High-Level Production of Catechol from Glucose by Engineered Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Plasmid Construction
2.3. Strain Construction
2.4. Media and Growth Conditions
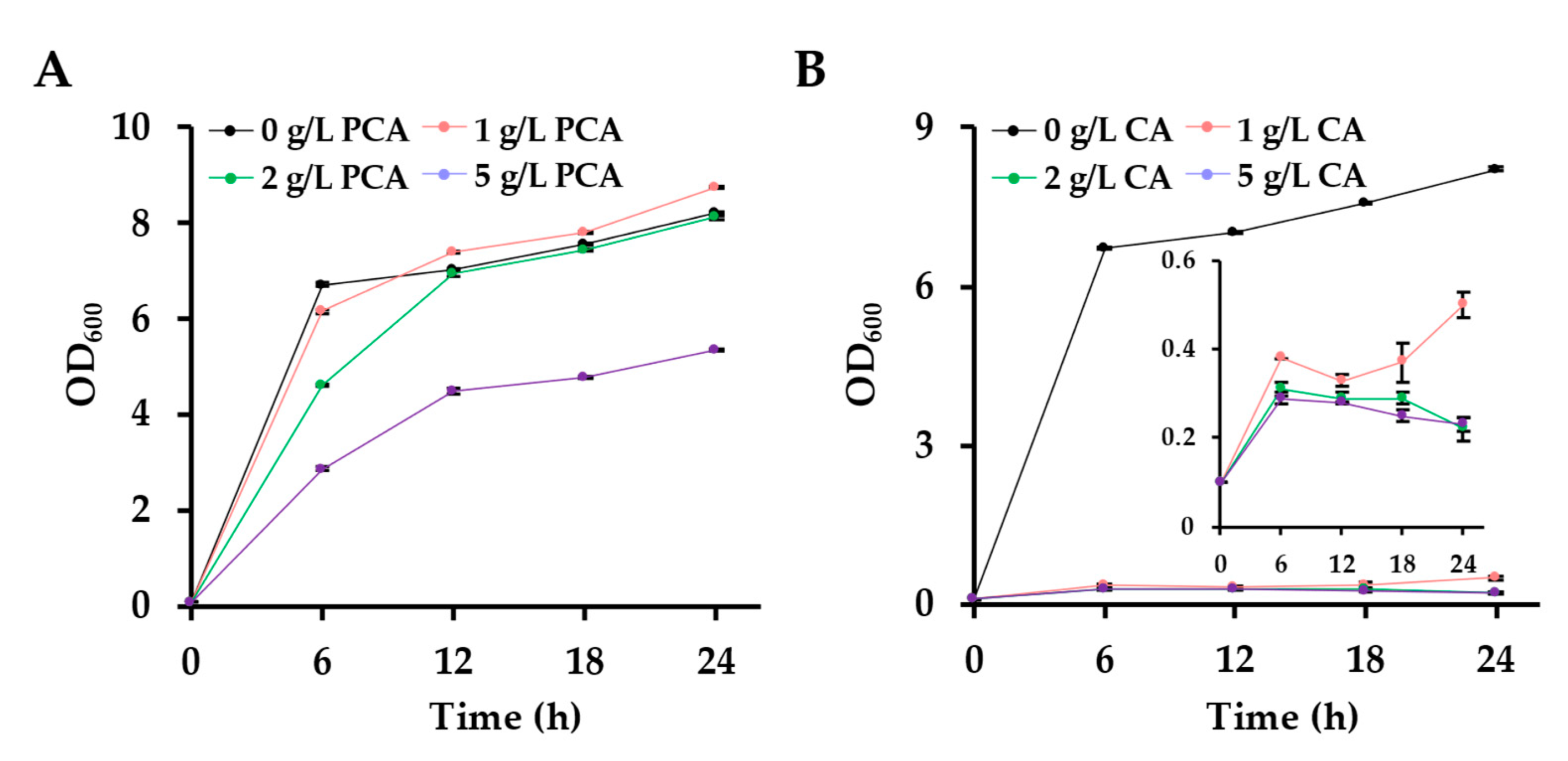
2.5. Tolerance Assays of E. coli DSM 1576 to CA and PCA
2.6. Whole-Cell Biocatalysis Conditions
2.7. Crude-Extract Biocatalysis Conditions
2.8. Analysis of OD600 and CA or PCA Accumulation
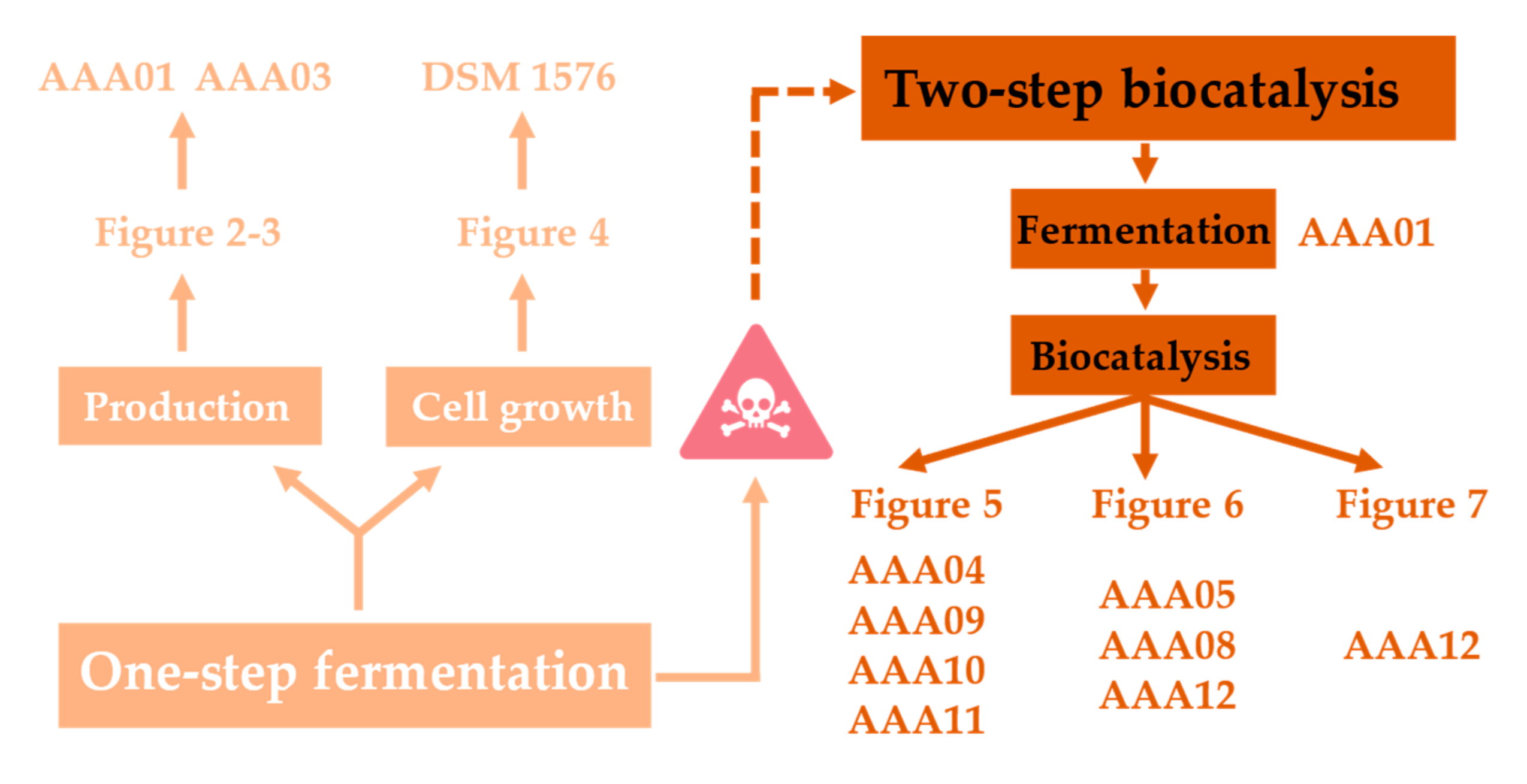
3. Results
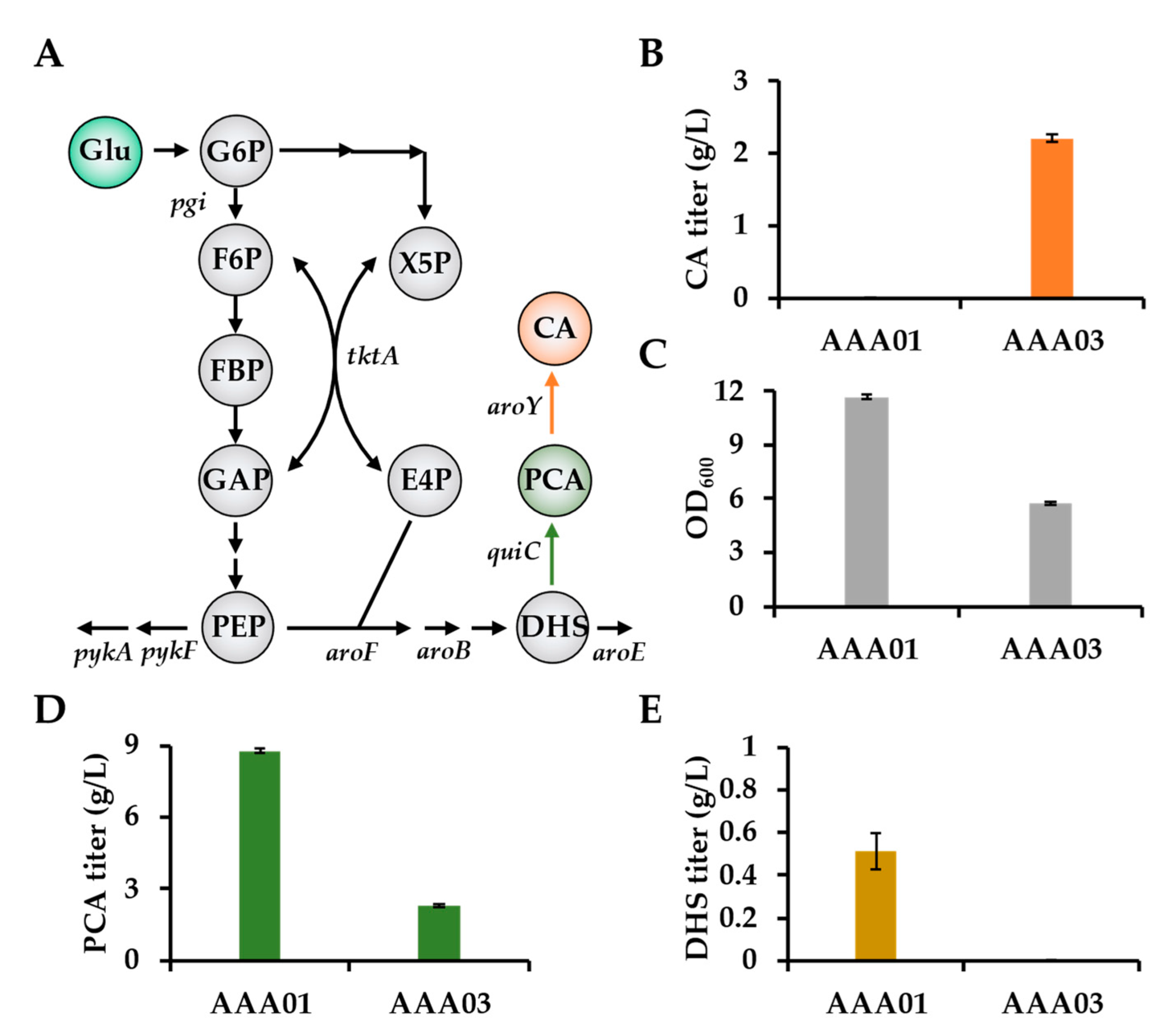
3.1. The Toxicity of CA Significantly Affects the CA Production via One-Step Fermentation
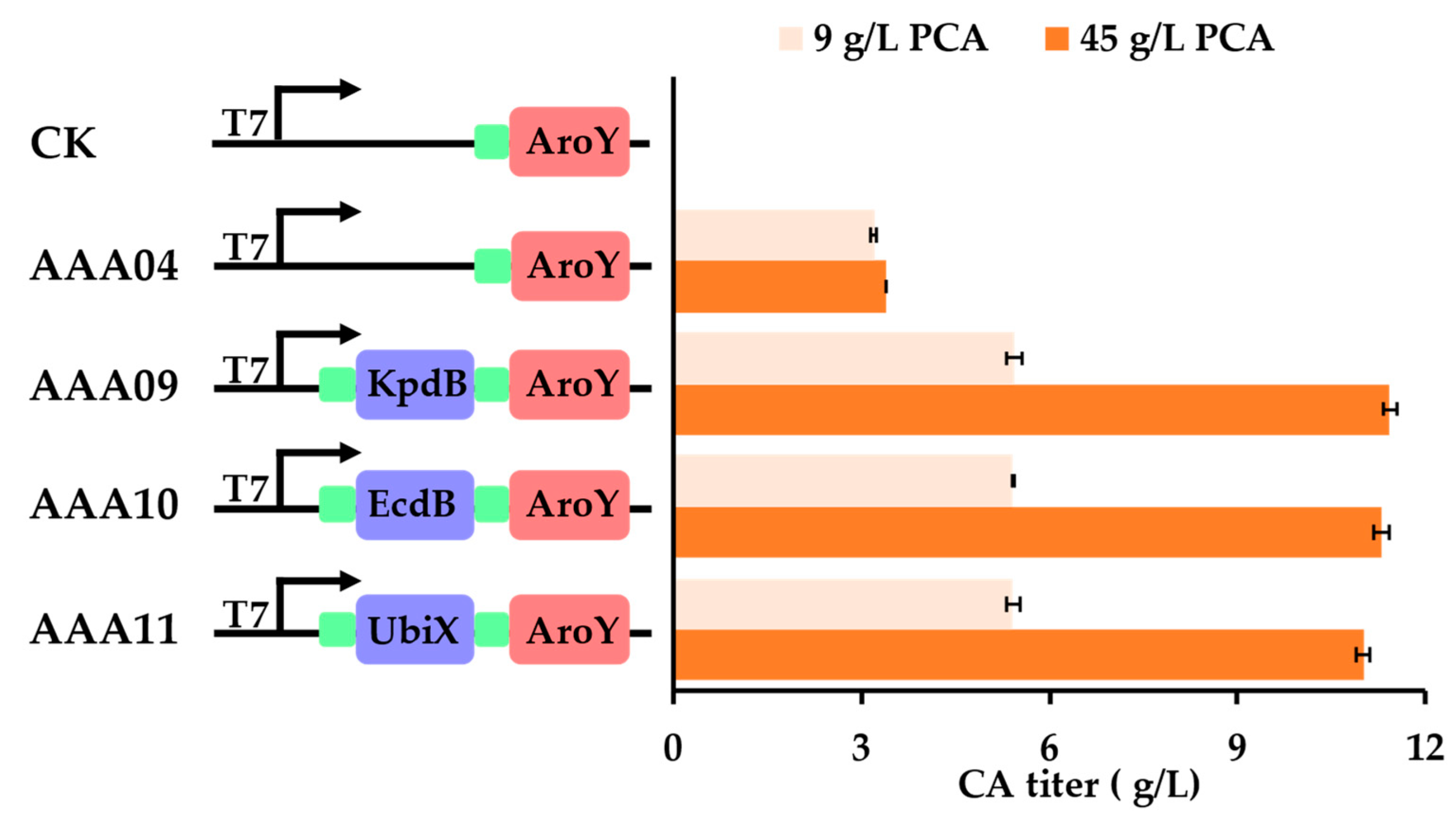
3.2. Introducing Flavin Isoprenyl Transferase Could Efficiently Improve the Conversion of PCA to CA
3.3. Optimization of Whole-Cell Biocatalysis Systems Could Improve the CA Production
3.4. Optimization of Whole-Cell Biocatalysis Reaction Conditions Could Further Improve the CA Production
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pugh, S.; McKenna, R.; Osman, M.; Thompson, B.; Nielsen, D.R. Rational engineering of a novel pathway for producing the aromatic compounds p-hydroxybenzoate, protocatechuate, and catechol in Escherichia coli. Process Biochem. 2014, 49, 1843–1850. [Google Scholar] [CrossRef]
- Balderas-Hernández, V.E.; Treviño-Quintanilla, L.G.; Hernández-Chávez, G.; Martinez, A.; Bolívar, F.; Gosset, G. Catechol biosynthesis from glucose in Escherichia coli anthranilate-overproducer strains by heterologous expression of anthranilate 1,2-dioxygenase from Pseudomonas aeruginosa PAO1. Microb. Cell Factories 2014, 13, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xie, D.; Frost, J.W. Benzene-free synthesis of catechol interfacing microbial and chemical catalysis. J. Am. Chem. Soc. 2005, 127, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Bentley, G.J.; Narayanan, N.; Jha, R.K.; Salvachúa, D.; Elmore, J.R.; Peabody, G.L.; Black, B.A.; Ramirez, K.; De Capite, A.; Michener, W.E.; et al. Engineering glucose metabolism for enhanced muconic acid production in Pseudomonas putida KT2440. Metab. Eng. 2020, 59, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, H.N.; Park, E.; Lee, S.J.; Kim, E.S. Recent advances in microbial production of cis,cis-muconic acid. Biomolecules 2020, 10, 1238. [Google Scholar] [CrossRef] [PubMed]
- Draths, D.K.; Frost, J.W. Environmentally compatible synthesis of catechol from D-glucose. J. Am. Chem. Soc. 1995, 117, 2395–2400. [Google Scholar] [CrossRef]
- Ding, D.; Li, J.; Bai, D.; Fang, H.; Lin, J.; Zhang, D. Biosensor-based monitoring of the central metabolic pathway metabolites. Biosens. Bioelectron. 2020, 167, 112456–112467. [Google Scholar] [CrossRef]
- Liu, X.; Niu, H.; Li, Q.; Gu, P. Metabolic engineering for the production of l-phenylalanine in Escherichia coli. 3 Biotech 2019, 9, 85–92. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Y.; Ding, D.; Wen, J.; Zhu, B.; Zhang, D. Genetic engineering of Escherichia coli to improve L-phenylalanine production. BMC Biotechnol. 2018, 18, 5–17. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, T.; Li, X.; Chen, Y.; Campbell, K.; Nielsen, J.; Chen, Y. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 2019, 10, 4976–4989. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.; Kildegaard, K.R.; Li, M.; Borodina, I.; Nielsen, J. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 2015, 31, 181–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitade, Y.; Hashimoto, R.; Suda, M.; Hiraga, K.; Inui, M. Production of 4-hydroxybenzoic acid by an aerobic growth-arrested bioprocess using metabolically engineered Corynebacterium glutamicum. Appl. Environ. Microbiol. 2018, 84, 2587–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.S.; Seo, S.Y.; Park, S.O.; Lee, H.N.; Song, J.S.; Kim, J.Y.; Park, J.H.; Kim, S.; Lee, S.J.; Chun, G.T.; et al. Cell factory design and culture process optimization for dehydroshikimate biosynthesis in Escherichia coli. Front. Bioeng. Biotechnol. 2019, 7, 241–252. [Google Scholar] [CrossRef]
- Kogure, T.; Suda, M.; Hiraga, K.; Inui, M. Protocatechuate overproduction by Corynebacterium glutamicum via simultaneous engineering of native and heterologous biosynthetic pathways. Metab. Eng. 2021, 65, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Mikola, M.R.; Draths, K.M.; Worden, R.M.; Frost, J.W. Fed-batch fermentor synthesis of 3-dehydroshikimic acid using recombinant Escherichia coli. Biotechnol. Bioeng. 1999, 64, 61–73. [Google Scholar] [CrossRef]
- Li, J.; Ye, B.C. Metabolic engineering of Pseudomonas putida KT2440 for high-yield production of protocatechuic acid. Bioresour. Technol. 2021, 319, 124239–124248. [Google Scholar] [CrossRef]
- Weber, C.; Bruckner, C.; Weinreb, S.; Lehr, C.; Essl, C.; Boles, E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 8421–8430. [Google Scholar] [CrossRef] [Green Version]
- Sonoki, T.; Morooka, M.; Sakamoto, K.; Otsuka, Y.; Nakamura, M.; Jellison, J.; Goodell, B. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J. Biotechnol. 2014, 192, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Payne, K.A.; White, M.D.; Fisher, K.; Khara, B.; Bailey, S.S.; Parker, D.; Rattray, N.J.; Trivedi, D.K.; Goodacre, R.; Beveridge, R.; et al. New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 2015, 522, 497–501. [Google Scholar] [CrossRef] [Green Version]
- White, M.D.; Payne, K.A.; Fisher, K.; Marshall, S.A.; Parker, D.; Rattray, N.J.; Trivedi, D.K.; Goodacre, R.; Rigby, S.E.; Scrutton, N.S.; et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 2015, 522, 502–506. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.W.; Salvachua, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Matson, M.M.; Cepeda, M.M.; Zhang, A.; Case, A.E.; Kavvas, E.S.; Wang, X.; Carroll, A.L.; Tagkopoulos, I.; Atsumi, S. Adaptive laboratory evolution for improved tolerance of isobutyl acetate in Escherichia coli. Metab. Eng. 2022, 69, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, X.; Wu, J.; Mu, S.; Wu, Z.; Jin, J.M.; Tang, S.Y. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab. Eng. 2020, 57, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.; Feng, D.; Xian, M.; Huang, W. De novo biosynthesis and whole-cell catalytic production of 2-acetamidophenol in Escherichia coli. J. Agric. Food Chem. 2022, 70, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Grubbe, W.S.; Rasor, B.J.; Kruger, A.; Jewett, M.C.; Karim, A.S. Cell-free styrene biosynthesis at high titers. Metab. Eng. 2020, 61, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Factories 2017, 16, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Yang, X.; Lin, B.; Huang, J.; Tao, Y. Cofactor self-sufficient whole-cell biocatalysts for the production of 2-phenylethanol. Metab. Eng. 2017, 44, 143–149. [Google Scholar] [CrossRef]
- Lu, J.; Tang, J.; Liu, Y.; Zhu, X.; Zhang, T.; Zhang, X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl. Microbiol. Biotechnol. 2012, 93, 2455–2462. [Google Scholar] [CrossRef]
- Thomason, L.C.; Sawitzke, J.A.; Li, X.; Costantino, N.; Court, D.L. Recombineering: Genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 2014, 106, 1.16.1–1.16.39. [Google Scholar] [CrossRef]
- Weber, H.E.; Gottardi, M.; Brückner, C.; Oreb, M.; Boles, E.; Tripp, J.; Vieille, C. Requirement of a functional flavin mononucleotide prenyltransferase for the activity of a bacterial decarboxylase in a heterologous muconic acid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017, 83, 3472–3488. [Google Scholar] [CrossRef] [Green Version]
- Brückner, C.; Oreb, M.; Kunze, G.; Boles, E.; Tripp, J. An expanded enzyme toolbox for production of cis, cis-muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, 17–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chichirau, A.; Flueraru, M.; Chepelev, L.L.; Wright, J.S.; Willmore, W.G.; Durst, T.; Hussain, H.H.; Charron, M. Mechanism of cytotoxicity of catechols and a naphthalenediol in PC12-AC cells: The connection between extracellular autoxidation and molecular electronic structure. Free Radic. Biol. Med. 2005, 38, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.P.P.; Tabelini, C.H.B.; Ramos, M.D.N.; Aguiar, A. Kinetic evaluation of Bismarck Brown Y Azo dye oxidation by Fenton processes in the presence of aromatic mediators. Water Air Soil Pollut. 2021, 232, 1–16. [Google Scholar] [CrossRef]
- Zhao, Z. Iron and oxidizing species in oxidative stress and Alzheimer’s disease. Aging Med (Milton) 2019, 2, 82–87. [Google Scholar] [CrossRef] [PubMed]



| Name. | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DSM 1576 | Wild type | Lab collection |
| E. coli AAA01 | DSM 1576 M1-12-aroETTG M1-30-aroFfbr ΔtyrR M1-93-tktA ΔptsI M1-12-galP M1-93-glk M1-12-pykFTTG M1-12-pykATTG M1-12-pgiTTG M1-93-aroBopt | Lab collection for PCA production |
| E. coli AAA03 | AAA01 ∆ ptsI :: M1-93 -aroY | This study |
| E. coli AAA04 | BL21 (DE3) with pET30a-T7-aroY | This study |
| E. coli AAA05 | BL21 (DE3) with pET30a-T7-kpdB | This study |
| E. coli AAA06 | BL21 (DE3) with pET30a-T7-ecdB | This study |
| E. coli AAA07 | BL21 (DE3) with pET30a-T7-ubiX | This study |
| E. coli AAA08 | BL21 (DE3) with pET30a-T7-EcaroY | This study |
| E. coli AAA09 | BL21 (DE3) with pET30a-T7-kpdB-RBS-aroY | This study |
| E. coli AAA10 | BL21 (DE3) with pET30a-T7-ecdB-RBS-aroY | This study |
| E. coli AAA11 | BL21 (DE3) with pET30a-T7-ubiX-RBS-aroY | This study |
| E. coli AAA12 | BL21 (DE3) with pET30a-T7-kpdB-RBS-EcaroY | This study |
| Plasmids | ||
| pET30a-T7-kpdB | pET30a expressing kpdB with T7 promoter | This study |
| pET30a-T7-ecdB | pET30a expressing ecdB with T7 promoter | This study |
| pET30a-T7-ubiX | pET30a expressing ubiX with T7 promoter | This study |
| pET30a-T7-aroY | pET30a expressing aroY with T7 promoter | This study |
| pET30a-T7-EcaroY | pET30a expressing EcaroY with T7 promoter | This study |
| pET30a-T7-kpdB-RBS-aroY | pET30a expressing kpdB and aroY with T7 promoter | This study |
| pET30a-T7-ecdB-RBS-aroY | pET30a expressing ecdB and aroY with T7 promoter | This study |
| pET30a-T7-ubiX-RBS-aroY | pET30a expressing ubiX and aroY with T7 promoter | This study |
| pET30a-T7-kpdB-RBS-EcaroY | pET30a expressing kpdB and EcaroY with T7 promoter | This study |
| pET30a-T7-ecdB-RBS-EcaroY | pET30a expressing ecdB and EcaroY with T7 promoter | This study |
| Strain | Carbon Source | Synthetic Method | Time (h) | Titer (g/L) | Reference |
|---|---|---|---|---|---|
| E. coli | Glucose | Resin-based extraction | 36 | 8.50 | [3] |
| E. coli | Glucose | Fed-batch fermentation | 48 | 2.00 | [6] |
| S. cerevisiae | Glucose | Shake-flask fermentation | 120 | 0.58 | [17] |
| E. coli | Glucose | Batch fermentation | 84 | 4.47 | [2] |
| E. coli | PCA | Whole-cell biocatalysis | 3 | 0.08 | [18] |
| E. coli | Glucose | Batch fermentation | 86 | 0.63 | [1] |
| S. cerevisiae | PCA | Whole-cell biocatalysis | 168 | 0.09 | [30] |
| S. cerevisiae | Glucose | Shake-flask fermentation | 96 | 0.62 | [31] |
| E. coli | Glucose | Fed-batch fermentation | 42 | 3.10 | This work |
| E. coli | Glucose | Whole-cell biocatalysis | 3 | 17.70 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, G.; Wu, F.; Peng, Y.; Jiang, X.; Wang, Q. High-Level Production of Catechol from Glucose by Engineered Escherichia coli. Fermentation 2022, 8, 344. https://doi.org/10.3390/fermentation8070344
Song G, Wu F, Peng Y, Jiang X, Wang Q. High-Level Production of Catechol from Glucose by Engineered Escherichia coli. Fermentation. 2022; 8(7):344. https://doi.org/10.3390/fermentation8070344
Chicago/Turabian StyleSong, Guotian, Fengli Wu, Yanfeng Peng, Xiaolong Jiang, and Qinhong Wang. 2022. "High-Level Production of Catechol from Glucose by Engineered Escherichia coli" Fermentation 8, no. 7: 344. https://doi.org/10.3390/fermentation8070344
APA StyleSong, G., Wu, F., Peng, Y., Jiang, X., & Wang, Q. (2022). High-Level Production of Catechol from Glucose by Engineered Escherichia coli. Fermentation, 8(7), 344. https://doi.org/10.3390/fermentation8070344