Bio-Fermentation Improved Rumen Fermentation and Decreased Methane Concentration of Rice Straw by Altering the Particle-Attached Microbial Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substrates Preparation
2.2. In Vitro Rumen Fermentation and Sample Collection
2.3. Analysis of In Vitro Fiber Digestibility of Substrates
2.4. Analysis of Substrate Fermentation Products
2.5. Analysis of Bacteria and Archaea Loosely or Tightly Attached to Substrates by Illumina Hiseq Sequencing of 16S Rrna Gene
2.6. Statistical Analysis
3. Results
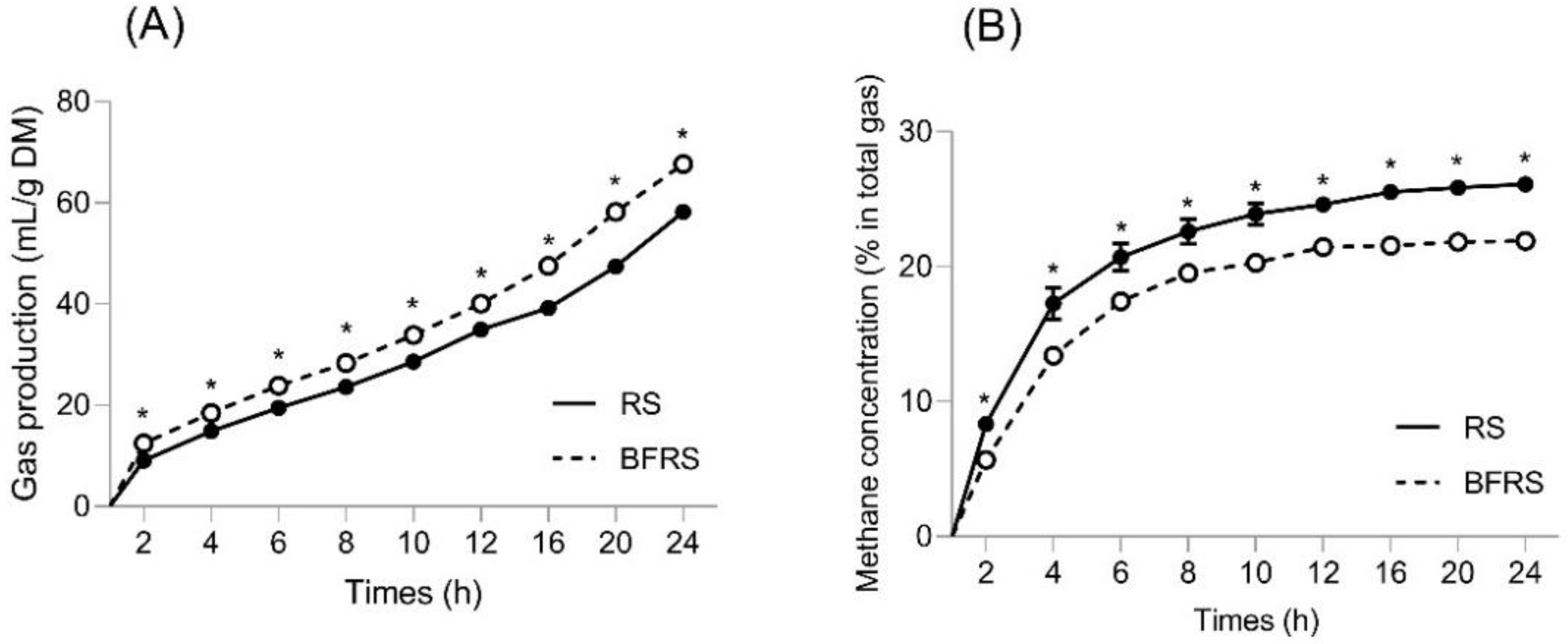
3.1. Effect of Bio-Fermentation on Gas Production, Methane Concentration, Fiber Degradation and In Vitro Fermentation Products of Rice Straw
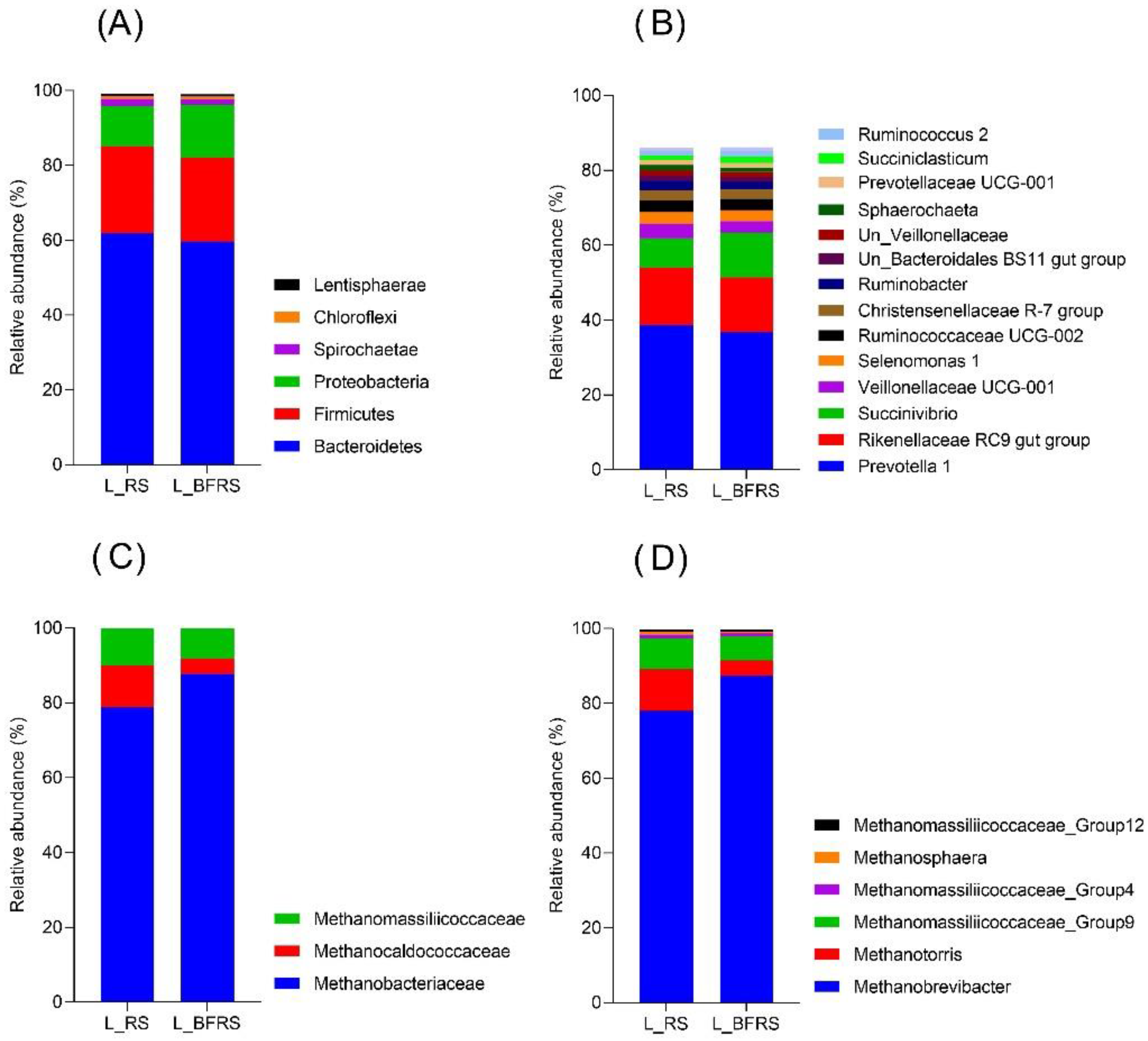
3.2. Effect of Bio-Fermentation on Loosely Attached Bacterial and Archaeal Community during In Vitro Rumen Fermentation
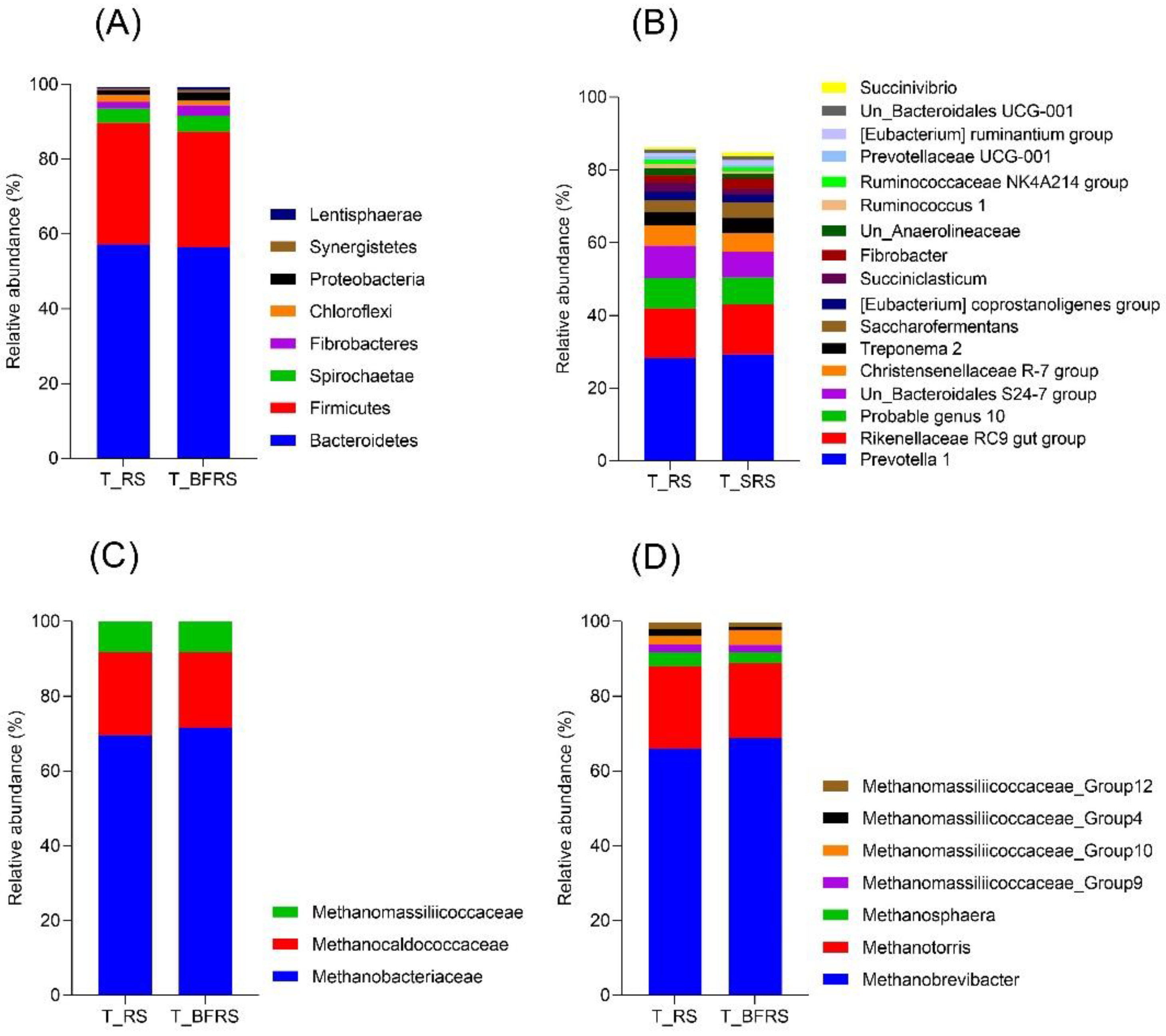
3.3. Effect of Bio-Fermentation on the Tightly Attached Bacterial and Archaeal Community during the In Vitro Rumen Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, Q.; Zhuang, X.S.; Wang, W.; Qi, W.; Wang, Q.; Tan, X.S.; Kong, X.Y.; Yuan, Z.H. Hemicellulose and lignin removal to improve the enzymatic digestibility and ethanol production. Biomass Bioenergy 2016, 94, 105–109. [Google Scholar] [CrossRef]
- Wanapat, M.; Polyorach, S.; Boonnop, K.; Mapato, C.; Cherdthong, A. Effects of treating rice straw with urea or urea and calcium hydroxide upon intake, digestibility, rumen fermentation and milk yield of dairy cows. Livest. Sci. 2009, 125, 238–243. [Google Scholar] [CrossRef]
- Bhutto, A.W.; Qureshi, K.; Harijan, K.; Abro, R.; Abbas, T.; Bazmi, A.A.; Karim, S.; Yu, G.R. Insight into progress in pre-treatment of lignocellulosic biomass. Energy 2017, 122, 724–745. [Google Scholar] [CrossRef]
- Tuyen, V.D.; Cone, J.W.; Baars, J.J.P.; Sonnenberg, A.S.M.; Hendriks, W.H. Fungal strain and incubation period affect chemical composition and nutrient availability of wheat straw for rumen fermentation. Bioresource Technol. 2012, 111, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Xing, L. Study on the Effects of Lactobacillus and Cellulase Additives on the Quality of Different Silages. Master’s Thesis, China Agricultural University, Beijing, China, 2004. (In Chinese). [Google Scholar]
- Li, J.; Shen, Y.X.; Cai, Y.M. Improvement of Fermentation Quality of Rice Straw Silage by Application of a Bacterial Inoculant and Glucose. Asian Australas. J. Anim. 2010, 23, 901–906. [Google Scholar] [CrossRef]
- Khota, W.; Pholsen, S.; Higgs, D.; Cai, Y.M. Fermentation quality and in vitro methane production of sorghum silage prepared with cellulase and lactic acid bacteria. Asian Australas. J. Anim. 2017, 30, 1568–1574. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. Fems Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Yanez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; McAllister, T.A. Rumen microbes, enzymes and feed digestion - A review. Asian Australas. J. Anim. 2002, 15, 1659–1676. [Google Scholar] [CrossRef]
- Mackie, R.I. Mutualistic fermentative digestion in the gastrointestinal tract: Diversity and evolution. Integr. Comp. Biol. 2002, 42, 319–326. [Google Scholar] [CrossRef]
- Jami, E.; Mizrahi, I. Composition and Similarity of Bovine Rumen Microbiota across Individual Animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.H.; Zhang, M.L.; Xue, C.X.; Zhu, W.Y.; Mao, S.Y. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J. Dairy Sci. 2016, 99, 9668–9681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.F.; Wang, Y.; Li, Y.F.; Zhang, Y.P.; Liu, T.Y.; Wang, Y.; Sharpton, T.J.; Zhu, W.Y. Progressive Colonization of Bacteria and Degradation of Rice Straw in the Rumen by Illumina Sequencing. Front. Microbiol. 2017, 8, 2165. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.M.; Forster, R.J.; Yang, W.; McKinnon, J.J.; McAllister, T.A. Characterization of rumen bacterial diversity and fermentation parameters in concentrate fed cattle with and without forage. J. Appl. Microbiol. 2012, 112, 1152–1162. [Google Scholar] [CrossRef]
- Yang, C.T.; Si, B.W.; Diao, Q.Y.; Jin, H.; Zeng, S.Q.; Tu, Y. Rumen fermentation and bacterial communities in weaned Chahaer lambs on diets with different protein levels. J. Integr. Agric. 2016, 15, 1564–1574. [Google Scholar] [CrossRef]
- Mould, F.L.; Kliem, K.E.; Morgan, R.; Mauricio, R.M. In vitro microbial inoculum: A review of its function and properties. Anim. Feed Sci. Technol. 2005, 123, 31–50. [Google Scholar] [CrossRef]
- Zapletalova, M.; Kasparovska, J.; Krizova, L.; Kasparovsky, T.; Sery, O.; Lochman, J. Bacterial community dynamics in a rumen fluid bioreactor during in-vitro cultivation. J. Biotechnol. 2016, 234, 43–49. [Google Scholar] [CrossRef]
- Menke, K.H.; Steingass, H. Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Dev. 1988, 28, 7–55. [Google Scholar]
- Theodorou, M.K.; Williams, B.A.; Dhanoa, M.S.; Mcallan, A.B.; France, J. A Simple Gas-Production Method Using a Pressure Transducer to Determine the Fermentation Kinetics of Ruminant Feeds. Anim. Feed Sci. Technol. 1994, 48, 185–197. [Google Scholar] [CrossRef]
- Larue, R.; Yu, Z.T.; Parisi, V.A.; Egan, A.R.; Morrison, M. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 2005, 7, 530–543. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of the Association of the Official Analysis Chemists, 15th ed.; AOAC International Publisher: Washington, DC, USA, 1990. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Weatherburn, M. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Jin, W.; Cheng, Y.F.; Zhu, W.Y. The community structure of Methanomassiliicoccales in the rumen of Chinese goats and its response to a high-grain diet. J. Anim. Sci. Biotechnol. 2017, 8, 47. [Google Scholar] [CrossRef]
- Li, Y.F.; Jin, W.; Cheng, Y.F.; Zhu, W.Y. Effect of the Associated Methanogen Methanobrevibacter thaueri on the Dynamic Profile of End and Intermediate Metabolites of Anaerobic Fungus Piromyces sp F1. Curr. Microbiol. 2016, 73, 434–441. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Akkermans, A.D.L.; De Vos, W.M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 1998, 64, 3854–3859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittelmann, S.; Seedorf, H.; Walters, W.A.; Clemente, J.C.; Knight, R.; Gordon, J.I.; Janssen, P.H. Simultaneous Amplicon Sequencing to Explore Co-Occurrence Patterns of Bacterial, Archaeal and Eukaryotic Microorganisms in Rumen Microbial Communities. PLoS ONE 2013, 8, e47879. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Shoskes, D.; Altemus, J.; Polackwich, A.; Tucky, B.; Wang, H.; Eng, C. Analysis of Gut Microbiome Reveals Significant Differences between Men with Chronic Prostatitis/Chronic Pelvic Pain Syndrome and Controls. J. Urol. 2016, 195, E451–E452. [Google Scholar] [CrossRef]
- De Boever, J.L.; Aerts, J.M.; Vanacker, J.M.; De Brabander, D.L. Evaluation of the nutritive value of maize silages using a gas production technique. Anim. Feed Sci. Technol. 2005, 123, 255–265. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Magaji, U.; Hussin, G.; Ramli, A.; Miah, G. Fermentation Quality and Additives: A Case of Rice Straw Silage. Biomed. Res. Int. 2016, 2016, 7985167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Soest, P.J. Fibre and physicochemical properties of feeds. In Nutritional Ecology of the Ruminant, 2nd ed.; Thomas, L.P., Ed.; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Blümmel, M.; Becker, K. The degradability characteristics of fifty-four roughages and roughage neutral-detergent fibres as described by in vitro gas production and their relationship to voluntary feed intake. Br. J. Nutr. 1997, 77, 757–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallam, S.; Abdalla, A.L.; Nozella, E.F.; Bueno, I.; Vitti, D.; Godoy, P.B. Nutritive value assessment of the artichoke (Cynara scolymus) by-product as an alternative feed resource for ruminants. Trop. Subtrop. Agroecosyst. 2008, 8, 181–189. [Google Scholar]
- Zhang, X.M.; Wang, M.; Wang, R.; Ma, Z.Y.; Long, D.L.; Mao, H.X.; Wen, J.N.; Bernard, L.A.; Beauchemin, K.A.; Tan, Z.L. Urea plus nitrate pretreatment of rice and wheat straws enhances degradation and reduces methane production in in vitro ruminal culture. J. Sci. Food Agric. 2018, 98, 5205–5211. [Google Scholar] [CrossRef]
- Gharechahi, J.; Vahidi, M.F.; Ding, X.Z.; Han, J.L.; Salekdeh, G.H. Temporal changes in microbial communities attached to forages with different lignocellulosic compositions in cattle rumen. Fems Microbiol. Ecol. 2020, 96, fiaa069. [Google Scholar] [CrossRef]
- Kafilzadeh, F.; Heidary, N. Chemical composition, in vitrodigestibility and kinetics of fermentation of whole-crop forage from 18 different varieties of oat (Avena sativa L.). J. Appl. Anim. Res. 2013, 41, 61–68. [Google Scholar] [CrossRef]
- Getachew, G.; Robinson, P.H.; DePeters, E.J.; Taylor, S.J. Relationships between chemical composition, dry matter degradation and in vitro gas production of several ruminant feeds. Anim. Feed Sci. Technol. 2004, 111, 57–71. [Google Scholar] [CrossRef]
- Patra, A.K.; Aschenbach, J.R. Ureases in the gastrointestinal tracts of ruminant and monogastric animals and their implication in urea-N/ammonia metabolism: A review. J. Adv. Res. 2018, 13, 39–50. [Google Scholar] [CrossRef]
- Jin, D.; Zhao, S.G.; Zheng, N.; Beckers, Y.; Wang, J.Q. Urea Metabolism and Regulation by Rumen Bacterial Urease in Ruminants - a Review. Ann. Anim. Sci. 2018, 18, 303–318. [Google Scholar] [CrossRef] [Green Version]
- Zened, A.; Combes, S.; Cauquil, L.; Mariette, J.; Klopp, C.; Bouchez, O.; Troegeler-Meynadier, A.; Enjalbert, F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. Fems Microbiol. Ecol. 2013, 83, 504–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzler-Zebeli, B.U.; Schmitz-Esser, S.; Klevenhusen, F.; Podstatzky-Lichtenstein, L.; Wagner, M.; Zebeli, Q. Grain-rich diets differently alter ruminal and colonic abundance of microbial populations and lipopolysaccharide in goats. Anaerobe 2013, 20, 65–73. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Naas, A.E.; Mackenzie, A.K.; Mravec, J.; Schuckel, J.; Willats, W.G.T.; Eijsink, V.G.H.; Pope, P.B. Do Rumen Bacteroidetes Utilize an Alternative Mechanism for Cellulose Degradation? mBio 2014, 5, e01401-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.; Zhang, L.L.; Jin, W.; Meng, Z.X.; Cheng, Y.F.; Wang, J.; Zhu, W.Y. Methane Emission, Rumen Fermentation, and Microbial Community Response to a Nitrooxy Compound in Low-Quality Forage Fed Hu Sheep. Curr. Microbiol. 2019, 76, 435–441. [Google Scholar] [CrossRef]
- Purushe, J.; Fouts, D.E.; Morrison, M.; White, B.A.; Mackie, R.I.; Coutinho, P.M.; Henrissat, B.; Nelson, K.E.; Bacteria, N.A.C.R. Comparative Genome Analysis of Prevotella ruminicola and Prevotella bryantii: Insights into Their Environmental Niche. Microbiol. Ecol. 2010, 60, 721–729. [Google Scholar] [CrossRef]
- Dai, X.; Tian, Y.; Li, J.T.; Su, X.Y.; Wang, X.W.; Zhao, S.G.; Liu, L.; Luo, Y.F.; Liu, D.; Zheng, H.J.; et al. Metatranscriptomic Analyses of Plant Cell Wall Polysaccharide Degradation by Microorganisms in the Cow Rumen. Appl. Environ. Microbiol. 2015, 81, 1375–1386. [Google Scholar] [CrossRef] [Green Version]
- Pitta, D.W.; Pinchak, W.E.; Dowd, S.E.; Osterstock, J.; Gontcharova, V.; Youn, E.; Dorton, K.; Yoon, I.; Min, B.R.; Fulford, J.D.; et al. Rumen Bacterial Diversity Dynamics Associated with Changing from Bermudagrass Hay to Grazed Winter Wheat Diets. Microbiol. Ecol. 2010, 59, 511–522. [Google Scholar] [CrossRef]
- Su, X.L.; Tian, Q.; Zhang, J.; Yuan, X.Z.; Shi, X.S.; Guo, R.B.; Qiu, Y.L. Acetobacteroides hydrogenigenes gen. nov., sp nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int. J. Syst. Evol. Microbiol. 2014, 64, 2986–2991. [Google Scholar] [CrossRef]
- Agarussi, M.C.N.; Pereira, O.G.; de Paula, R.A.; da Silva, V.P.; Roseira, J.P.S.; Silva, F.F.E. Novel lactic acid bacteria strains as inoculants on alfalfa silage fermentation. Sci. Rep.-UK 2019, 9, 8007. [Google Scholar] [CrossRef] [Green Version]
- Gharechahi, J.; Salekdeh, G.H. A metagenomic analysis of the camel rumen’s microbiome identifies the major microbes responsible for lignocellulose degradation and fermentation. Biotechnol. Biofuels 2018, 11, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapebie, P.; Lombard, V.; Drula, E.; Terrapon, N.; Henrissat, B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019, 10, 2043. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Huang, B.L.; Fernandez-Garcia, V.; Miesel, J.; Yan, L.; Lv, C.Q. Biochar and Rhizobacteria Amendments Improve Several Soil Properties and Bacterial Diversity. Microorganisms 2020, 8, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suen, G.; Weimer, P.J.; Stevenson, D.M.; Aylward, F.O.; Boyum, J.; Deneke, J.; Drinkwater, C.; Ivanova, N.N.; Mikhailova, N.; Chertkov, O.; et al. The Complete Genome Sequence of Fibrobacter succinogenes S85 Reveals a Cellulolytic and Metabolic Specialist. PLoS ONE 2011, 6, e18814. [Google Scholar] [CrossRef]
- Fukuma, N.M.; Koike, S.; Kobayashi, Y. Monitoring of gene expression in Fibrobacter succinogenes S85 under the co-culture with non-fibrolytic ruminal bacteria. Arch. Microbiol. 2015, 197, 269–276. [Google Scholar] [CrossRef]
- Huws, S.A.; Edwards, J.E.; Creevey, C.J.; Stevens, P.R.; Lin, W.C.; Girdwood, S.E.; Pachebat, J.A.; Kingston-Smith, A.H. Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. Fems Microbiol. Ecol. 2016, 92, fiv137. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.Y.; Niu, L.L.; Zhang, Y.X. Saccharofermentans acetigenes gen. nov., sp nov., an anaerobic bacterium isolated from sludge treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 2010, 60, 2735–2738. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.Q.; Hou, Z.S.; Shi, Q.C.; Cheng, Y.F.; Zhu, W.Y. Methane Production From Different Parts of Corn Stover via a Simple Co-culture of an Anaerobic Fungus and Methanogen. Front. Bioeng. Biotechnol. 2020, 8, 314. [Google Scholar] [CrossRef]
- Koike, S.; Kobayashi, Y. Fibrolytic Rumen Bacteria: Their Ecology and Functions. Asian Australas. J. Anim. 2009, 22, 131–138. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T.; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; DeSilva, U. Rumen Microbial Population Dynamics during Adaptation to a High-Grain Diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.A.; Johnson, D.E. Methane Emissions from Cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Hwang, M.H.; Jang, N.J.; Hyun, S.H.; Lee, S.T. Effect of low pH on the activity of hydrogen utilizing methanogen in bio-hydrogen process. Int. J. Hydrog. Energ 2004, 29, 1133–1140. [Google Scholar] [CrossRef]
- Huang, J.Q.; Li, Y.J. Rumen methanogen and protozoal communities of Tibetan sheep and Gansu Alpine Finewool sheep grazing on the Qinghai-Tibetan Plateau, China. BMC Microbiol. 2018, 18, 212. [Google Scholar] [CrossRef] [Green Version]
- Janssen, P.H.; Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.L.; Singh, P.K.; Singh, M.P. Biomethanization of coal to obtain clean coal energy: A review. Energy Explor. Exploit. 2012, 30, 837–852. [Google Scholar] [CrossRef]
- Maspolim, Y.; Zhou, Y.; Guo, C.H.; Xiao, K.K.; Ng, W.J. Comparison of single-stage and two-phase anaerobic sludge digestion systems - Performance and microbial community dynamics. Chemosphere 2015, 140, 54–62. [Google Scholar] [CrossRef]
- Mauerhofer, L.M.; Zwirtmayr, S.; Pappenreiter, P.; Bernacchi, S.; Seifert, A.H.; Reischl, B.; Schmider, T.; Taubner, R.S.; Paulik, C.; Rittmann, S.K.M.R. Hyperthermophilic methanogenic archaea act as high-pressure CH4 cell factories. Commun. Biol. 2021, 4, 289. [Google Scholar] [CrossRef]
- Lang, K.; Schuldes, J.; Klingl, A.; Poehlein, A.; Daniel, R.; Brune, A. New Mode of Energy Metabolism in the Seventh Order of Methanogens as Revealed by Comparative Genome Analysis of “Candidatus Methanoplasma termitum”. Appl. Environ. Microbiol. 2015, 81, 1338–1352. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.D.; Tan, H.Y.; Long, R.J.; Liang, J.B.; Wright, A.D.G. Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai-Tibetan plateau, China. BMC Microbiol. 2012, 12, 237. [Google Scholar] [CrossRef] [Green Version]


| Items | Groups | |
|---|---|---|
| RS | BFRS | |
| Fiber digestibility | ||
| DMD, % | 26.44 ± 0.64 b | 31.27 ± 1.11 a |
| NDFD, % | 32.93 ± 0.86 b | 38.50 ± 1.31 a |
| ADFD, % | 23.94 ± 0.98 b | 31.76 ± 2.10 a |
| ADLD, % | 50.26 ± 1.95 | 47.06 ± 2.28 |
| HD, % | 41.65 ± 0.83 b | 45.81 ± 1.00 a |
| CD, % | 23.66 ± 1.40 b | 32.81 ± 1.03 a |
| Fermentation products | ||
| pH | 6.76 ± 0.03 | 6.75 ± 0.02 |
| Total VFA (mmol/L) | 51.78 ± 0.70 b | 56.16 ± 0.27 a |
| Acetate (mmol/L) | 30.65 ± 0.50 b | 33.52 ± 0.43 a |
| Propionate (mmol/L) | 11.65 ± 0.18 b | 13.03 ± 0.19 a |
| Acetate: Propionate | 2.63 ± 0.04 | 2.57 ± 0.04 |
| Isobutyrate (mmol/L) | 0.99 ± 0.01 | 0.97 ± 0.02 |
| Butyrate (mmol/L) | 6.52 ± 0.16 | 6.57 ± 0.19 |
| Isovalerate (mmol/L) | 1.50 ± 0.04 | 1.47 ± 0.08 |
| Valerate (mmol/L) | 0.48 ± 0.01 b | 0.60 ± 0.01 a |
| Lactate (mmol/L) | 0.16 ± 0.01 b | 0.21 ± 0.01 a |
| NH3-N (mg/dL) | 9.81 ± 0.16 b | 11.02 ± 0.26 a |
| MCP (mg/dL) | 10.36 ± 0.37 b | 12.05 ± 0.31 a |
| Items | Loosely Attached Fraction | Tightly Attached Fraction | ||
|---|---|---|---|---|
| RS | BFRS | RS | BFRS | |
| Bacteria | ||||
| Number of ASV | 355.75 ± 2.06 | 350.50 ± 20.34 | 448.75 ± 8.59 | 461.50 ± 3.59 |
| Evenness | 0.74 ± 0.00 | 0.72 ± 0.03 | 0.75 ± 0.00 b | 0.76 ± 0.00 a |
| Faith_pd | 27.10 ± 0.22 | 25.78 ± 0.85 | 29.99 ± 0.35 | 30.56 ± 0.18 |
| Shannon | 6.25 ± 0.03 | 6.08 ± 0.31 | 6.55 ± 0.04 b | 6.72 ± 0.04 a |
| Archaea | ||||
| Number of ASV | 55.00 ± 12.08 | 49.00 ± 8.69 | 96.50 ± 16.66 | 96.25 ± 16.75 |
| Evenness | 0.60 ± 0.02 | 0.51 ± 0.07 | 0.58 ± 0.01 | 0.58 ± 0.01 |
| Faith_pd | 24.96 ± 6.69 | 22.98 ± 5.25 | 42.93 ± 10.16 | 45.55 ± 8.85 |
| Shannon | 3.43 ± 0.26 | 2.78 ± 0.36 | 3.81 ± 0.18 | 3.79 ± 0.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Aung, M.; Sun, Z.; Zhou, Y.; Cheng, Y.; Hao, L.; Padmakumar, V.; Zhu, W. Bio-Fermentation Improved Rumen Fermentation and Decreased Methane Concentration of Rice Straw by Altering the Particle-Attached Microbial Community. Fermentation 2022, 8, 72. https://doi.org/10.3390/fermentation8020072
Xu Y, Aung M, Sun Z, Zhou Y, Cheng Y, Hao L, Padmakumar V, Zhu W. Bio-Fermentation Improved Rumen Fermentation and Decreased Methane Concentration of Rice Straw by Altering the Particle-Attached Microbial Community. Fermentation. 2022; 8(2):72. https://doi.org/10.3390/fermentation8020072
Chicago/Turabian StyleXu, Yao, Min Aung, Zhanying Sun, Yaqi Zhou, Yanfen Cheng, Lizhuang Hao, Varijakshapanicker Padmakumar, and Weiyun Zhu. 2022. "Bio-Fermentation Improved Rumen Fermentation and Decreased Methane Concentration of Rice Straw by Altering the Particle-Attached Microbial Community" Fermentation 8, no. 2: 72. https://doi.org/10.3390/fermentation8020072
APA StyleXu, Y., Aung, M., Sun, Z., Zhou, Y., Cheng, Y., Hao, L., Padmakumar, V., & Zhu, W. (2022). Bio-Fermentation Improved Rumen Fermentation and Decreased Methane Concentration of Rice Straw by Altering the Particle-Attached Microbial Community. Fermentation, 8(2), 72. https://doi.org/10.3390/fermentation8020072








