |
| Ace | 158.82 ± 12.28 a | 124.04 ± 21.44 b | 100.31 ± 9.69 b | 118.00 ± 20.43 b | 0.016 |
| Shannon | 1.65 ± 0.20 a | 1.27 ± 0.24 b | 1.24 ± 0.05 b | 1.13 ± 0.17 b | 0.034 |
| Simpson | 0.35 ± 0.08 | 0.42 ± 0.15 | 0.46 ± 0.04 | 0.50 ± 0.12 | 0.444 |
| Coverage (%) | 99.94 ± 0.01 | 99.94 ± 0.02 | 99.93 ± 0.02 | 99.93 ± 0.01 | 0.861 |
| Items 2 | a 1 (%) | b (%) | c (%) | a + b (%) | ED (%) | |
|---|---|---|---|---|---|---|
| DM | UTR FTR | 16.05 ± 0.32 b | 58.45 ± 0.68 a | 0.07 ± 0.002 | 74.50 ± 0.60 b | 56.13 ± 0.16 b |
| 23.61 ± 0.32 a | 53.50 ± 0.45 b | 0.06 ± 0.003 | 77.11 ± 0.55 a | 59.80 ± 0.39 a | ||
| CP | UTR FTR | 9.61 ± 0.29 b | 80.05 ± 0.78 a | 0.06 ± 0.002 | 89.66 ± 0.80 b | 62.67 ± 0.05 b |
| 19.13 ± 0.52 a | 73.26 ± 1.00 b | 0.06 ± 0.001 | 92.40 ± 0.48 a | 67.16 ± 0.20 a | ||
| ADF | UTR FTR | 5.60 ± 0.73 | 36.30 ± 0.28 b | 0.08 ± 0.010 a | 41.90 ± 1.01 b | 31.52 ± 0.14 b |
| 6.31 ± 0.63 | 48.45 ± 1.81 a | 0.06 ± 0.003 b | 54.76 ± 1.33 a | 37.67 ± 0.23 a | ||
| NDF | UTR FTR | 6.90 ± 1.24 b | 44.53 ± 0.47 | 0.11 ± 0.010 a | 51.42 ± 1.05 b | 41.41 ± 0.35 b |
| 11.36 ± 1.28 a | 44.27 ± 1.78 | 0.09 ± 0.009 b | 55.63 ± 1.37 a | 43.83 ± 0.46 a | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Xu, Y.; Wu, X.; Ding, Y.; Fan, C.; Xue, Y.; Zhuo, Z.; Cheng, J. Mixed Fermentation of Lactiplantibacillus plantarum and Bacillus licheniformis Changed the Chemical Composition, Bacterial Community, and Rumen Degradation Rate of Tea Residue. Fermentation 2022, 8, 380. https://doi.org/10.3390/fermentation8080380
Huang X, Xu Y, Wu X, Ding Y, Fan C, Xue Y, Zhuo Z, Cheng J. Mixed Fermentation of Lactiplantibacillus plantarum and Bacillus licheniformis Changed the Chemical Composition, Bacterial Community, and Rumen Degradation Rate of Tea Residue. Fermentation. 2022; 8(8):380. https://doi.org/10.3390/fermentation8080380
Chicago/Turabian StyleHuang, Xiaoyun, Yinying Xu, Xinyan Wu, Yuhang Ding, Caiyun Fan, Yanfeng Xue, Zhao Zhuo, and Jianbo Cheng. 2022. "Mixed Fermentation of Lactiplantibacillus plantarum and Bacillus licheniformis Changed the Chemical Composition, Bacterial Community, and Rumen Degradation Rate of Tea Residue" Fermentation 8, no. 8: 380. https://doi.org/10.3390/fermentation8080380
APA StyleHuang, X., Xu, Y., Wu, X., Ding, Y., Fan, C., Xue, Y., Zhuo, Z., & Cheng, J. (2022). Mixed Fermentation of Lactiplantibacillus plantarum and Bacillus licheniformis Changed the Chemical Composition, Bacterial Community, and Rumen Degradation Rate of Tea Residue. Fermentation, 8(8), 380. https://doi.org/10.3390/fermentation8080380







