Engineered Microbial Cell Factories for Sustainable Production of L-Lactic Acid: A Critical Review
Abstract
1. Introduction
2. Synthetic Pathways and Microorganisms for L-LA Production
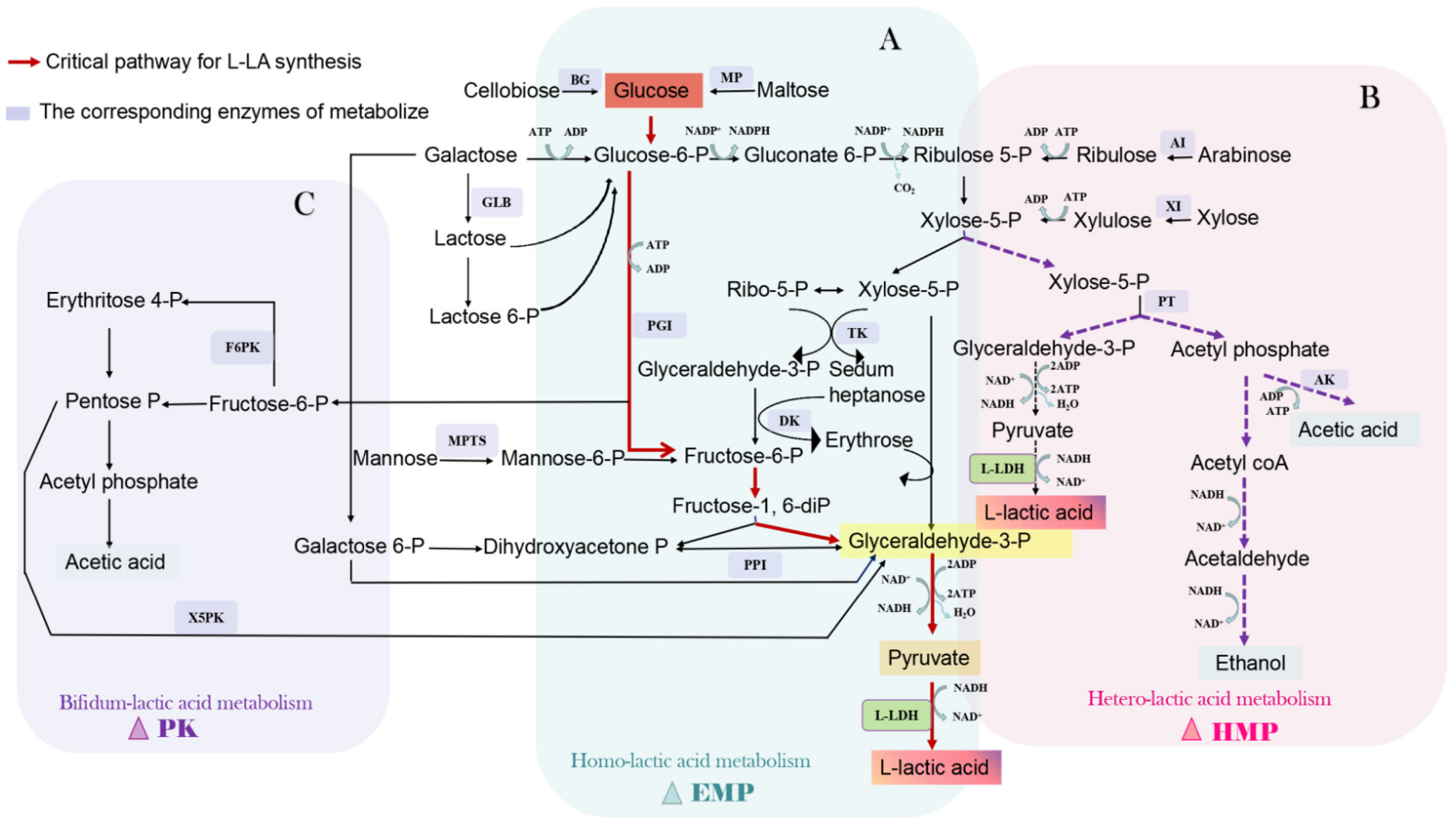
2.1. Biosynthetic Pathway of L-LA
2.1.1. Homolactic Fermentation
2.1.2. Heterolactic Fermentation
2.1.3. Bifidobacterium Fermentation
2.2. Strains for L-LA Production
3. Metabolic Engineering Strategies for Improving L-LA Production
3.1. Mutation Breeding
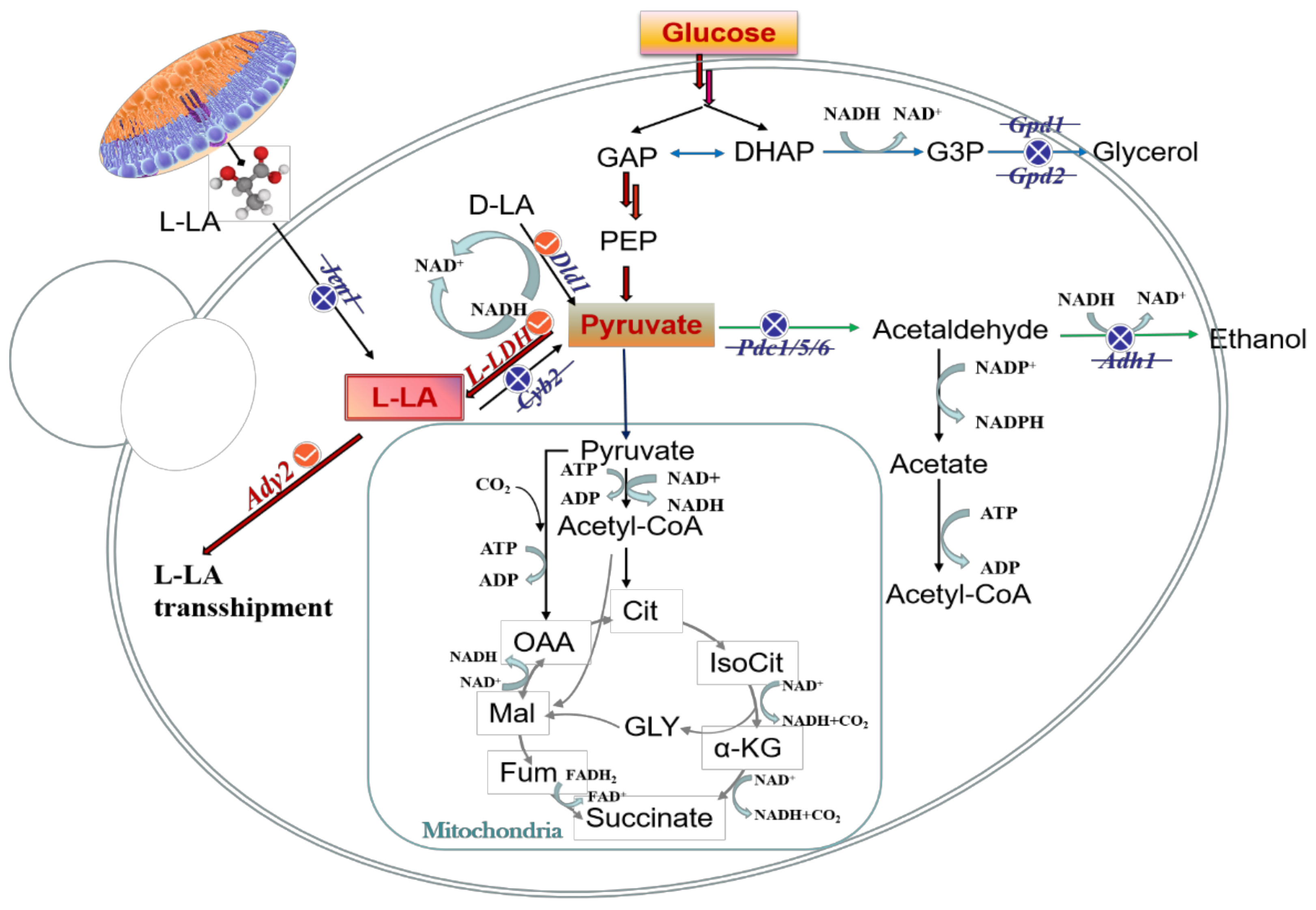
3.2. Strain Improvement by Metabolic Engineering
3.2.1. Expression of Exogenous L-LA Dehydrogenase
3.2.2. Pyr Metabolic Pathway
3.2.3. Cofactor Engineering Strategies
3.2.4. Intracellular and Extracellular Transport of L-LA
3.2.5. Genome Editing Tools
4. Utilization of Raw Materials and Renewable Resources
4.1. Whey
4.2. Molasses
4.3. Starch
4.4. Other Wastes
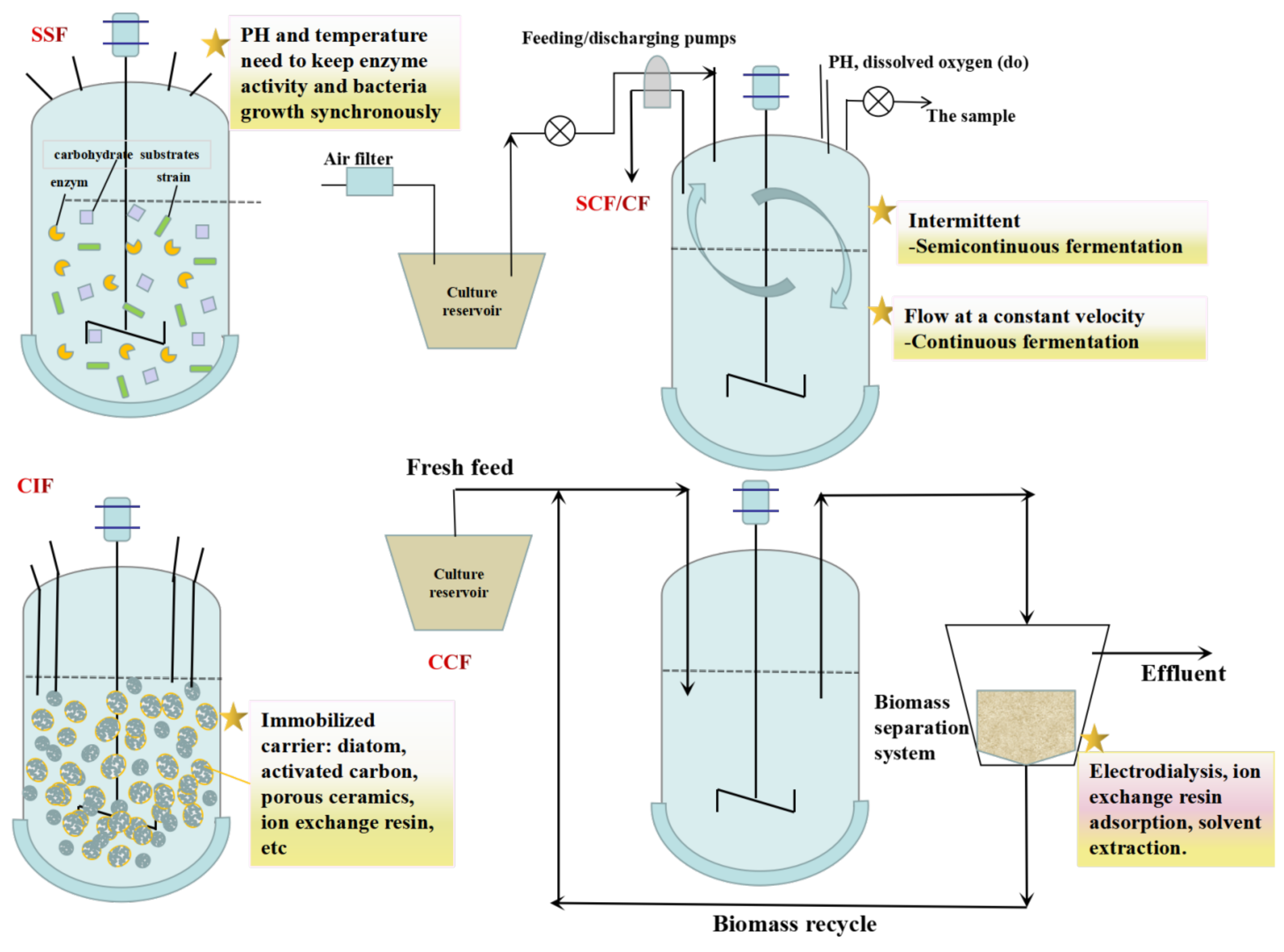
5. Fermentation Modes for L-LA Production
5.1. SSF
5.2. SCF/CF
5.3. CIF
5.4. CCF
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Douglas, R.D.; Arne, N.W. Metabolism of lactic acid in the intact rabbit. Am. J. Physiol. 1956, 184, 304–308. [Google Scholar]
- Nduko, J.M.; Matsumoto, K.; Ooi, T.; Taguchi, S. Enhanced production of poly(lactate-co-3-hydroxybutyrate) from xylose in engineered Escherichia coli overexpressing a galactitol transporter. Appl. Microbiol. Biotechnol. 2014, 98, 2453–2460. [Google Scholar] [CrossRef]
- Younes, M.; Aquilina, G.; Castle, L.; Engel, K.H.; Fowler, P.; Frutos Fernandez, M.J.; Furst, P.; Gurtler, R.; Gundert-Remy, U.; Husoy, T.; et al. Re-evaluation of acetic acid, lactic acid, citric acid, tartaric acid, mono- and diacetyltartaric acid, mixed acetic and tartaric acid esters of mono- and diglycerides of fatty acids (E 472a-f) as food additives. EFSA J. 2020, 18, e06032. [Google Scholar] [PubMed]
- Chen, L.; Song, Z.; Tan, S.Y.; Zhang, H.; Yuk, H.G. Application of bacteriocins produced from lactic acid bacteria for microbiological food safety. Curr. Top. Lact. Acid Bact. Probiotics 2020, 6, 1–8. [Google Scholar] [CrossRef]
- Zhang, G.M.; Wang, X.F.; Tan, H.; Lin, X.U.; Zhao, C. Lactic acid and saturated hydrogen saline could attenuate myocardial apoptosis by mimicking ischemic postconditioning. Chin. J. Evid.-Based Cardiovasc. Med. 2016, 3, 533–538. [Google Scholar]
- Venkatesan, K.B.; Karkhanis, S.S.; Matuana, L. Microcellular foaming of poly(lactic acid) branched with food-grade chain extenders. J. Appl. Polym. Sci. 2021, 138, 121–134. [Google Scholar] [CrossRef]
- Huang, Y.; Dapeng, L.I.; Zuo, H.; Shen, T.; Zou, J.D.L. Effect of plla/β-tcp abosorable plate by internal fixation on fracture healing of dog middle bitia. Clin. Med. Eng. 2011, 18, 15–18. [Google Scholar]
- Maki-Arvela, P.; Aho, A.; Murzin, D.Y. Heterogeneous catalytic synthesis of methyl lactate and lactic acid from sugars and their derivatives. ChemSusChem 2020, 13, 4833–4855. [Google Scholar] [CrossRef] [PubMed]
- Würmel, J.; Somers, K.P.; Simmie, J.M. Selections Reprinted from Mathematical Reviews. Bull. Am. Math. Soc. 2006, 43, 405–414. [Google Scholar]
- Yusoff, N.H.; Pal, K.; Narayanan, T.; de Souza, F.G. Recent trends on bioplastics synthesis and characterizations: Polylactic acid (PLA) incorporated with tapioca starch for packaging applications. J. Mol. Struct. 2021, 1232, 129954. [Google Scholar] [CrossRef]
- Fabio, A.C.M.; Eduardo, M.B.; José, M.S.; José, M.D.G.; Attilio, C.; Ricardo, P.D.S.O. Lactic acid properties, applications and production: A review. Trends Food Sci. Technol. 2013, 30, 70–83. [Google Scholar]
- Wasewar, K.L.; Pangarkar, V.G.; Bert, A.; Heesink, M.; Versteeg, G.F. Intensification of enzymatic conversion of glucose to lactic acid by reactive extraction. Chem. Eng. Sci. 2003, 15, 3385–3393. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Sonomoto, K. Lactic acid production from lignocellulose-derived sugars using lactic acid bacteria: Overview and limits. J. Biotechnol. 2011, 156, 286–301. [Google Scholar] [CrossRef] [PubMed]
- Amini, Z.; Self, R.; Strong, J.; Speight, R.; Harrison, M.D. Valorization of sugarcane biorefinery residues using fungal biocatalysis. Biomass Convers. Biorefin. 2022, 12, 997–1011. [Google Scholar] [CrossRef]
- Chung, M.; Tan, I.S.; Foo, H.; Lam, M.K.; Lim, S. Potential of macroalgae-based biorefinery for lactic acid production from exergy aspect. Biomass Convers. Biorefin. 2021, 1, 1–31. [Google Scholar] [CrossRef]
- Shahri, S.Z.; Vahabzadeh, F.; Mogharei, A. Lactic acid production by loofah-immobilized Rhizopus oryzae through one-step fermentation process using starch substrate. Bioproc. Biosyst. Eng. 2020, 43, 333–345. [Google Scholar] [CrossRef]
- Razi, F.; Yuwono, S.D. Batch and Continuous Lactic Acid Production from Cassava by Streptococcus bovis. J. Rekayasa Kimia Lingkungan 2006, 5, 17–20. [Google Scholar]
- Peng, L.; Wang, L.; Che, C.; Yang, G.; Yu, B.; Ma, Y. Bacillus sp. strain P38: An efficient producer of L-lactate from cellulosic hydrolysate, with high tolerance for 2-furfural. Bioresour. Technol. 2013, 149, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Vuyst, L.D.; Leroy, F. Functional role of yeasts, lactic acid bacteria, and acetic acid bacteria in cocoa fermentation processes. FEMS Microbiol. Rev. 2020, 44, 432–453. [Google Scholar] [CrossRef]
- Ma, X.; Gao, M.; Yin, Z.; Zhu, W.; Wang, Q. Lactic acid and animal feeds production from sophora flavescens residues by Rhizopus oryzae fermentation. Process Biochem. 2020, 92, 401–408. [Google Scholar] [CrossRef]
- Xu, P.; Zhao, B.; Qin, J.Y. A Method for Producing L-lactic Acid and Its Special Lactobacillus rhamnosus. Patent CN/2007/10176057, 7 May 2008. Available online: https://d.wanfangdata.com.cn/patent/CN200710176057.7 (accessed on 23 May 2022).
- Tsuneo, Y.; Ryohsuke, T. Highly accumulative production of L(+)-lactate from glucose by crystallization fermentation with immobilized Rhizopus oryzae. J. Biosci. Bioeng. 2013, 115, 90–95. [Google Scholar]
- PMD Oliveira, S.L.P.C. Production of L(+) Lactic Acid by Lactobacillus casei Ke11: Fed Batch Fermentation Strategies. Fermentation 2021, 7, 151. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Laffend, H.; Jiang, S.L.S. Optimization of immobilized lactobacillus pentosus cell fermentation for lactic acid production. Bioresour. Bioprocess. 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Cubas-Cano, E.; Gonzalez-Fernandez, C.; Tomas-Pejo, E. Evolutionary engineering of Lactobacillus pentosus improves lactic acid productivity from xylose-rich media at low pH. Bioresour. Technol. 2019, 288, 121540. [Google Scholar] [CrossRef] [PubMed]
- Liguori, R.; Soccol, C.R.; Vandenberghe, L.P.; Woiciechowski, A.L.; Ionata, E.; Marcolongo, L.; Faraco, V. Selection of the strain Lactobacillus acidophilus atcc 43121 and its application to brewers’ spent grain conversion into lactic acid. Biomed. Res. Int. 2015, 2015, 240231. [Google Scholar] [CrossRef]
- Kato, H.; Shiwa, Y.; Oshima, K.; Machii, M.; Araya-Kojima, T.; Zendo, T.; Shimizu-Kadota, M.; Hattori, M.; Sonomoto, K.; Yoshikawa, H. Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of L-lactic acid. J. Bacteriol. 2012, 194, 2102–2103. [Google Scholar] [CrossRef]
- Su, J.H. Kinetics study on fermentation of l-lactic acid by lactobacillus thermophilus(T-1). Mod. Food Sci. Technol. 2007, 23, 30–32. [Google Scholar]
- Zhang, L.; Li, X.; Yong, Q.; Yang, S.T.; Ouyang, J.; Yu, S. Impacts of lignocellulose-derived inhibitors on L-lactic acid fermentation by Rhizopus oryzae. Bioresour. Technol. 2016, 203, 173–180. [Google Scholar] [CrossRef]
- Takano, M.; Hoshino, K. Lactic acid production from paper sludge by SSF with thermotolerant Rhizopus sp. Bioresour. Bioproc. 2016, 3, 1–10. [Google Scholar] [CrossRef]
- Kwan, T.H.; Vlysidis, A.; Wu, Z.; Hu, Y.; Koutinas, A.; Lin, C. Lactic acid fermentation modelling of Streptococcus thermophilus YI-B1 and Lactobacillus casei Shirota using food waste derived media. Biochem. Eng. J. 2017, 6, 97–109. [Google Scholar] [CrossRef]
- Regiane, A.; Schneider, R.; Rossell, C.V.; Filho, R.M.V.J. Polymer grade L-lactic acid production from sugarcane bagasse hemicellulosic hydrolysate using Bacillus coagulans. Bioresour. Technol. Rep. 2019, 11, 26–31. [Google Scholar]
- Wang, L.; Cai, Y.; Zhu, L.; Guo, H.; Yu, B. Major role of NAD-dependent lactate dehydrogenases in the production of L-lactic acid with high optical purity by the thermophile Bacillus coagulans. Appl. Environ. Microbiol. 2014, 80, 7134–7141. [Google Scholar] [CrossRef] [PubMed]
- Thomasser, C.; Danner, H.; Neureiter, M.; Saidi, B.; Braun, R. Thermophilic lactic acid production on hemicellulose hydrolysate. Meded. Rijksuniv. Gent. Fak. Landbouwkd. Toegep. Biol. Wet. 2001, 66, 325–328. [Google Scholar] [PubMed]
- Juturu, V.; Wu, J.C. Production of high concentration of l-lactic acid from oil palm empty fruit bunch by thermophilic Bacillus coagulans JI12. Biotechnol. Appl. Biochem. 2018, 65, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Eldarov, M.A.; Kishkovskaia, S.A.; Tanaschuk, T.N.; Mardanov, a.v. Genomics and biochemistry of Saccharomyces cerevisiae wine yeast strains. Biochem. Biokhimiia 2016, 81, 1650–1668. [Google Scholar] [CrossRef] [PubMed]
- Colombie, S.; Sablayrolles, J.M. Nicotinic acid controls lactate production by K1-LDH: A Saccharomyces cerevisiae strain expressing a bacterial LDH gene. J. Ind. Microbiol. Biotechnol. 2004, 31, 209–215. [Google Scholar] [CrossRef]
- Novy, V.; Brunner, B.; Nidetzky, B. L-Lactic acid production from glucose and xylose with engineered strains of Saccharomyces cerevisiae: Aeration and carbon source influence yields and productivities. Microb. Cell Fact. 2018, 17, 59. [Google Scholar] [CrossRef]
- Zhang, L.H.; Chen, X.Z.; Shen, W.; Fan, Y. Progresses in the metabolic engineering of Escherichia coli for lactic acid production. Chin. J. Bioprocess Eng. 2017, 15, 48–56. [Google Scholar]
- Niu, D.; Tian, K.; Prior, B.A.; Wang, M.; Wang, Z.; Lu, F.; Singh, S. Highly efficient L-lactate production using engineered Escherichia coli with dissimilar temperature optima for L-lactate formation and cell growth. Microb. Cell Fact. 2014, 13, 78. [Google Scholar] [CrossRef]
- Le, X.J.; Wang, C.L.; Gu, X.B. Breeding of L-lactic Acid Producing Bacteria. China Food Addit. 2004, 5, 67–71. [Google Scholar]
- Gu, S.B.; Ge, C.M.; Wang, Q.H. Characteristics of L-lactic acid producing strain Rhizopus oryzae PW352 and high productive mutant by low energy ions implantation. Ind. Microbiol. 2004, 34, 12–16. [Google Scholar]
- Xian, M.; Zou, H.B.; Liu, W. The Invention Discloses Lactobacillus casei Producing High Photopurity L-lactic acid and a Fermentation Method. Patent CN/2013/10474231, 12 October 2013. Available online: https://d.wanfangdata.com.cn/patent/CN201310474231.1 (accessed on 23 May 2022).
- Jiang, A.L.; Hu, W.; Li, W.J.; Liu, L.; Tian, X.J.; Liu, J.; Wang, S.Y.; Lu, D.; Chen, J.H. Enhanced production of l-lactic acid by Lactobacillus thermophilus SRZ50 mutant generated by high-linear energy transfer heavy ion mutagenesis. Eng. Life Sci. 2018, 18, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Ishida, N.; Saitoh, S.; Tokuhiro, K.; Nagamori, E.; Matsuyama, T.; Kitamoto, K.; Takahashi, H. Efficient production of L-Lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Appl. Environ. Microb. 2005, 71, 1964–1970. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhang, B.; Hua, Y.; Zhu, Y.; Li, W.; Wang, D.; Hong, J. Efficient L-lactic acid production from corncob residue using metabolically engineered thermo-tolerant yeast. Bioresour. Technol. 2019, 273, 220–230. [Google Scholar] [CrossRef]
- Redden, H.; Alper, H.S. The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 2015, 6, 7810. [Google Scholar] [CrossRef]
- Flagfeldt, D.B.; Siewers, V.; Huang, L.; Nielsen, J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 2009, 26, 545–551. [Google Scholar] [CrossRef]
- Reider, A.A.; D’Espaux, L.; Wehrs, M.; Sachs, D.; Li, R.A.; Tong, G.J.; Garber, M.; Nnadi, O.; Zhuang, W.; Hillson, N.J.; et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 45, 496–508. [Google Scholar] [CrossRef]
- Nobuhiro, I.; Satoshi, S.; Toru, O.; Kenro, T.; Eiji, N.; Katsuhiko, K.; Haruo, T. The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci. Biotechnol. Biochem. 2006, 70, 1148–1153. [Google Scholar]
- Kenro, T.; Nobuhiro, I.; Eiji, N.; Satoshi, S.; Toru, O.; Akihiko, K.; Haruo, T. Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl. Microbiol. Biotechnol. 2009, 82, 883–890. [Google Scholar]
- Mazumdar, S.; Blankschien, M.D.; Clomburg, J.M.; Gonzalez, R. Efficient synthesis of L-lactic acid from glycerol by metabolically engineered Escherichia coli. Microb. Cell Fact. 2013, 12, 7–12. [Google Scholar] [CrossRef]
- Kim, J.W.; Lee, Y.G.; Kim, S.J.; Jin, Y.S.; Seo, J.H. Deletion of glycerol-3-phosphate dehydrogenase genes improved 2,3-butanediol production by reducing glycerol production in pyruvate decarboxylase-deficient Saccharomyces cerevisiae. J. Biotechnol. 2019, 304, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Pontes, R.; Romani, A.; Michelin, M.; Domingues, L.; Teixeira, J.; Nunes, J. L-lactic acid production from multi-supply autohydrolyzed economically unexploited lignocellulosic biomass. Ind. Crop. Prod. 2021, 170, 113775. [Google Scholar] [CrossRef]
- Li, J.; Shi, S.; Wang, Y.; Jiang, Z. Integrated production of optically pure L-lactic acid from paper mill sludge by simultaneous saccharification and co-fermentation (SSCF). Waste Manag. 2021, 129, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.T.; Zhang, Q.; Guo, X.P. Effect of ptsG and mglB genes knock-out on the production of L-lactic acid by Escherichia coli from mixed sugars. China Brew. 2021, 40, 82–86. [Google Scholar]
- Lopez-Gomez, J.P.; Alexandri, M.; Schneider, R.; Latorre-Sanchez, M.; Coll Lozano, C.; Venus, J. Organic fraction of municipal solid waste for the production of L-lactic acid with high optical purity. J. Clean. Prod. 2020, 247, 119165. [Google Scholar] [CrossRef]
- Unban, K.; Khanongnuch, R.; Kanpiengjai, A.; Shetty, K.; Khanongnuch, C. Utilizing gelatinized starchy waste from rice noodle factory as substrate for L(+)-lactic acid production by amylolytic lactic acid bacterium Enterococcus faecium k-1. Appl. Biochem. Biotechnol. 2020, 192, 353–366. [Google Scholar] [CrossRef]
- Bae, J.; Kim, H.; Kim, M.; Sung, B.H.; Jeon, J.; Kim, H.; Jin, Y.; Kweon, D.; Sohn, J. Direct fermentation of Jerusalem artichoke tuber powder for production of L-lactic acid and d-lactic acid by metabolically engineered Kluyveromyces marxianus. J. Biotechnol. 2018, 266, 27–33. [Google Scholar] [CrossRef]
- Okano, K.; Uematsu, G.; Hama, S.; Tanaka, T.; Noda, H.; Kondo, A.; Honda, K. Metabolic engineering of lactobacillus plantarum for direct L-lactic acid production from raw corn starch. Biotechnol. J. 2018, 13, e1700517. [Google Scholar] [CrossRef]
- Lee, J.W.; In, J.H.; Park, J.B.; Shin, J.; Park, J.H.; Sung, B.H.; Kweon, D.H. Co-expression of two heterologous lactate dehydrogenases genes in Kluyveromyces marxianus for L-lactic acid production. J. Biotechnol. 2017, 241, 81–86. [Google Scholar] [CrossRef]
- Novy, V.; Brunner, B.; Muller, G.; Nidetzky, B. Toward “homolactic” fermentation of glucose and xylose by engineered Saccharomyces cerevisiae harboring a kinetically efficient L-lactate dehydrogenase within pdc1-pdc5 deletion background. Biotechnol. Bioeng. 2017, 114, 163–171. [Google Scholar] [CrossRef]
- De Lima, P.B.; Mulder, K.C.; Melo, N.T.; Carvalho, L.S.; Menino, G.S.; Mulinari, E.; de Castro, V.H.; Dos, R.T.; Goldman, G.H.; Magalhaes, B.S.; et al. Novel homologous lactate transporter improves L-lactic acid production from glycerol in recombinant strains of Pichia pastoris. Microb. Cell Fact. 2016, 15, 158. [Google Scholar] [CrossRef]
- Timothy, L.T.; Guo, C.Z.; Eun, J.O.; Vijay, S.; Andrew, A.; Vimal, S.; Christopher, D.S.; Ji, Y.J.; Byung, J.Y.; In, P.; et al. Lactic acid production from cellobiose and xylose by engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 2016, 113, 1075–1083. [Google Scholar]
- Garrett, B.G.; Srinivas, K.; Ahring, B.K. Performance and stability of Amberlite (TM) IRA-67 ion exchange resin for product extraction and pH control during homolactic fermentation of corn stover sugars. Biochem. Eng. J. 2015, 94, 1–8. [Google Scholar] [CrossRef]
- Xu, K.; Xu, P. Efficient production of L-lactic acid using co-feeding strategy based on cane molasses/glucose carbon sources. Bioresour. Technol. 2014, 153, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Y.; Zhang, G.Y. Construction of Commercial Saccharomyces cerevisiae with L-lactic acid Efficient Production Ability. Food Food Res. Dev. 2012, 33, 164–168. [Google Scholar]
- Gao, T.; Wong, Y.; Ng, C.; Ho, K. L-lactic acid production by Bacillus subtilis MUR1. Bioresour. Technol. 2012, 121, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Zhou, J.; Liu, L.; Du, G.; Chen, J. Modification of carbon flux in Sacchromyces cerevisiae to improve L-lactic acid production. Acta Microbiol. Sin. 2011, 51, 50–58. [Google Scholar]
- Zhang, Q.; Zhang, L.; Ding, Z.; Wang, Z.; Shi, G. Metabolic engineering of wild acid-resistant yeast for L-lactic acid production. Chin. J. Biotechnol. 2011, 27, 1024–1031. [Google Scholar]
- Ikushima, S.; Fujii, T.; Kobayashi, O.; Yoshida, S.; Yoshida, A. Genetic engineering of Candida utilis yeast for efficient production of L-lactic acid. Biosci. Biotechnol. Biochem. 2009, 73, 1818–1824. [Google Scholar] [CrossRef]
- Osawa, F.; Fujii, T.; Nishida, T.; Tada, N.; Ohnishi, T.; Kobayashi, O.; Komeda, T.; Yoshida, S. Efficient production of L-lactic acid by Crabtree-negative yeast Candida boidinii. Yeast 2009, 26, 485–496. [Google Scholar] [CrossRef]
- Saitoh, S.; Ishida, N.; Onishi, T.; Tokuhiro, K.; Nagamori, E.; Kitamoto, K.; Takahashi, H. Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl. Environ. Microb. 2005, 71, 2789–2792. [Google Scholar] [CrossRef]
- Hou, J.; Suo, F.; Wang, C.; Li, X.; Shen, Y.; Bao, X. Fine-tuning of NADH oxidase decreases byproduct accumulation in respiration deficient xylose metabolic Saccharomyces cerevisiae. BMC Biotechnol. 2014, 14, 13. [Google Scholar] [CrossRef]
- Heux, S.; Cachon, R.; Dequin, S. Cofactor engineering in Saccharomyces cerevisiae: Expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab. Eng. 2006, 8, 303–314. [Google Scholar] [CrossRef]
- Bhatt, S.M.; Srivastava, S. Mannitol increases lactic acid production by shifting NADH/NAD+ ratio inhibiting ethanol production. J. Biotechnol. 2008, 136, 22–27. [Google Scholar] [CrossRef]
- Kozak, B.U.; van Rossum, H.M.; Benjamin, K.R.; Wu, L.; Daran, J.M.; Pronk, J.T.; van Maris, A.J. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab. Eng. 2014, 21, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Si, T.; Nair, N.U.; Zhao, H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab. Eng. 2014, 24, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Talaia, G.; Sa-Pessoa, J.; Bessa, D.; Goncalves, M.J.; Moreira, R.; Paiva, S.; Casal, M.; Queiros, O. Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2. FEMS Yeast Res. 2012, 12, 375–381. [Google Scholar] [CrossRef]
- Paiva, S.; Vieira, N.; Nondier, I.; Haguenauer-Tsapis, R.; Casal, M.; Urban-Grimal, D. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: Role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 2009, 284, 19228–19236. [Google Scholar] [CrossRef]
- Wakamatsu, M.; Tomitaka, M.; Tani, T.; Taguchi, H.; Kida, K.; Akamatsu, T. Improvement of ethanol production from D-lactic acid by constitutive expression of lactate transporter Jen1p in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2013, 77, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.P.; Casal, M. Expression of the lactate permease gene JEN1 from the yeast Saccharomyces cerevisiae. Fungal Genet. Biol. 2001, 32, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Rothstein, R. Cell biology of mitotic recombination. Cold Spring Harb. Perspect. Biol. 2015, 7, a16535. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, D.; Zhang, Z.; Liu, T.; Hu, G.; He, M.; Zhao, S.; Peng, N. High-efficiency genome editing based on endogenous crispr-cas system enhances cell growth and lactic acid production in Pediococcus acidilactici. Appl. Environ. Microb. 2021, 87, e94821. [Google Scholar] [CrossRef] [PubMed]
- Tavares, T.G.; Malcata, F. Whey proteins as source of bioactive peptides against hypertension. Bioact. Food Pept. Health Dis. 2017, 4, 75–84. [Google Scholar]
- Juodeikiene, G.; Zadeike, D.; Bartkiene, E.; Klupsaite, D. Application of acid tolerant Pedioccocus strains for increasing the sustainability of lactic acid production from cheese whey. LWT-Food Sci. Technol. 2016, 72, 399–406. [Google Scholar] [CrossRef]
- Turner, T.L.; Kim, E.; Hwang, C.; Zhang, G.C.; Liu, J.J.; Jin, Y.S. Short communication: Conversion of lactose and whey into lactic acid by engineered yeast. J. Dairy Sci. 2017, 100, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Huimin, L.I.; Wang, X.; Wang, J.; Zhang, X.; Liu, Q. Efficient production of lactic acid and other organic acids from sugarcane molasses by Lactic acid bacteria. Food Ind. 2019, 1, 109–112. [Google Scholar]
- Liu, Y.T.; Wu, M.Y.; Jin, Y.L.; Shen, W.L.; Fang, Y.Z.H. Lactic acid fermentation by Lactobacillus rhamnosus from sweet potato residue. Sci. Agric. Sin. 2016, 9, 1767–1777. [Google Scholar]
- Nurkhamidah, S.; Altway, A.; Susianto; Rahmawati, Y.; Ramadhani, A. Utilization of molasses to produce lactic acid by using Lactobacillus delbrueckii and Lactobacillus plantarum. IOP Conf. Ser. Mater. Sci. Eng. 2019, 1, 5–12. [Google Scholar] [CrossRef]
- Trakarnpaiboon, S.; Srisuk, N.; Piyachomkwan, K.; Yang, S.; Kitpreechavanich, V. L-Lactic acid production from liquefied cassava starch by thermotolerant Rhizopus microsporus: Characterization and optimization. Process Biochem. 2017, 63, 26–34. [Google Scholar] [CrossRef]
- Panesar, P.S.; Kaur, S. Screenning of media components and process parameters for production of L(+) lactic acid from potato wast liquid using amylolytic Rhizopus oryzae. Acta Aliment. 2017, 46, 312–322. [Google Scholar] [CrossRef]
- Zacharof, M.P. Industrial Symbiosis: Beer Brewery Wastewater-Based Biorefinery. Circ. Econ. Sustain. 2021, 1, 593–609. [Google Scholar] [CrossRef]
- Gui, X.; Luo, Y.; Li, Z.; Nie, M.; Yang, Y.; Zhang, C.; Liu, J. Co-fermentation of kitchen waste and excess sludge for organic acid production: A review. Chin. J. Biotechnol. 2021, 37, 448–460. [Google Scholar] [CrossRef]
- Buenavista, R.; Siliveru, K.; Zheng, Y. Utilization of distiller’s dried grains with solubles: A review. J. Agric. Food Res. 2021, 1, 100195. [Google Scholar] [CrossRef]
- Alexandri, M.; Neu, A.K.; Schneider, R.; Lopez-Gomez, J.P.; Venus, J. Evaluation of various bacillus coagulans isolates for the production of high purity L-lactic acid using defatted rice bran hydrolysates. Int. J. Food Sci. Technol. 2018, 54, 1321–1329. [Google Scholar] [CrossRef]
- Nalawade, K.; Saikia, P.; Behera, S.; Konde, K.; Patil, S. Assessment of multiple pretreatment strategies for 2G L-lactic acid production from sugarcane bagasse. Biomass Convers. Biorefin. 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Hu, J.L. High-Efficient and Titer Lactic acid Production from Corn Stover by Bacillus coagulans LA204 Using Simultaneous Saccharification and Fermentation. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2015. [Google Scholar]
- Li, X.; Chen, Y.G.; Zhao, S.; Chen, H.; Zheng, X.; Luo, J.Y.; Liu, Y.N. Efficient production of optically pure L-lactic acid from food waste at ambient temperature by regulating key enzyme activity. Water Res. 2015, 70, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Mendes, F.; Sieuwerts, S.; de Hulster, E.; Almering, M.J.H.; Luttik, M.A.H.; Pronk, J.T.; Smid, E.J.; Bron, P.A.; Daran-Lapujade, P. Transcriptome-based characterization of interactions between Saccharomyces cerevisiae and Lactobacillus delbrueckii subsp bulgaricus in lactose-grown chemostat cocultures. Appl. Environ. Microbiol. 2013, 79, 5949–5961. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Murata, Y.; Yamazumi, H.; Tau, Y.; Mori, M.; Moriguch, M.; Shira, R.Y. Selective proliferation of lactic acid bacteria and accumulation of lactic acid during open fermentation of kitchen refuse with intermittent pH adjustment. Food Sci. Technol. Res. 2000, 6, 140–145. [Google Scholar] [CrossRef]
- Tashiro, Y.; Inokuchi, S.; Poudel, P.; Okugawa, Y.; Miyamoto, H.; Miayamoto, H.; Sakai, K. Novel pH control strategy for efficient production of optically active L-lactic acid from kitchen refuse using a mixed culture system. Bioresour. Technol. 2016, 216, 52–59. [Google Scholar] [CrossRef]
- Policastro, G.; Carraturo, F.; Compagnone, M.; Giugliano, M.; Guida, M.; Luongo, V.; Napolitano, R.; Fabbricino, M. A preliminary study on a novel bioaugmentation technique enhancing lactic acid production by mixed cultures fermentation. Bioresour. Technol. 2021, 340, 125595. [Google Scholar] [CrossRef]
- Ma, X.Y.; Gao, M.; Li, C.L.; Wang, N.H.; Wang, Q.H.; Sun, X.H. Effects of different lignocellulosic wastes on alleviating acidification of L-lactic acid production from food waste fermentation. Bioresour. Technol. 2021, 342, 126043. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huo, K.; Gao, L.; Zhao, G.; Wang, B.; Liu, J. Open simultaneous saccharification and fermentation of l-lactic acid by complete utilization of sweet sorghum stalk: A water-saving process. RSC Adv. 2021, 11, 5284–5290. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Primary Study on Prodution of L-Lactic Acid by High Cell Density and Semi-Continuous Fermentation with Rhizopus oryzae. Master’s Thesis, Hefei University of Technology, Hefei, China, 2009. [Google Scholar]
- Zhao, L.; Jiang, X.W.; Bian, J.W. Process conditions for L-lactic acid production by corncob-immobilized Rhizopus oryzae. Food Sci. 2013, 34, 201–205. [Google Scholar]
- Luongo, V.; Policastro, G.; Ghimire, A.; Pirozzi, F.F.M. Repeated-batch fermentation of cheese whey for semi-continuous lactic acid production using mixed cultures at uncontrolled pH. Sustainability 2019, 11, 3330. [Google Scholar] [CrossRef]
- Radosavljevic, M.; Levic, S.; Belovic, M.; Pejin, J.; Djukic-Vukovic, A.; Mojovic, L.; Nedovic, V. Immobilization of Lactobacillus rhamnosus in polyvinyl alcohol/calcium alginate matrix for production of lactic acid. Bioproc. Biosyst. Eng. 2020, 43, 315–322. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Zhang, J.; Pan, J. Using tobacco waste extract in pre-culture medium to improve xylose utilization for L-lactic acid production from cellulosic waste by Rhizopus oryzae. Bioresour. Technol. 2016, 218, 344–350. [Google Scholar] [CrossRef]
- Danner, H.; Madzingaidzo, L.; Thomasser, C.; Neureiter, M.; Braun, R. Thermophilic production of Lactic acid using integrated membrane bioreactor systems coupled with monopolar electrodialysis. Appl. Microbiol. Biotechnol. 2002, 59, 160–169. [Google Scholar] [CrossRef]
- Russo, F.; Luongo, V.; Mattei, M.R.; Frunzo, L. Mathematical modeling of metal recovery from E-waste using a dark-fermentation-leaching process. Sci. Rep. 2022, 12, 4274. [Google Scholar] [CrossRef]
- Gadhamshetty, V.; Arudchelvam, Y.; Nirmalakhandan, N.; Johnson, D.C. Modeling dark fermentation for biohydrogen production: ADM1-based model vs. Gompertz model. Int. J. Hydrogen Energy 2010, 35, 479–490. [Google Scholar] [CrossRef]
- Do Nascimento, T.R.; Cavalcante, W.A.; de Oliveira, G.H.D.; Zaiat, M.; Ribeiro, R. Modeling dark fermentation of cheese whey for H2 and n-butyrate production considering the chain elongation perspective. Bioresour. Technol. Rep. 2022, 17, 100940. [Google Scholar] [CrossRef]
- Kim, M.S.; Cho, K.H.; Park, K.H.; Jang, J.; Hahn, J.S. Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae. Nucleic Acids Res. 2019, 47, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Heredia, M.Y.; Ikeh, M.; Gunasekaran, D.; Conrad, K.A.; Filimonava, S.; Marotta, D.H.; Nobile, C.J.; Rauceo, J.M. An expanded cell wall damage signaling network is comprised of the transcription factors Rlm1 and Sko1 in Candida albicans. PLoS Genet. 2020, 16, e1008908. [Google Scholar] [CrossRef] [PubMed]
| Strains | Fermentation Temperature (°C) | Raw Materials | L-LA (g/L) | Characteristic | Reference |
|---|---|---|---|---|---|
| LAB genera | Heterotrophic anaerobic type | ||||
| L. casei, L. plantarum | 28–32 | Sucrose | 175.8 | Facultative anaerobic; no need for ventilation; energy-saving economy; a variety of biological resources | [23] |
| Lactobacillus pentosus | 30–32 | Glu | 108.1 | [24] | |
| Lactobacillus xylose | 30–40 | Xylose | 40.3 | [25] | |
| Lactobacillus sake, Lactobacillus acidophilus, Lactobacillus amyloidus | 35–38 | Brewers’ spent grain | 22.1 | [26] | |
| Lactococcus lactis | 36–45 | Xylose | 58.3 | [27] | |
| L. thermophilus | 50–60 | Glu | 97.5 | [28] | |
| Rhizopus | Aerobic | ||||
| Rhizopus nigra, Aspergillus triticum, R. chinensis, Rhizopus sweet potato, R. oryzae, Rhizopus tuberosus, Rhizopus japonica, Rhizopus paucus, Rhizopus meilis | 30 | Paper sludge; rice straw | 88.9 | Abundant biomass resources | [29,30] |
| Streptococcus | Microanaerobic | ||||
| Streptococcus thermophilus, Streptococcus lactobacillus, Streptococcus salivary | 40–45 | Glu, fructose | 49.9 | Has a certain tolerance | [31] |
| Bacillus | Facultative and anaerobic | ||||
| B. coagulans | 50–60 | Lignocellulose | 55.9 | High optical purity and conversion rate | [32,33,34,35] |
| Thermophilic adipose bacillus | 55–60 | Oil palm empty fruit bunch | 105.4 |
| Strain | Carbon Source | Exogenous LDH Source | Genotype/Methods | L-LA (g/L) | Fermentation Time (h) | Reference |
|---|---|---|---|---|---|---|
| L. rhamnosus | Mixture of lignocellulosic biomass | - | SSF | 61.74 | 44 | [54] |
| B. coagulans | Papermill sludge | - | SCF | 82.4 | 120 | [55] |
| E. coli | Glu, xylose | - | ΔptsG, ΔmglB | 53.2 | 60 | [56] |
| B. coagulans | Organic fraction of municipal solid waste | - | Monopolar electrodialysis membranes | 61.1 | 36 | [57] |
| Enterococcus faecium | GSW, corn steep liquor | - | CH3COONa, MgSO4,MnSO4, K2HPO4, CaCl2, and Tween 80 | 93.1 | 48 | [58] |
| K. marxianus | Corncob | P. falciparum, B. subtilis | Overexpressing PFK, ΔDld1 | 103.0 | 50 | [46] |
| K. marxianus | Jerusalem artichoke tuber powder | L. plantarum | Δpdc1, Δcyb2, ΔDld1 | 130.0 | 66 | [59] |
| L. plantarum | Raw starch | - | ΔldhD, ΔlarA-E | 87.0 | 72 | [60] |
| K. marxianus | Glu | Staphylococcus epidermidis, L. acidophilus, Bos taurus | LaLDH is coexpressed with SeLDH | 24.0 | 60 | [61] |
| S. cerevisiae | Glu, xylose | P. falciparum | LDH insert Δpdc1, Δpdc5 | 50.0 | 140 | [62] |
| Pichia pastoris | Glycerol | B. taurus | Expressing transporter PAS | 47.0 | 105 | [63] |
| S. cerevisiae | Cellobiose and xylose | R. oryzae | Expressing cdt-1, gh1-1, XYL1, XYL2, XYL3, ldhA | 83.0 | 80 | [64] |
| B. coagulans | Raw hemp hurd | - | Organosolv pretreatment and enzymatic hydrolysis | 141 | 148 | [65] |
| S. cerevisiae | Glu | Bovine | Δpdc1, Δpdc5 | 82.3 | 216 | [50] |
| E. coli | Glu | L. casei, S. bovis, B. coagulans | ΔldhA::diflldD::Pldh-ldhBcoa | 142.2 | 40 | [40] |
| B. coagulans | Glu, cane molasses | - | Cofeeding fermentation | 168.3 | 100 | [66] |
| R. oryzae | Glu | - | Immobilized in cubic particles | 231.0 | 130 | [22] |
| S. cerevisiae | Molasses, corn paste wastewater | Lactobacillus helveticus | LDH insert Δpdc1 | 52.2 | 96 | [67] |
| B. subtilis | Glu, corn syrup | - | Batch and fed-batch culture | 183.2 | 96 | [68] |
| S. cerevisiae | Glu | Bovine | Δpdc1, regulatory cofactor | 20.0 | 100 | [69] |
| Candida magnolia | Glu | R. oryzae | pH 2.5 | 40.0 | 48 | [70] |
| Candida utilis | Glu | B. taurus | Δpdc1 | 103.3 | 33 | [71] |
| Candida boidinii | Glu | Bovine | LDH insert Δpdc1 | 85.9 | 48 | [72] |
| L. rhamnosus | Glu | - | - | 235 | 60 | [18] |
| S. cerevisiae | Glu | Bovine | Δpdc5, Δpdc6 | 122.0 | 48 | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Xu, X.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Engineered Microbial Cell Factories for Sustainable Production of L-Lactic Acid: A Critical Review. Fermentation 2022, 8, 279. https://doi.org/10.3390/fermentation8060279
Liu T, Xu X, Liu Y, Li J, Du G, Lv X, Liu L. Engineered Microbial Cell Factories for Sustainable Production of L-Lactic Acid: A Critical Review. Fermentation. 2022; 8(6):279. https://doi.org/10.3390/fermentation8060279
Chicago/Turabian StyleLiu, Tiantian, Xianhao Xu, Yanfeng Liu, Jianghua Li, Guocheng Du, Xueqin Lv, and Long Liu. 2022. "Engineered Microbial Cell Factories for Sustainable Production of L-Lactic Acid: A Critical Review" Fermentation 8, no. 6: 279. https://doi.org/10.3390/fermentation8060279
APA StyleLiu, T., Xu, X., Liu, Y., Li, J., Du, G., Lv, X., & Liu, L. (2022). Engineered Microbial Cell Factories for Sustainable Production of L-Lactic Acid: A Critical Review. Fermentation, 8(6), 279. https://doi.org/10.3390/fermentation8060279









