Abstract
Several studies have shown the ability of yeast to consume peptides as a nitrogen source in single-peptide containing media. However, a suitable and cost-effective methodology to study the utilization of peptides by yeast and other microorganisms in a complex peptide mixture has yet to be put forward. This article addresses this issue by presenting a screening methodology for tracking the consumption of peptides by yeast during alcoholic fermentation. As a peptide source, the methodology makes use of an in-house prepared peptide-mapped bovine serum albumin (BSA) proteolytic digest, which was applied to a synthetic grape must. The peptide uptake was analyzed using high-throughput ultra-high-pressure liquid chromatography coupled to data-independent acquisition-based ion mobility separation-enabled high-resolution mass spectrometry (UPLC-DIA-IMS-HRMS) analysis. The relative changes of abundance of 123 di- to hexapeptides were monitored and reported during fermentations with three commercial wine strains, demonstrating different uptake kinetics for individual peptides. Using the same peptide-mapped BSA hydrolysate, the applicability of an untargeted workflow was additionally assessed for peptide profiling in unelucidated matrixes. The comparison of the results from peptide mapping and untargeted analysis experiments highlighted the ability of untargeted analysis to consistently identify small molecular weight peptides on the length and amino acid composition. The proposed method, in combination with other analytical techniques, such as gene or protein expression analysis, can be a useful tool for different metabolic studies related to the consumption of complex nitrogen sources by yeast or other microorganisms.
1. Introduction
Sufficient assimilable nitrogen content is a prerequisite for successful alcoholic fermentation. Saccharomyces cerevisiae, the main microorganism used in the production of alcoholic beverages and bioethanol, is known to utilize ammonia, free amino acids and short peptides as nitrogen sources [1]. While the consumption preferences of S. cerevisiae are well studied for ammonia and free amino acids, the role of peptides as a yeast nitrogen source requires further elucidation.
Many feedstocks used in alcoholic fermentation contain a significant proportion of peptides that could be considered as an important nitrogen source for yeast. Peptides thus have a significant technological value. For example, in grape must, about 17% of the total nitrogen can be attributed to oligopeptides, while for beer wort, this number is estimated at around 40% [2,3]. The nitrogen levels in grape must are dependent on the grape variety and grape cultivation conditions used. Due to these factors, the nitrogen levels in some cases can be deficient [4]. Similarly, high gravity fermentation with substantial content of adjuncts in beer and whisky production is a process that may suffer from nitrogen limitation [2,5].
In any fermentation, nitrogen deficiency can result in premature cessation or even complete arrest of the fermentation process, as well as in poor flavor properties of the product [6,7,8]. Fermentation nutrients, such as yeast autolysates, rich in free amino acids and peptides, are often used in industry to overcome this problem. Regardless of whether the peptides are present in the fermentation feedstock naturally or are supplemented with nutrients, a suitable screening methodology is required to characterize yeast strains for their ability to take up different peptides. The information obtained from such a screening method would be useful to better anticipate nitrogen deficiency and to avoid related fermentation problems through either specific nitrogen supplementation or the selection of yeast strains with broader peptide consumption capabilities.
S. cerevisiae can internalize peptides of different sizes through multiple membrane peptide transporters [9]. Di- and tripeptide transport is facilitated by Ptr2p, Dal5p and fungal oligopeptide transporters (Fot1-3p) found in oenological yeast [10,11,12,13,14]. However, it is not excluded that FOT transporters can also transport longer peptides [9]. Oligopeptide transporters (Opt1p and Opt2p) have been shown to be involved in tetra- and pentapeptide transport [15,16,17].
The current knowledge on functionality of different yeast peptide transporters has been mainly obtained using experiments with single synthetic peptides as the sole nitrogen source [10,14,15,16,17,18]. This type of experiment provides accurate information about the ability of yeast strains to utilize the selected peptide as a nitrogen source. However, they do not provide information on the kinetics of peptide utilization throughout different time points during fermentation on complex peptide-rich media, such as wort, grain mash and grape must. Moreover, the characterization of peptide transporters in more complex matrices and peptide utilization capability by different yeasts is challenging due to the lack of suitable cost-efficient screening media with defined and diverse peptide composition. Another challenge is the lack of accurate semi-quantitative analyses for monitoring the uptake of these peptides during fermentation.
Liquid chromatography coupled to mass spectrometry (LC-MS) is currently one of the most promising techniques for short peptide analysis [19,20,21,22]. The two biggest analytical challenges are insufficient short peptide identification accuracy for unambiguous identity assignment and throughput limitation caused by data-dependent acquisition (DDA). Unlike longer peptides (pentapeptides and longer) that, due to a higher number of peptide bonds, tend to ionize with a higher charge state (≥2+), shorter peptides (di- to tetrapeptides) most commonly ionize as singly charged ions. The singly charged peptides produce fewer selective fragments, which leads to reduced identification accuracy [23].
Even though DDA is known to produce higher quality fragmentation spectra due to active quadrupole-based selection of precursor ions prior to fragmentation, it comes at he cost of a reduced acquisition rate that often results in missing data and prolonged analytical gradients [24]. In contrast, data-independent acquisition (DIA) minimizes the likelihood of missing data by rapidly alternating between low and high collision energies and assigning fragments based on the chromatographic elution profile [25]. For fermentation matrices with highly diverse peptide composition, the main drawback of such data acquisition is the generation of complex fragmentation spectra that are often caused by simultaneous fragmentation of closely eluting precursor ions.
A rapid gas-phase separation of ions, also commonly referred to as ion mobility (IM) separation, is often used as an orthogonal method of separation that complements chromatography and mass spectrometry [26]. Hyphenation of travelling wave ion mobility separation (TWIMS) with DIA results in similar drift times for both fragment and precursor ions, allowing alignment by both the chromatographic and ion mobility elution profiles and thus improving the spectral clarity. Moreover, the “trap–release” mode of operation of the TWIMS results in not only increased peak capacity but also in increased duty cycle of the mass spectrometer. This allows for shorter analytical gradients and the use of the collisional cross section (CCS) as an additional identification qualifier.
This study reports a screening methodology for tracking the consumption of a wide range of peptides by yeast during alcoholic fermentation. The methodology makes use of an in-house prepared peptide-mapped (against a known protein sequence) proteolytic digest of bovine serum albumin (BSA) as a peptide source that was applied to a synthetic grape must. The peptide composition in the hydrolysate as well as the consumption of peptides by yeast in the synthetic grape must were analyzed using high-throughput ultra-high-pressure liquid chromatography coupled with data-independent acquisition-based ion mobility separation-enabled high-resolution mass spectrometry (UPLC-DIA-IMS-HRMS).
To simulate the analysis of an unelucidated matrix, untargeted data analysis (that does not rely on a protein sequence) of the BSA hydrolysate was additionally performed and compared to the results of peptide mapping to assess the identification accuracy of the untargeted analysis.
2. Materials and Methods
An LC-MS methodology was developed for analyzing peptide composition in BSA proteolytic digest (peptide mapping), as well as for monitoring the relative concentration changes of these peptides during fermentation with three commercial wine yeast strains (screening). A modified synthetic grape must with a reduced concentration of free amino acids and ammonia was used as a base fermentation medium and was supplemented with additional nitrogen in the form of peptides from proteolytic digest of BSA. The relative changes of peptide abundances during fermentation were monitored and reported.
Peptide mapping requires a protein sequence to identify peptides. However, when analysing fermentation feedstocks with unknown protein/peptide content, such information is not always available. Therefore, the peptide identification accuracy of an untargeted workflow should be investigated. For this purpose, the peptide identification results obtained by untargeted data analysis against a short peptide database were compared to the results obtained by peptide mapping.
2.1. Chemicals
Hi3 E.coli STD (p/n: 186006012) and Leucine-Enkephalin (p/n: WT186006013) were purchased from Waters Corporation (Milford, MA, USA). Ultrapure water (18.2 MΩ×cm) was prepared with MilliQ® IQ 7000 equipped with LC-Pak (Merck KGaA, Darmstadt, Germany). Acetonitrile (MeCN; LiChrosolv, hypergrade for LC-MS), formic acid (FA; LC-MS grade), bovine serum albumin (lyophilized powder, ≥96%), fructose, cobalt chloride, boric acid, ammonium molybdate tetrahydrate, ergosterol, oleic acid, all vitamins and cysteine were acquired from Sigma-Aldrich (Darmstadt, Germany).
Histidine, methionine, glutamic acid and arginine were acquired from SERVA Electrophoresis GmbH (Heidelberg, Germany). Alanine, aspartic acid, cysteine, glutamine, glycine, isoleucine, leucine, lysine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine and valine were acquired from Thermo Fisher Scientific (Waltham, MA, USA). Hydrogen chloride (HCl, 36%) was acquired from Labbox Labware (Barcelona, Spain). Liquified phenol was acquired from Avantor® (Wayne, PA, USA).
2.2. Fermentation Media
2.2.1. BSA Hydrolysate
Hydrolysis of BSA (40 g/L dH2O) was conducted in a 1 L benchtop fermenter (Applikon, Delft, The Netherlands) using the industrial protease COROLASE®7089 (AB Enzymes, Darmstadt, Germany), which was selected based on its endoproteolytic activity. This facilitated the greater production of low molecular weight peptides, which were potentially assimilable by yeast. The protease dose rate was 0.5% w/w of BSA, and hydrolysis was performed at pH 7 (maintained by titration using 2 M NaOH) for 20 h at 50 °C. After completion of the BSA hydrolysis, the hydrolysate was filtered with a Vivaflow®200 10 kDa cut-off Hydrosart crossflow cassette (Sartorius, Göttingen, Germany) to remove the protease as well as larger peptides. It was then freeze-dried and stored at −20 °C until further use.
As the scope of this research was to develop a methodology for following small molecular weight peptide consumption by yeast during alcoholic fermentation, the nitrogen content of peptides smaller than 1 kDa in the fraction was determined from the bound amino acid content and used to calculate the amount of total hydrolysate to be added into the nitrogen deficient synthetic grape must. The sub-1-kDa fraction for amino acid analysis was prepared by rehydrating the BSA hydrolysate (1 g/L dH2O) and filtering through a Pall® Microsep Advance 1K Omega™ centrifugal filter device (Pall® Corporation, Port Washington, NY, USA).
The amounts of free and total amino acids in this fraction were analyzed on a Waters ACQUITY UPLC® system (Waters Corporation, Milford, MA, USA) that was coupled to a TUV detector after derivatization using Waters AccQ-Tag chemistry as described by Fiechter and Mayer [27]. For the total amino acids, the samples were first hydrolyzed with 6M HCl + 1% (v/v) phenol for 22 h in a vacuum using an Eldex H/D workstation (Eldex Laboratories Inc., Napa, CA, USA). The amount of bound amino acids was calculated by subtracting the free amino acids from the total amino acid concentration.
2.2.2. Synthetic Grape Must
A synthetic grape must (MS300) as described by Salmon and Barre [28] was prepared with a reduced initial nitrogen content. The free amino acid and NH3 concentrations were both reduced by 75% to provide 116 mg/L of nitrogen, which was considered insufficient to ferment 210 g/L of fermentable sugars in the synthetic grape must [29,30]. The amino acid and NH3 concentrations can be found in Table A2. An additional 136 mg/L of nitrogen was then supplemented in the form of peptides from the BSA hydrolysate, resulting in 252 mg/L nitrogen in the synthetic grape must.
2.3. Fermentation
Three commercial S. cerevisiae wine yeast strains (LalvinTM ICV Opale 2.0, LalvinTM Persy and LalvinTM QA23), provided by Lallemand Inc. (Montreal, QC, Canada), were used in this study. All strains were obtained as active dry yeast cultures. Inoculum was prepared by rehydrating 1 g of dry culture in 10 mL of sterile 0.9 % NaCl for 15 min at room temperature. The synthetic grape must was then pitched to give an initial yeast concentration of about 5∙106 cells/mL.
All fermentations were performed at 24 °C in 250 mL Pyrex™ bottles equipped with a GL45 open top PBT screw cap and PYREX™ Media Bottle Septum (Corning Inc., Corning, NY, USA). A sampling port was assembled with size 13 Masterflex™ Norprene™ Food L/S™ Precision Pump Tubing (Masterflex SE, Gelsenkirchen, Germany), connected with a metal tubing piercing through the septum. A gas outlet was installed to prevent overpressure by piercing the septum with a Sterican® Ø 0.8 × 40 mm single-use hypodermic needle (B. Braun, Melsungen, Germany) attached to a Millex-FG 0.2 µm hydrophobic PTFE filter (Merck KGaA, Darmstadt, Germany).
The initial volume of synthetic grape must was 200 mL. Samples (3 mL) were collected at 0, 24, 48, 72 and 168 h. For each strain, a control fermentation was performed in parallel without the addition of BSA hydrolysate.
2.4. Sample Preparation
Prior to peptide analysis, samples (200 µL) were mixed with MeCN (200 µL) in an Eppendorf® tube (Eppendorf AG, Hamburg, Germany) to precipitate proteins, vortexed for 30 s and centrifuged at 11,200 × g for 15 min. The supernatant (100 µL) and MilliQ® water (900 µL) were then transferred into a new Eppendorf® tube and vortexed for 30 s. The obtained mixture (100 µL) was transferred into a low-volume insert and spiked with 20 µL of 1 pmol/µL of Hi3 E.coli STD.
Before and after injection of the batch of samples, a blank (MilliQ® water) and a blank (100 µl MilliQ® water) spiked with 20 µL of 1 pmol/µL of Hi3 E.coli STD were injected to determine the baseline and system variability. A pooled sample (100 µL) spiked with 20 µL of 1 pmol/µL of Hi3 E.coli STD was also injected three times at the beginning of the experiment and one time after each strain to determine the intra-batch variability.
2.5. Liquid Chromatography Mass Spectrometry
Samples were analyzed using Waters I-Class Plus (SM-FL) UPLC® system (Waters Corporation, Milford, MA, USA) coupled with a Waters Vion IMS-QTof Mass Spectrometer equipped with LockSpray II Exact Mass source enclosure and ESI Tool-free Mk3 UPC2 probe (50 µm × 750 mm, p/n: 700011376) directly connected to the column outlet. Nitrogen was used as collision gas. The instrument was controlled by Waters UNIFI 1.9.4 (3.1.0, Waters Corporation, Milford, MA, USA).
Mobile phases (MP) were as follows: (A) MilliQ® + 0.1% formic acid and (B) MeCN + 0.1% formic acid. The weak needle wash was 90:10 (MilliQ®:MeCN), the strong needle wash was 10:90 (MilliQ®:MeCN), and the seal wash was 50:50 (MilliQ®:MeCN). Injection volume was 2 µL. Samples were analyzed using Acquity UPLC® HSS T3 Column (1.8 µm, 1 × 150 mm, Waters Corporation, Milford, MA, USA) kept at 40 °C. The initial flow rate was 0.1 mL/min. The gradient was as follows: a 0–0.5 min hold at 1% B, 0.5–10.5 min linear gradient 1–50% B, 10.5–12.5 min linear gradient, 50–99% B accompanied by a linear flow rate increase of 0.1–0.2 mL/min, 12.5–14.5 min hold at 99% B, 14.5–15 min linear gradient 99–1% B accompanied by a linear flow rate decrease of 0.2–0.1 mL/min, and 15–18.5 min hold at 1% B.
The instrument was operated in positive polarity, sensitivity mode (33,000 FWHM at 556.2766 m/z) and labile ion mobility tune. The analysis type was set as Peptide Map (IMS) and the experiment type was set to MSe. Data was acquired in HDMSe mode with a scan time of 0.2 s. The following manual quadrupole profile was used: mass 150/300/600 (m/z), dwell time 50/30 (%scan time), ramp time 10/10 (%scan time). The recorded mass range was from 50 to 1000 m/z for both low and high energy spectra. The collision energy was ramped from 10 to 63 V.
The cone voltage was set to 30 V, capillary voltage was set to 0.5 kV and source offset was set to 50 V. Source temperature was set to 120 °C and desolvation temperature set to 500 °C. Cone gas flow rate was set to 50 L/h and desolvation gas flow rate was set to 700 L/h. Leucine-enkephalin (50 pg/µL at 15 µL/min) was used as LockMass for mass axis correction and was acquired before and after each acquisition as well as every 5 min during the acquisition.
2.6. Data Processing
For peptide identification in the BSA hydrolysate, peptide mapping was conducted using UNIFI Large Molecule Package (LMP) by searching the peptides against the sequence of bovine serum albumin (ALBU_BOVIN; P02769). Low energy intensity threshold of 2000 counts and high energy intensity threshold of 200 counts were used. The retention time range was set between 1 and 12 min. Peptide search was set to non-specific digest with no missed cleavages allowed, and the minimum sequence length was set to 2.
The following filters were applied: mass error between −3 and 3 ppm, matched first gen primary ions greater than or equal to 1, fragment label not containing in-source fragments, loss of H2O and loss of NH3. For relative quantification (screening of peptide uptake by yeast) and untargeted analysis of short peptides the data was exported using UNIFI Export Package (UEP) to Progenesis QI (PQI) software (Nonlinear Dynamics, Newcastle, UK). During the import, masses were lock mass corrected with 556.2766 m/z, corresponding to singly charged Leucine-enkephalin. Data was subjected to normalization of compound abundances based on a set of reference housekeeping peptides (spiked Hi3 E.coli STD). Automatic sensitivity at level 2 was used.
Automatic retention time alignment was conducted, and the retention time limits were set between 1 and 12 min. Fragment sensitivity of 1% of the base peak was used. The following adducts were used for peptide mapping: M+H, M+2H and M+3H. ChemSpider data source: peptides (PQI_CS_Peptides), containing in-silico database of short peptides (2-4 AA), were used for de novo identification. The precursor ion tolerance was set to 3 ppm and fragment ion tolerance was set to 5 ppm. One or more fragment ions per peptide were required for ion matching.
2.7. Data Analysis
Peptide peaks identified by peptide mapping and untargeted analysis of short peptides were aligned using observed mass to charge ratio, charge, observed retention time (min) as well as collision cross section (Ų). Peptides without a distinct consumption trend were filtered out using PQI software. Peptides with up to six amino acids in length and an initial abundance higher than 0.1% of the summed abundance of all peptides were considered and reported for each fermentation experiment.
The aligned data was further used to construct a data matrix for the aforementioned data processing methods. The comparison between two identification methods was performed with the help of in-house data analysis and visualization scripts written in the PythonTM programming language (Python Software Foundation, Wilmington, DE, USA). The identification accuracy and reproducibility assessment between peptide mapping and untargeted analysis was performed based on four categories: absolute amino acid sequence match (Absolute), amino acid sequence match while leucine (L) and isoleucine (I) were not differentiated and annotated as “J” (J Absolute), and amino acid composition match (J Composition) and peptide length match (Length).
3. Results
3.1. BSA Hydrolysate Composition
The total nitrogen in the BSA hydrolysate was determined in three fractions: free amino acids, peptides with molecular weight (MW) smaller than 1 kDa and peptides with MW higher than 1 kDa. Most of the nitrogen (69%; 81.4 mg/g) came from peptides with MW below 1 kDa. Endoproteolytic activity of COROLASE® 7089 was apparent as only 7% of total nitrogen could be assigned to free amino acids (8.1 mg/g). The remaining 24% of nitrogen (29 mg/g) was present in the higher than 1 kDa fraction. The degree of hydrolysis was calculated, as described by Adler-Nissen [31] and was 40%.
3.2. LC-MS Method Reproducibility
We observed that the default settings of the ion optics and quadrupole of Vion IMS-Qtof resulted in a lower response of the lower molecular weight peptides. Combination of the labile ion mobility tune and a manual short peptide specific quadrupole transmission profile resulted in an increased response of di- and tripeptides. The system performance was found to be highly reproducible as illustrated by Figure 1 (overlay of three technical replicates). The standard deviation of the peptide retention times was below 0.1 min, and the PQI alignment score was over 95% for the six QC samples and over 92% for all 36 samples.
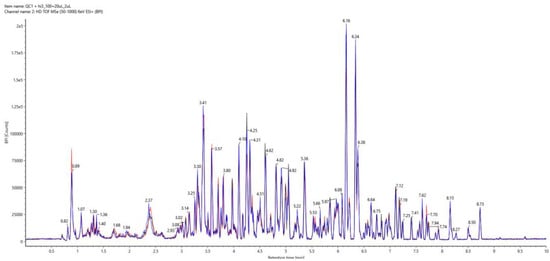
Figure 1.
Overlay of triplicate injection of Hi3 E.coli STD spiked pooled-sample base peak intensity (BPI) chromatograms.
Moreover, the starting points of all three fermentations display an average relative standard deviation of 5.27% for the intensities of the peptides of interest. The average peak width at 10% of peak height was 8 s, thus, resulting in not only enough data points for reliable qualitative and quantitative integration but also in a sufficient number of data points for accurate tracking of the peak’s lift-off, apex and touch-down points for improved high-to-low energy spectra association.
3.3. Fermentation and Peptide Uptake Kinetics
The maximum CO2 production rate was reached at about 48 h in case of all strains. However, the fermentations supplemented with the BSA hydrolysate showed increased sugar consumption, higher CO2 production and faster biomass accumulation compared to the corresponding controls without the hydrolysate (Figure 2). At the final experimental point (168 h), up to 95% of sugars were consumed in fermentations with added BSA hydrolysate, whereas up to 83% of sugars were consumed without hydrolysate addition (Table A1).
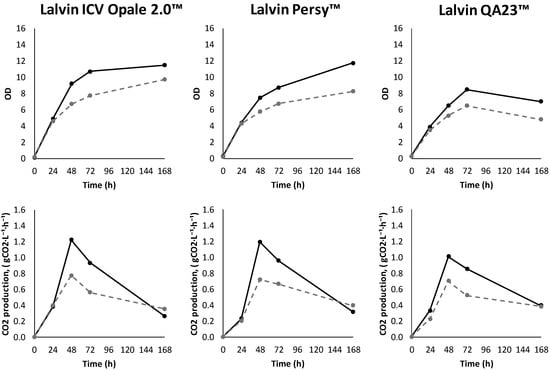
Figure 2.
The optical density (OD) and CO2 production rate in fermentations with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must with (solid line) and without (dashed line) added BSA hydrolysate as the peptide source.
In total, 123 peptide candidates complying with data analysis filtration criteria (19 dipeptides, 41 tripeptides, 31 tetrapeptides, 19 pentapeptides and 13 hexapeptides) were monitored throughout the fermentation experiments. The relative changes of individual peptide candidate intensities during the fermentation of the synthetic grape must are shown in Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7 and Table A4, Table A5, Table A6, Table A7 and Table A8.
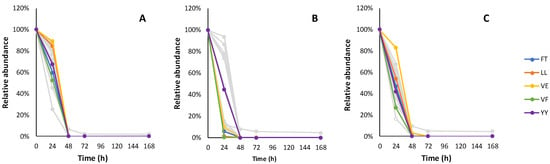
Figure 3.
The relative consumption trends of 19 dipeptides in fermentation with Lalvin ICV Opale 2.0™ (A), Lalvin Persy™ (B) and Lalvin QA23™ (C) of the modified synthetic must with added BSA hydrolysate as the peptide source. The five dipeptide candidates with the highest abundance are shown in colour. Other dipeptide candidate sequences and their observed relative consumption trends are shown in Table A4.
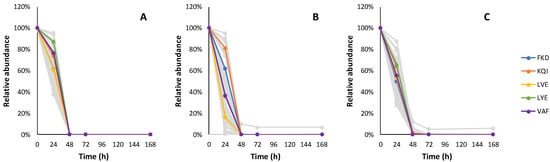
Figure 4.
The relative consumption trends of 41 tripeptides in fermentation with Lalvin ICV Opale 2.0™ (A), Lalvin Persy™ (B) and Lalvin QA23™ (C) of the modified synthetic must with added BSA hydrolysate as the peptide source. The five tripeptide candidates with the highest abundance are shown in colour. Other tripeptide candidate sequences and their observed relative consumption trends are shown in Table A5.
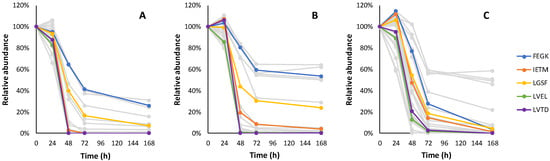
Figure 5.
The relative consumption trends of 31 tetrapeptides in fermentation with Lalvin ICV Opale 2.0™ (A), Lalvin Persy™ (B) and Lalvin QA23™ (C) of the modified synthetic must with added BSA hydrolysate as the peptide source. The five tetrapeptide candidates with the highest abundance are shown in colour. Other tetrapeptide candidate sequences and their observed relative consumption trends are shown in Table A6.
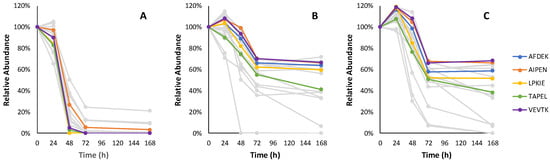
Figure 6.
The relative consumption trends of 19 pentapeptides in fermentation with Lalvin ICV Opale 2.0™ (A), Lalvin Persy™ (B) and Lalvin QA23™ (C) of the modified synthetic must with added BSA hydrolysate as the peptide source. The five pentapeptide candidates with the highest abundance are shown in colour. Other pentapeptide candidate sequences and their observed relative consumption trends are shown in Table A7.
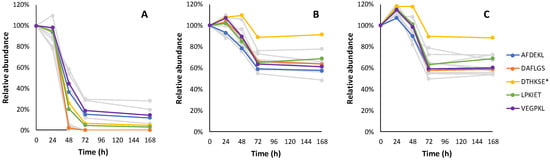
Figure 7.
The relative consumption trends of 13 hexapeptides in fermentation with Lalvin ICV Opale 2.0™ (A), Lalvin Persy™ (B) and Lalvin QA23™ (C) of the modified synthetic must with added BSA hydrolysate as the peptide source. The five hexapeptide candidates with the highest abundance are shown in colour. Other hexapeptide candidate sequences and their observed relative consumption trends are shown in Table A8. * DTHKSE elutes within the void volume.
All three strains demonstrated similar uptake trends of di-, tri- and tetrapeptides. For all strains, di- and tripeptides were consumed during the first 48 h of fermentation, simultaneously with the free amino acids (Figure 8). The uptake rate of di- and tripeptides was higher by Lalvin Persy™ compared to Lalvin ICV Opale 2.0™ and Lalvin QA23™, as several peptides were consumed by this strain already at 24 h.
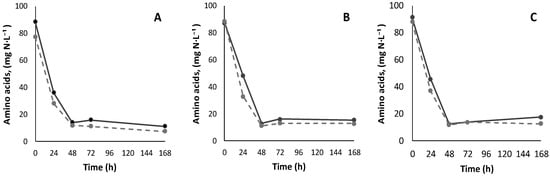
Figure 8.
Free amino acid consumption trends in fermentations with Lalvin ICV Opale 2.0™ (A), Lalvin Persy™ (B) and Lalvin QA23™ (C) of the modified synthetic must with (solid line) and without (dashed line) added BSA hydrolysate as the peptide source.
Unlike di- and tripeptides, not all tetra- to hexapeptides were fully depleted (signal to noise ratio < 3:1), and their uptake ceased when the growth of cells entered the stationary phase (Lalvin ICV Opale 2.0™ and Lalvin QA23™) or slowed down remarkably (Lalvin Persy™) after 72 h. The differences in peptide uptake between strains were most notable for penta- and hexapeptides. Thus, the relative abundances of pentapeptides at 168 h decreased on average by 49% for Lalvin Persy™ and 58% for Lalvin QA23™, and hexapeptides decreased by 35% for both strains. The respective values for Lalvin ICV Opale 2.0™ were 98% for pentapeptides and 93% for hexapeptides.
3.4. Untargeted Peptide Analysis
The results of the untargeted data analysis (using the PQI_CS_Peptides database) were compared against the results obtained by peptide mapping to assess the identification accuracy of the untargeted workflow.
Figure 9 highlights the ability of the untargeted approach to identify all di- to tetrapeptides in the BSA hydrolysate in comparison to the peptide mapping. A trend emerged highlighting how the number of peptides assigned identically by both search engines depends on the identification specificity. Only 27.4% of the peptides were matched on the level of absolute amino acid sequence and 37.5% on the level of “J Absolute” where leucine (L) and isoleucine (I) were not differentiated and annotated as “J”. On the level of “J Composition”, where the order of the amino acids in the sequence was ignored and only the amino acid compositions of the peptide was considered, 71.4% of peptides were matched. Lastly, on the level of “Length”, where only the length of the peptide was considered, 90.5% of peptides were matched.
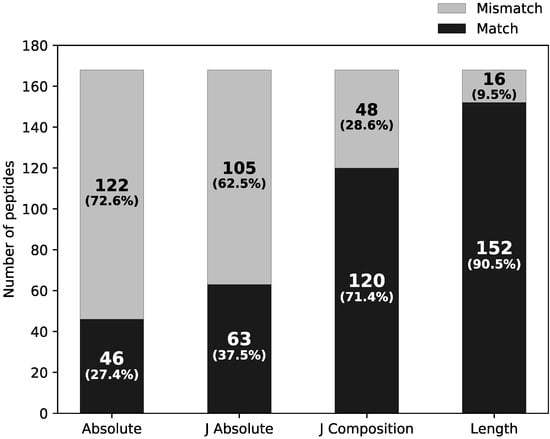
Figure 9.
The dependence of the peptide matching specificity criteria on the peptide identification assignment discrepancy: absolute amino acid sequence match (Absolute), amino acid sequence match, where leucine (L) and isoleucine (I) were not differentiated and are annotated as “J” (J Absolute), amino acid composition match (J Composition) and peptide length match (Length).
Figure 10 displays the identification discrepancies between the two methods regarding the peptide length assignment for all di- to tetrapeptides. For dipeptides, the match was 100%: all 29 dipeptides were assigned by the untargeted method and the peptide mapping. The tripeptide length assignment displayed minimal discrepancy: out of 62 tripeptides identified by the untargeted method, only one (1.6%) was assigned as a tetrapeptide by the peptide mapping.
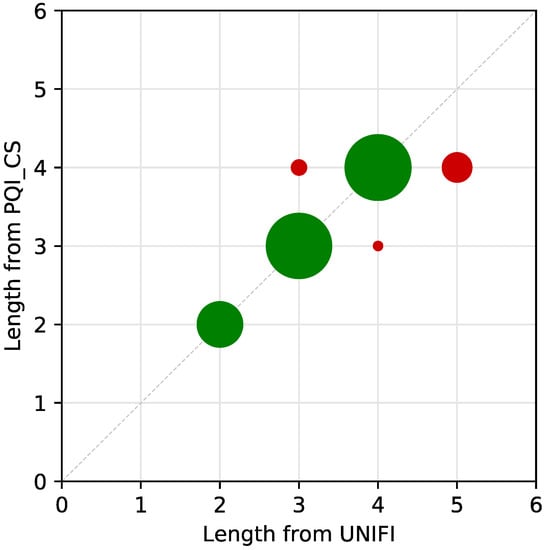
Figure 10.
Length assignment discrepancies by the untargeted method in relation to peptide mapping. Green circles represent peptide identification overlap between the two identification methods, whereas red circles represent mismatches between peptide identifications. Circle areas are scaled to represent the ratios between matched and mismatched peptides.
A larger discrepancy was observed for tetrapeptides: out of 77 tetrapeptides assigned by the untargeted method, three (3.9%) were assigned as tripeptides by peptide mapping, and 12 (15.6%) were assigned as pentapeptides by peptide mapping. The larger mismatch for tetrapeptides can be partly explained by the fact that the PQI_CS database contains sequences only up to tetrapeptides. While the peptide mapping approach resulted in a higher number of total identifications (not limited to di- to hexapeptides), a consistently low number (<10%) of unique (picked only by PQI) peptides were detected by the untargeted approach.
4. Discussion
In this study, we explored the application of an in-house produced BSA protein hydrolysate as a cost-efficient model mixture of peptides for yeast studies. The results indicate the potential of this method for screening yeast strains for their ability to consume different peptides during alcoholic fermentation, for studying peptide transporters, as well as for evaluating the effect of peptides as a nitrogen source on fermentation kinetics. The hydrolysis of BSA with COROLASE® 7089 resulted in 123 di- to hexapeptides, followed by their consumption by different wine yeast strains during fermentation experiments.
The results highlight differences in penta- and hexapeptide consumption trends, indicating the need for further research. Until now, the studies of peptide consumption by S. cerevisiae have focused on di- to pentapeptides, mainly in single peptide-based systems and small volumes, mostly due to the higher cost of synthetic peptides. An example of such a system is the Biolog (Hayward, CA, USA) Phenotype MicroArrays (PM) used by Becerra-Rodríguez et al. [10] to characterize FOT transporter knock-out strains for their ability to utilize 270 dipeptides and 14 tripeptides as nitrogen sources using a 96-well plate system. The ability of yeast to consume an individual peptide was assessed based on the growth in the medium containing a single peptide as the nitrogen source.
Despite being a high-throughput method for screening, this approach, due to its set-up (one single peptide per experiment) and very small volume, imposes several limitations for more in-depth studies of peptide consumption. These limitations exclude the possibility to determine fermentation and peptide uptake kinetics as well as peptide transporter expression analysis in the continuously changing environment of fermentation processes. The application of in-house-prepared protein hydrolysates allows for a more versatile and cost-effective approach to studying yeasts and other microorganisms for their ability to utilize various peptides in environments with more complex peptide compositions, which resemble more natural fermentation environments.
For this study, COROLASE® 7089 was chosen due to its high and broad endoproteolytic activity, which resulted in a higher number of di- to hexapeptides, which is important when peptide consumption by yeast is studied. However, the endoprotease and the protein source could be easily substituted with alternatives producing hydrolysates with different peptide compositions, allowing the tailor-made production of peptide mixtures based on various (micro-)organism-specific requirements [32,33,34,35].
For peptide composition analysis, it is typically recommended to hydrolyze a single protein with a known amino acid sequence. The identification of peptides against a known protein sequence allows for an additional degree of confidence in the identification accuracy. However, a protein sequence is not always available, and thus de novo identification should be used. The challenge of characterizing peptide mixtures using industrial proteases is often accompanied by unelucidated cleavage specificity of the proteases. This is also evident when peptide composition in natural fermentation feedstocks is to be analyzed, emphasizing the importance of the identification accuracy.
The results generated in this work by the untargeted analysis suggest that the method does not allow for small peptide unambiguous absolute sequence identification, which might be crucial for studying peptide transporters specificities. However, the method allows for peptide identification at the level of peptide length (“Length”, 90.5%) and amino acid composition, where isoleucine or leucine are not differentiated (“J Composition”, 71.4%), suggesting its potential for characterization of the composition of unelucidated natural peptide-containing fermentation matrices, such as grain mash, wort or grape must.
Even though the TWIMS used in this study increased the peak capacity and allowed for a shorter gradient time, the IMS resolution of the system was insufficient to reliably distinguish between isobaric peptides of the same length and different sequences. However, the collision cross section values were found to provide an extra qualifier when aligning features between two peak picking methods.
Quantitative representation of the content and consumption of all peptides remains a topic of further research. Due to the variation in the ionization efficiency of different peptides [36], consumption trends could only be used for tracking individual peptides across different samples, setting a limitation regarding the analysis of specific consumption rates of individual peptides by yeast in complex environments. The absolute quantitation of peptides remains reliant on labelled isotope dilution or tandem-mass-tag labelling [37,38,39].
5. Conclusions
We developed a methodology for characterization of the peptide composition in enzymatically produced hydrolysates of bovine serum albumin and implemented high-throughput monitoring of the peptide consumption trends of yeast during the alcoholic fermentation of a BSA-hydrolysate-supplemented synthetic grape must.
A correlation between the identification results of peptide mapping and untargeted analysis of di- to tetrapeptides (limited by the PQI_CS_Peptides database) suggests the potential of the untargeted analysis for following the peptide composition in unelucidated matrices, such as grain mash, wort or grape must. The method can additionally be used for profiling various enzymatically produced protein hydrolysates. However, consumption trends can only be used for tracking individual peptides across different samples. For the absolute quantitation of short peptides, the applications of tandem-mass-tag labelling must be further studied. The proposed methods, in combination with other analytical techniques, such as gene or protein expression analysis, can be a useful tool for different metabolic studies related to the consumption of complex nitrogen sources by yeast or other microorganisms.
Author Contributions
Conceptualization, G.A., H.Y.B. and I.N.; methodology, G.A. and H.Y.B.; software, G.A.; validation, G.A., H.Y.B. and T.L.; formal analysis, G.A. and H.Y.B.; investigation, G.A. and H.Y.B.; resources, I.N.; data curation, G.A. and H.Y.B.; writing—original draft preparation, G.A. and H.Y.B.; writing—review and editing, T.L. and I.N.; visualization, H.Y.B. and T.L.; supervision, I.N.; project administration, G.A.; funding acquisition, G.A., H.Y.B. and I.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “TUT Institutional Development Program for 2016–2022” Graduate School in Biomedicine and Biotechnology receiving funding from the European Regional Development Fund under program ASTRA 2014–2020.4.01.16-0032 in Estonia and the Estonian Research Council via project RESTA13.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Acknowledgments
The authors would like to acknowledge Signe Saarmets for performing the fermentations.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Sugars and ethanol were measured using the Waters Alliance 2695 system (Waters Corporation, Milford, MA, USA). The flow rate used was 0.6 mL/min. The mobile phase was isocratic for 25 min on 100% v/v 0.5 mmol H2SO4 in MilliQ water. The column used for analyses was Bio-Rad HPX-87H column (Bio-Rad Laboratories Inc, Hercules, CA, USA) with the dimensions 7.8 × 300 mm and 9 μm particle size, coupled to a Micro-Guard Cation H Cartridge pre-column (Bio-Rad Laboratories Inc, Hercules, CA, USA). The analytes were detected by the refractive index detector.

Table A1.
Glucose, fructose and ethanol measurements at the start and end of fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. ‘Control’ is without added peptides. ‘BSA’ is with added peptides.
Table A1.
Glucose, fructose and ethanol measurements at the start and end of fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. ‘Control’ is without added peptides. ‘BSA’ is with added peptides.
| Strain | Glucose Consumed (g⋅L−1) | Fructose Consumed (g⋅L−1) | Ethanol Produced (g⋅L−1) | |
|---|---|---|---|---|
| Lalvin ICV Opale 2.0™ | Control | 97.3 | 72.4 | 77.5 |
| BSA | 102.8 | 97.3 | 95.4 | |
| Lalvin Persy™ | Control | 97.4 | 76.8 | 81.4 |
| BSA | 104.4 | 101.2 | 95.7 | |
| Lalvin QA23™ | Control | 90.1 | 66.9 | 75.5 |
| BSA | 100.0 | 92.1 | 96.0 |
Appendix B
Free amino acids and ammonium chloride in a modified synthetic must

Table A2.
Free amino acid and ammonium chloride concentrations in a modified synthetic must.
Table A2.
Free amino acid and ammonium chloride concentrations in a modified synthetic must.
| Compound | Amount (mg⋅L−1) | Compound | Amount (mg⋅L−1) |
|---|---|---|---|
| Tryptophan | 38.7 | Alanine | 31.4 |
| Phenylalanine | 8.2 | Threonine | 16.4 |
| Isoleucine | 7.1 | Glutamic acid | 26.0 |
| Leucine | 10.5 | Aspartic acid | 9.6 |
| Valine | 9.6 | Glycine | 4.0 |
| Methionine | 6.8 | Arginine | 80.8 |
| Tyrosine | 4.0 | Glutamine | 109.1 |
| Lysine | 3.7 | Serine | 17.0 |
| Cysteine | 2.8 | Asparagine | 0.0 |
| Proline | 132.2 | Histidine | 7.1 |
| NH4CL | 102.5 |
Appendix C
Observed nitrogen consumption

Table A3.
The observed nitrogen consumption of free amino acids (FAA), bound amino acids (BAA) and ammonia during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. ‘Control’ is without added peptides, ‘BSA’ is with added peptides. Standard deviations represent measurements in three technical replicates.
Table A3.
The observed nitrogen consumption of free amino acids (FAA), bound amino acids (BAA) and ammonia during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. ‘Control’ is without added peptides, ‘BSA’ is with added peptides. Standard deviations represent measurements in three technical replicates.
| Strain | N from FAA (mg·L−1) | N from BAA (mg·L−1) | N from NH4 (mg·L−1) | N Total (mg·L−1) | |
|---|---|---|---|---|---|
| Lalvin ICV Opale 2.0™ | Control | 70.20 ± 0.07 | - | 18.08 ± 6.87 | 88.2 ± 6.87 |
| BSA | 77.44 ± 0.22 | 74.4 ± 2.12 | 22.49 ± 2.23 | 174.3 ± 3.08 | |
| Lalvin Persy™ | Control | 75.8 ± 0.17 | - | 26.96 ± 6.35 | 102.8 ± 6.65 |
| BSA | 72.0 ± 0.18 | 57.3 ± 0.53 | 25.41 ± 5.07 | 154.7 ± 5.10 | |
| Lalvin QA23™ | Control | 75.50 ± 0.20 | - | 25.13 ± 6.19 | 100.6 ± 6.49 |
| BSA | 73.79 ± 0.16 | 51.7 ± 5.87 | 25.41 ± 5.07 | 150.9 ± 7.76 |
Appendix D
The proposed peptide candidates and their relative consumption by yeast

Table A4.
The proposed dipeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
Table A4.
The proposed dipeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
| Peptide Candidate | Retention Time (min) | Observed m/z | Charge | CCS (Å2) | Average Original Intensity | RSD | Lalvin ICV Opale 2.0™ (Consumption, %) | Lalvin Persy™ (Consumption, %) | Lalvin QA23™ (Consumption, %) |
|---|---|---|---|---|---|---|---|---|---|
| AH | 2.68 | 227.1134 | 1 | 150.41 | 807 | 4.86 | 99.97 | 100.00 | 99.98 |
| AW | 5.40 | 276.1338 | 1 | 160.10 | 1914 | 5.03 | 100.00 | 100.00 | 100.00 |
| AY | 4.56 | 253.1177 | 1 | 157.67 | 1470 | 6.44 | 100.00 | 100.00 | 100.00 |
| FT | 3.72 | 267.1335 | 1 | 163.99 | 5487 | 3.53 | 100.00 | 100.00 | 100.00 |
| FW | 7.03 | 352.1652 | 1 | 156.33 | 2617 | 5.18 | 100.00 | 100.00 | 100.00 |
| FY | 5.38 | 329.1487 | 1 | 179.32 | 698 | 3.61 | 97.94 | 95.49 | 95.50 |
| JT | 3.18 | 233.1492 | 1 | 156.95 | 2875 | 4.95 | 99.99 | 100.00 | 100.00 |
| KQ | 2.39 | 275.1709 | 1 | 160.14 | 1164 | 2.32 | 99.99 | 99.82 | 99.87 |
| KT | 1.40 | 248.1601 | 1 | 156.18 | 2201 | 8.75 | 99.97 | 99.98 | 100.00 |
| LL | 5.84 | 245.1856 | 1 | 166.83 | 20,475 | 3.04 | 100.00 | 100.00 | 100.00 |
| LY | 4.88 | 295.1648 | 1 | 171.64 | 4360 | 3.64 | 100.00 | 100.00 | 100.00 |
| TF | 4.78 | 267.1335 | 1 | 160.50 | 1507 | 3.99 | 100.00 | 99.87 | 100.00 |
| TK | 2.51 | 248.1600 | 1 | 156.18 | 2070 | 2.46 | 100.00 | 100.00 | 100.00 |
| VE | 2.14 | 247.1285 | 1 | 154.51 | 6009 | 6.03 | 99.96 | 99.98 | 99.95 |
| VF | 5.50 | 265.1542 | 1 | 164.08 | 5044 | 4.39 | 100.00 | 100.00 | 100.00 |
| VN | 1.37 | 232.1288 | 1 | 150.14 | 1012 | 2.78 | 99.98 | 100.00 | 100.00 |
| VR | 3.30 | 274.1866 | 1 | 167.20 | 1039 | 5.77 | 100.00 | 100.00 | 100.00 |
| YE | 3.72 | 311.1229 | 1 | 172.79 | 1091 | 4.24 | 99.67 | 100.00 | 100.00 |
| YY | 4.55 | 345.1441 | 1 | 184.22 | 5445 | 5.16 | 100.00 | 100.00 | 100.00 |

Table A5.
The proposed tripeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
Table A5.
The proposed tripeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
| Peptide Candidate | Retention Time (min) | Observed m/z | Charge | CCS (Å2) | Average Original Intensity | %RSD | Lalvin ICV Opale 2.0™ (Consumption, %) | Lalvin Persy™ (Consumption, %) | Lalvin QA23™ (Consumption, %) |
|---|---|---|---|---|---|---|---|---|---|
| AEF | 5.27 | 366.1656 | 1 | 181.69 | 5873 | 4.08 | 100.00 | 100.00 | 100.00 |
| AWS | 4.68 | 363.1658 | 1 | 181.79 | 2470 | 4.50 | 100.00 | 100.00 | 100.00 |
| DAF | 5.21 | 352.1496 | 1 | 176.71 | 1100 | 2.70 | 100.00 | 100.00 | 100.00 |
| DLL | 5.76 | 360.2124 | 1 | 189.23 | 2953 | 6.90 | 100.00 | 100.00 | 100.00 |
| DQF | 4.88 | 409.1712 | 1 | 195.14 | 728 | 3.28 | 100.00 | 100.00 | 100.00 |
| ELT | 4.01 | 362.1919 | 1 | 187.32 | 3543 | 2.99 | 99.98 | 99.62 | 99.92 |
| FKD | 3.53 | 205.1072 | 2 | 249.17 | 7981 | 3.00 | 100.00 | 100.00 | 100.00 |
| FKG | 3.33 | 176.1043 | 2 | 242.20 | 2685 | 2.41 | 100.00 | 100.00 | 100.00 |
| FLG | 5.65 | 336.1910 | 1 | 182.71 | 734 | 3.52 | 100.00 | 100.00 | 100.00 |
| FQE | 4.32 | 423.1866 | 1 | 198.49 | 658 | 3.50 | 100.00 | 100.00 | 100.00 |
| FSA | 4.73 | 324.1549 | 1 | 174.09 | 3223 | 2.23 | 100.00 | 100.00 | 100.00 |
| FTF | 7.23 | 414.2020 | 1 | 194.99 | 3313 | 6.62 | 100.00 | 100.00 | 100.00 |
| FVE | 4.88 | 394.1970 | 1 | 190.00 | 3242 | 3.60 | 100.00 | 100.00 | 100.00 |
| GSF | 4.64 | 310.1391 | 1 | 167.50 | 1014 | 6.12 | 100.00 | 100.00 | 100.00 |
| IAE | 3.72 | 332.1811 | 1 | 175.60 | 2444 | 4.33 | 100.00 | 100.00 | 100.00 |
| IAF | 6.63 | 350.2071 | 1 | 180.40 | 5291 | 6.60 | 100.00 | 100.00 | 100.00 |
| IAH | 2.66 | 340.1973 | 1 | 182.57 | 1628 | 7.50 | 100.00 | 100.00 | 100.00 |
| IAR | 3.08 | 359.2396 | 1 | 185.57 | 2132 | 6.89 | 100.00 | 100.00 | 100.00 |
| ISL | 6.42 | 332.2170 | 1 | 182.85 | 933 | 5.48 | 100.00 | 100.00 | 100.00 |
| IVR | 3.31 | 194.1387 | 2 | 253.10 | 4260 | 4.53 | 99.92 | 99.99 | 99.80 |
| KIE | 3.26 | 195.1228 | 2 | 243.74 | 2444 | 6.39 | 99.61 | 99.86 | 99.84 |
| KQI | 2.40 | 194.6309 | 2 | 246.86 | 8652 | 6.83 | 100.00 | 100.00 | 100.00 |
| LAK | 2.50 | 331.2332 | 1 | 181.06 | 482 | 3.99 | 100.00 | 100.00 | 100.00 |
| LEE | 3.90 | 390.1876 | 1 | 188.27 | 1557 | 5.17 | 100.00 | 93.10 | 94.29 |
| LFT | 5.92 | 380.2172 | 1 | 190.43 | 706 | 3.45 | 100.00 | 100.00 | 100.00 |
| LLF | 8.00 | 392.2539 | 1 | 197.53 | 1399 | 5.91 | 100.00 | 100.00 | 100.00 |
| LSQ | 3.30 | 347.1919 | 1 | 178.69 | 3179 | 5.20 | 100.00 | 100.00 | 100.00 |
| LVE | 4.37 | 360.2126 | 1 | 183.71 | 18,437 | 5.06 | 100.00 | 100.00 | 100.00 |
| LVN | 3.77 | 345.2125 | 1 | 180.57 | 990 | 4.59 | 100.00 | 100.00 | 100.00 |
| LYE | 4.68 | 424.2076 | 1 | 196.58 | 9328 | 3.77 | 100.00 | 100.00 | 100.00 |
| LYY | 6.08 | 458.2284 | 1 | 207.03 | 3200 | 6.61 | 100.00 | 99.99 | 100.00 |
| SQY | 3.78 | 397.1714 | 1 | 186.22 | 3568 | 5.93 | 100.00 | 100.00 | 100.00 |
| SVL | 5.63 | 318.2013 | 1 | 176.11 | 265 | 7.45 | 100.00 | 100.00 | 99.94 |
| TEF | 5.23 | 396.1756 | 1 | 189.94 | 620 | 3.45 | 100.00 | 100.00 | 100.00 |
| TLV | 4.44 | 332.2174 | 1 | 181.03 | 1307 | 2.71 | 100.00 | 100.00 | 100.00 |
| VAF | 6.01 | 336.1915 | 1 | 175.46 | 10,184 | 2.70 | 100.00 | 99.96 | 100.00 |
| VNE | 3.12 | 361.1714 | 1 | 178.22 | 5809 | 4.62 | 100.00 | 100.00 | 100.00 |
| VTF | 5.96 | 366.2019 | 1 | 179.87 | 1740 | 6.69 | 100.00 | 100.00 | 100.00 |
| VTK | 1.40 | 347.2286 | 1 | 180.50 | 3469 | 6.77 | 100.00 | 100.00 | 100.00 |
| YEY | 5.06 | 474.1869 | 1 | 208.53 | 3085 | 5.50 | 99.98 | 100.00 | 99.99 |
| YNG | 3.07 | 353.1448 | 1 | 183.95 | 737 | 3.00 | 100.00 | 100.00 | 100.00 |

Table A6.
The proposed tetrapeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
Table A6.
The proposed tetrapeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
| Peptide Candidate | Retention Time (min) | Observed m/z | Charge | CCS (Å2) | Average Original Intensity | %RSD | Lalvin ICV Opale 2.0™ (Consumption, %) | Lalvin Persy™ (Consumption, %) | Lalvin QA23™ (Consumption, %) |
|---|---|---|---|---|---|---|---|---|---|
| ALVE | 5.13 | 431.2496 | 1 | 200.15 | 3448 | 7.14 | 100.00 | 98.07 | 99.34 |
| FAKT | 3.49 | 233.6360 | 2 | 266.06 | 2319 | 7.68 | 100.00 | 100.00 | 100.00 |
| FEGK | 5.44 | 269.1441 | 2 | 279.98 | 9697 | 5.25 | 74.05 | 46.42 | 96.43 |
| FEKL | 5.27 | 268.6570 | 2 | 276.78 | 1106 | 8.84 | 96.81 | 95.23 | 93.37 |
| FHAD | 3.66 | 245.1077 | 2 | 262.19 | 5008 | 5.24 | 100.00 | 100.00 | 99.99 |
| FKDL | 5.26 | 261.6491 | 2 | 277.18 | 3826 | 7.25 | 100.00 | 100.00 | 99.40 |
| FLGS | 5.62 | 423.2233 | 1 | 198.49 | 1214 | 1.92 | 100.00 | 100.00 | 100.00 |
| FSQY | 5.07 | 544.2400 | 1 | 222.40 | 5451 | 4.56 | 99.89 | 99.65 | 99.77 |
| GERA | 1.71 | 216.6131 | 2 | 254.59 | 646 | 5.90 | 84.37 | 71.04 | 78.31 |
| IETM | 5.11 | 493.2326 | 1 | 215.78 | 10,279 | 5.05 | 100.00 | 95.85 | 98.39 |
| IKQN | 1.91 | 251.6520 | 2 | 261.81 | 745 | 8.35 | 100.00 | 95.20 | 98.59 |
| ISSK | 1.99 | 217.6335 | 2 | 257.65 | 1951 | 10.49 | 100.00 | 99.71 | 99.07 |
| KDAF | 4.45 | 480.2451 | 1 | 204.55 | 4169 | 6.61 | 93.70 | 37.87 | 41.44 |
| LEKS | 2.52 | 238.6388 | 2 | 262.58 | 3371 | 5.37 | 100.00 | 100.00 | 99.97 |
| LGSF | 6.08 | 423.2236 | 1 | 196.61 | 7284 | 4.72 | 93.04 | 76.01 | 95.77 |
| LILN | 6.24 | 472.3125 | 1 | 218.28 | 2581 | 5.87 | 100.00 | 100.00 | 100.00 |
| LIVR | 4.82 | 250.6806 | 2 | 277.82 | 1165 | 3.78 | 75.70 | 49.87 | 50.76 |
| LLEK | 4.13 | 251.6648 | 2 | 271.33 | 4134 | 3.64 | 91.38 | 48.34 | 49.20 |
| LRET | 3.30 | 259.6495 | 2 | 267.68 | 915 | 7.21 | 100.00 | 100.00 | 100.00 |
| LTAD | 3.81 | 419.2132 | 1 | 194.85 | 2055 | 6.78 | 100.00 | 100.00 | 100.00 |
| LTEF | 6.28 | 509.2602 | 1 | 217.34 | 2520 | 6.27 | 100.00 | 100.00 | 100.00 |
| LVEL | 6.85 | 473.2970 | 1 | 214.35 | 7655 | 5.36 | 100.00 | 99.36 | 99.86 |
| LVNE | 4.15 | 474.2558 | 1 | 208.53 | 6118 | 6.06 | 99.95 | 99.71 | 99.98 |
| LVTD | 4.33 | 447.2446 | 1 | 201.61 | 8109 | 3.72 | 100.00 | 99.72 | 99.85 |
| MENF | 5.75 | 540.2122 | 1 | 222.49 | 2536 | 6.75 | 100.00 | 100.00 | 100.00 |
| TQTA | 2.58 | 420.2083 | 1 | 196.70 | 1160 | 6.33 | 100.00 | 100.00 | 100.00 |
| VASL | 2.93 | 195.1228 | 2 | 253.02 | 1457 | 5.55 | 99.77 | 99.68 | 99.85 |
| VEVS | 4.02 | 433.2284 | 1 | 198.21 | 2150 | 6.29 | 98.70 | 97.71 | 97.77 |
| VFDK | 4.02 | 254.6413 | 2 | 267.97 | 1976 | 5.98 | 69.07 | 35.82 | 53.99 |
| VSEK | 1.95 | 231.6310 | 2 | 259.87 | 2212 | 7.45 | 99.97 | 97.52 | 92.49 |
| VVST | 3.70 | 405.2340 | 1 | 191.53 | 3913 | 3.23 | 100.00 | 100.00 | 100.00 |

Table A7.
The proposed pentapeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
Table A7.
The proposed pentapeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
| Peptide Candidate | Retention Time (min) | Observed m/z | Charge | CCS (Å2) | Average Original Intensity | RSD | Lalvin ICV Opale 2.0™ (Consumption, %) | Lalvin Persy™ (Consumption, %) | Lalvin QA23™ (Consumption, %) |
|---|---|---|---|---|---|---|---|---|---|
| AFDEK | 3.91 | 305.1472 | 2 | 274.91 | 15,458 | 6.56 | 99.93 | 36.21 | 41.18 |
| AIPEN | 4.24 | 543.2772 | 1 | 218.50 | 8777 | 4.98 | 96.76 | 33.49 | 33.68 |
| ALVEL | 7.32 | 544.3338 | 1 | 230.33 | 912 | 7.41 | 99.99 | 40.31 | 48.36 |
| FDEKL | 5.50 | 326.1703 | 2 | 300.05 | 5931 | 5.12 | 100.00 | 58.69 | 61.54 |
| FLGSF | 7.55 | 570.2920 | 1 | 231.74 | 934 | 6.74 | 99.96 | 33.44 | 31.64 |
| FYAPE | 5.74 | 626.2814 | 1 | 244.72 | 2299 | 4.18 | 100.00 | 66.49 | 66.40 |
| GFQNA | 4.50 | 536.2458 | 1 | 220.62 | 1214 | 7.16 | 78.76 | 34.47 | 38.77 |
| KFWGK | 5.00 | 333.1909 | 2 | 296.41 | 878 | 4.19 | 100.00 | 99.98 | 100.00 |
| LAKEY | 4.23 | 312.1727 | 2 | 277.79 | 4602 | 2.52 | 100.00 | 61.22 | 91.98 |
| LFGDE | 6.00 | 580.2609 | 1 | 227.53 | 825 | 7.83 | 99.79 | 28.10 | 35.07 |
| LGEYG | 4.95 | 538.2504 | 1 | 218.61 | 831 | 3.51 | 90.99 | 32.33 | 35.78 |
| LILNR | 5.33 | 314.7094 | 2 | 287.40 | 797 | 7.07 | 99.75 | 30.78 | 47.93 |
| LPKIE | 5.42 | 300.1912 | 2 | 284.85 | 8843 | 6.91 | 100.00 | 60.78 | 99.87 |
| LVELL | 7.98 | 586.3803 | 1 | 241.49 | 707 | 5.49 | 90.22 | 33.34 | 64.02 |
| LVEVS | 5.21 | 546.3130 | 1 | 224.32 | 4239 | 0.63 | 99.26 | 39.13 | 54.87 |
| TAPEL | 6.26 | 592.2976 | 1 | 235.28 | 15,546 | 5.29 | 100.00 | 93.48 | 93.12 |
| TVFDK | 4.61 | 305.1650 | 2 | 284.60 | 2748 | 4.80 | 99.98 | 66.96 | 61.71 |
| VEVTK | 3.65 | 288.1730 | 2 | 275.74 | 7041 | 8.04 | 98.83 | 39.91 | 48.04 |
| VVSTQ | 3.53 | 533.2928 | 1 | 218.72 | 3638 | 5.90 | 98.97 | 43.60 | 46.92 |

Table A8.
The proposed hexapeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). * Elution within void volume. Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
Table A8.
The proposed hexapeptide sequences and observed relative consumption during fermentation with Lalvin ICV Opale 2.0™, Lalvin Persy™ and Lalvin QA23™ of the modified synthetic must. Peptide sequences were acquired by peptide mapping to bovine serum albumin protein sequence (UniProtKB–P02769 (ALBU_BOVIN)). * Elution within void volume. Additionally presented are the retention time, observed m/z, collision cross section values, as well as the averaged peptide intensities at the starting point and associated relative standard deviations.
| Peptide Candidate | Retention Time (min) | Observed m/z | Charge | CCS (Å2) | Average Original Intensity | %RSD | Lalvin ICV Opale 2.0™ (Consumption, %) | Lalvin Persy™ (Consumption, %) | Lalvin QA23™ (Consumption, %) |
|---|---|---|---|---|---|---|---|---|---|
| AFDEKL | 5.70 | 361.6892 | 2 | 288.60 | 36,673 | 6.08 | 88.2 | 42.5 | 40.9 |
| DAFLGS | 3.91 | 305.1472 | 2 | 274.91 | 15,458 | 6.56 | 99.9 | 36.2 | 41.2 |
| DTHKSE * | 1.19 | 358.6637 | 2 | 292.00 | 4537 | 2.90 | 95.4 | 8.3 | 11.6 |
| KDAIPE | 4.55 | 336.6810 | 2 | 286.40 | 952 | 7.69 | 71.7 | 35.6 | 27.2 |
| KFGERA | 3.66 | 354.1946 | 2 | 298.79 | 1133 | 3.60 | 93.9 | 22.1 | 27.9 |
| KFPKAE | 4.01 | 360.2070 | 2 | 308.56 | 2291 | 5.69 | 100.0 | 39.4 | 44.3 |
| LFGDEL | 7.43 | 693.3452 | 1 | 253.69 | 902 | 10.18 | 100.0 | 34.2 | 32.5 |
| LLPKIE | 6.10 | 356.7331 | 2 | 315.47 | 1720 | 7.65 | 100.0 | 41.7 | 46.1 |
| LPKIET | 5.60 | 350.7149 | 2 | 292.34 | 7905 | 6.27 | 96.9 | 31.0 | 31.1 |
| NLPPLT | 6.57 | 654.3815 | 1 | 248.25 | 1517 | 7.63 | 80.2 | 51.5 | 43.0 |
| TVFDKL | 6.40 | 361.7072 | 2 | 295.17 | 2718 | 5.65 | 100.0 | 35.9 | 40.2 |
| VEGPKL | 5.33 | 321.6941 | 2 | 280.58 | 9864 | 5.33 | 85.6 | 38.7 | 39.6 |
| VSTPTL | 6.14 | 617.3495 | 1 | 238.79 | 674 | 5.56 | 100.0 | 43.6 | 46.0 |
References
- Cruz, S.H.; Cilli, E.M.; Ernandes, J.R. Structural Complexity of the Nitrogen Source and Influence on Yeast Growth and Fer-mentation. J. Inst. Brew. 2002, 108, 54–61. [Google Scholar] [CrossRef]
- Lei, H.; Zhao, H.; Zhao, M. Proteases supplementation to high gravity worts enhances fermentation performance of brewer’s yeast. Biochem. Eng. J. 2013, 77, 1–6. [Google Scholar] [CrossRef]
- Verbelen, P.J.; Delvaux, F.R. Brewing yeast in action: Beer fermentation. Appl. Mycol. 2009, 7, 110–135. [Google Scholar] [CrossRef]
- Nisbet, M.A.; Martinson, T.E.; Mansfield, A.K. Accumulation and Prediction of Yeast Assimilable Nitrogen in New York Winegrape Cultivars. Am. J. Enol. Vitic. 2014, 65, 325–332. [Google Scholar] [CrossRef]
- Gibson, B.R.; Lawrence, S.; LeClaire, J.P.R.; Powell, C.; Smart, K.A. Yeast responses to stresses associated with industrial brewery handling: Figure 1. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [CrossRef]
- Malfeito-Ferreira, M. Yeasts and wine off-flavours: A technological perspective. Ann. Microbiol. 2010, 61, 95–102. [Google Scholar] [CrossRef]
- Vilanova, M.; Ugliano, M.; Varela, C.; Siebert, T.; Pretorius, I.S.; Henschke, P.A. Assimilable Nitrogen Utilisation and Produc-tion of Volatile and Non-Volatile Compounds in Chemically Defined Medium by Saccharomyces Cerevisiae Wine Yeasts. Appl. Microbiol. Biotechnol. 2007, 77, 145–157. [Google Scholar] [CrossRef]
- Bely, M.; Rinaldi, A.; Dubourdieu, D. Influence of Assimilable Nitrogen on Volatile Acidity Production by Saccharomyces Cerevisiae during High Sugar Fermentation. J. Biosci. Bioeng. 2003, 96, 507–512. [Google Scholar] [CrossRef]
- Becerra-Rodríguez, C.; Marsit, S.; Galeote, V. Diversity of Oligopeptide Transport in Yeast and Its Impact on Adaptation to Winemaking Conditions. Front. Genet. 2020, 11, 602. [Google Scholar] [CrossRef]
- Becerra-Rodríguez, C.; Taghouti, G.; Portier, P.; Dequin, S.; Casal, M.; Paiva, S.; Galeote, V. Yeast Plasma Membrane Fungal Oligopeptide Transporters Display Distinct Substrate Preferences despite Their High Sequence Identity. J. Fungi 2021, 7, 963. [Google Scholar] [CrossRef]
- Ganapathy, V.; Leibach, F.H. Proton-coupled solute transport in the animal cell plasma membrane. Curr. Opin. Cell Biol. 1991, 3, 695–701. [Google Scholar] [CrossRef]
- Marsit, S.; Mena, A.; Bigey, F.; Sauvage, F.-X.; Couloux, A.; Guy, J.; Legras, J.-L.; Barrio, E.; Dequin, S.; Galeote, V. Evolutionary Advantage Conferred by an Eukaryote-to-Eukaryote Gene Transfer Event in Wine Yeasts. Mol. Biol. Evol. 2015, 32, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.W.; Smith, M.W. Peptide Transport by Micro-Organisms. Adv. Microb. Physiol. 1994, 36, 1–80. [Google Scholar] [CrossRef] [PubMed]
- Damon, C.; Vallon, L.; Zimmermann, S.; Haider, M.Z.; Galeote, V.; Dequin, S.; Luis, P.; Fraissinet-Tachet, L.; Marmeisse, R. A novel fungal family of oligopeptide transporters identified by functional metatranscriptomics of soil eukaryotes. ISME J. 2011, 5, 1871–1880. [Google Scholar] [CrossRef]
- Hauser, M.; Donhardt, A.M.; Barnes, D.; Naider, F.; Becker, J.M. Enkephalins Are Transported by a Novel Eukaryotic Peptide Uptake System. J. Biol. Chem. 2000, 275, 3037–3041. [Google Scholar] [CrossRef]
- Lubkowitz, M.A.; Barnes, D.; Breslav, M.; Burchfield, A.; Naider, F.; Becker, J.M. Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol. Microbiol. 2002, 28, 729–741. [Google Scholar] [CrossRef]
- Lubkowitz, M.A.; Hauser, L.; Breslav, M.; Naider, F.; Becker, J.M. An oligopeptide transport gene from Candida albicans. Microbiology 1997, 143, 387–396. [Google Scholar] [CrossRef]
- Wiles, A.M.; Cai, H.; Naider, F.; Becker, J.M. Nutrient regulation of oligopeptide transport in Saccharomyces cerevisiae. Microbiology 2006, 152, 3133–3145. [Google Scholar] [CrossRef]
- Harscoat-Schiavo, C.; Nioi, C.; Ronat-Heit, E.; Paris, C.; Vanderesse, R.; Fournier, F.; Marc, I. Hydrophilic properties as a new contribution for computer-aided identification of short peptides in complex mixtures. Anal. Bioanal. Chem. 2012, 403, 1939–1949. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Dias, F.F.G.; de Moura, J.M.L.N.; Barile, D. A complete workflow for discovering small bioactive peptides in foods by LC-MS/MS: A case study on almonds. Food Chem. 2021, 369, 130834. [Google Scholar] [CrossRef]
- Le Maux, S.; Nongonierma, A.B.; Murray, B.; Kelly, P.M.; FitzGerald, R.J. Identification of short peptide sequences in the nanofiltration permeate of a bioactive whey protein hydrolysate. Food Res. Int. 2015, 77, 534–539. [Google Scholar] [CrossRef]
- Piovesana, S.; Capriotti, A.L.; Cerrato, A.; Crescenzi, C.; La Barbera, G.; Laganà, A.; Montone, C.M.; Cavaliere, C. Graphitized Carbon Black Enrichment and UHPLC-MS/MS Allow to Meet the Challenge of Small Chain Peptidomics in Urine. Anal. Chem. 2019, 91, 11474–11481. [Google Scholar] [CrossRef] [PubMed]
- Sonsmann, G.; Römer, A.; Schomburg, D. Investigation of the influence of charge derivatization on the fragmentation of multiply protonated peptides. J. Am. Soc. Mass Spectrom. 2002, 13, 47–58. [Google Scholar] [CrossRef][Green Version]
- Hernández-Mesa, M.; Escourrou, A.; Monteau, F.; Le Bizec, B.; Dervilly-Pinel, G. Current applications and perspectives of ion mobility spectrometry to answer chemical food safety issues. TrAC Trends Anal. Chem. 2017, 94, 39–53. [Google Scholar] [CrossRef]
- Arju, G.; Taivosalo, A.; Pismennoi, D.; Lints, T.; Vilu, R.; Daneberga, Z.; Vorslova, S.; Renkonen, R.; Joenvaara, S. Application of the UHPLC-DIA-HRMS Method for Determination of Cheese Peptides. Foods 2020, 9, 979. [Google Scholar] [CrossRef]
- Campuzano, I.D.G.; Giles, K. Historical, Current and Future Developments of Travelling Wave Ion Mobility Mass Spectrom-etry: A Personal Perspective. TrAC Trends Anal. Chem. 2019, 120, 115620. [Google Scholar] [CrossRef]
- Fiechter, G.; Mayer, H. UPLC analysis of free amino acids in wines: Profiling of on-lees aged wines. J. Chromatogr. B 2011, 879, 1361–1366. [Google Scholar] [CrossRef]
- Salmon, J.-M.; Barre, P. Improvement of Nitrogen Assimilation and Fermentation Kinetics under Enological Conditions by Derepression of Alternative Nitrogen-Assimilatory Pathways in an Industrial Saccharomyces cerevisiae Strain. Appl. Environ. Microbiol. 1998, 64, 3831–3837. [Google Scholar] [CrossRef]
- Bell, S.-J.; Henschke, P.A. Implications of Nitrogen Nutrition for Grapes, Fermentation and Wine. Aust. J. Grape Wine Res. 2008, 11, 242–295. [Google Scholar] [CrossRef]
- Bely, M.; Sablayrolles, J.-M.; Barre, P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 1990, 70, 246–252. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Enzymic Hydrolysis of Food Proteins; Elsevier Science Limited: London, UK, 1986. [Google Scholar]
- Pasupuleti, V.K.; Demain, A.L. Protein Hydrolysates in Biotechnology; Springer: Cham, Switzerland, 2010. [Google Scholar] [CrossRef]
- Aloo, S.O.; Oh, D.-H. The Functional Interplay between Gut Microbiota, Protein Hydrolysates/Bioactive Peptides, and Obesity: A Critical Review on the Study Advances. Antioxidants 2022, 11, 333. [Google Scholar] [CrossRef] [PubMed]
- Colla, G.; Hoagland, L.; Ruzzi, M.; Cardarelli, M.; Bonini, P.; Canaguier, R.; Rouphael, Y. Biostimulant Action of Protein Hy-drolysates: Unraveling Their Effects on Plant Physiology and Microbiome. Front. Plant Sci. 2017, 8, 2202. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.J.; Kies, A.K.; Saris, W.H. Protein and protein hydrolysates in sports nutrition. Int. J. Sport Nutr. Exerc. Metab. 2007, 17, S1–S4. [Google Scholar] [CrossRef] [PubMed]
- Liigand, P.; Kaupmees, K.; Kruve, A. Influence of the amino acid composition on the ionization efficiencies of small peptides. Biol. Mass Spectrom. 2019, 54, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson-Bunch, C.; Sanford, J.A.; Hansen, J.R.; Gritsenko, M.A.; Rodland, K.D.; Piehowski, P.D.; Qian, W.-J.; Adkins, J.N. Assessment of TMT Labeling Efficiency in Large-Scale Quantitative Proteomics: The Critical Effect of Sample pH. ACS Omega 2021, 6, 12660–12666. [Google Scholar] [CrossRef]
- Waliczek, M.; Kijewska, M.; Rudowska, M.; Setner, B.; Stefanowicz, P.; Szewczuk, Z. Peptides Labeled with Pyridinium Salts for Sensitive Detection and Sequencing by Electrospray Tandem Mass Spectrometry. Sci. Rep. 2016, 6, 37720. [Google Scholar] [CrossRef]
- Zecha, J.; Satpathy, S.; Kanashova, T.; Avanessian, S.C.; Kane, H.; Clauser, K.; Mertins, P.; Carr, S.A.; Kuster, B. TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach. Mol. Cell. Proteom. 2019, 18, 1468–1478. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).