Molecular Characterization of a Stable and Robust L-Asparaginase from Pseudomonas sp. PCH199: Evaluation of Cytotoxicity and Acrylamide Mitigation Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Bacterial Strains, and Cell Line
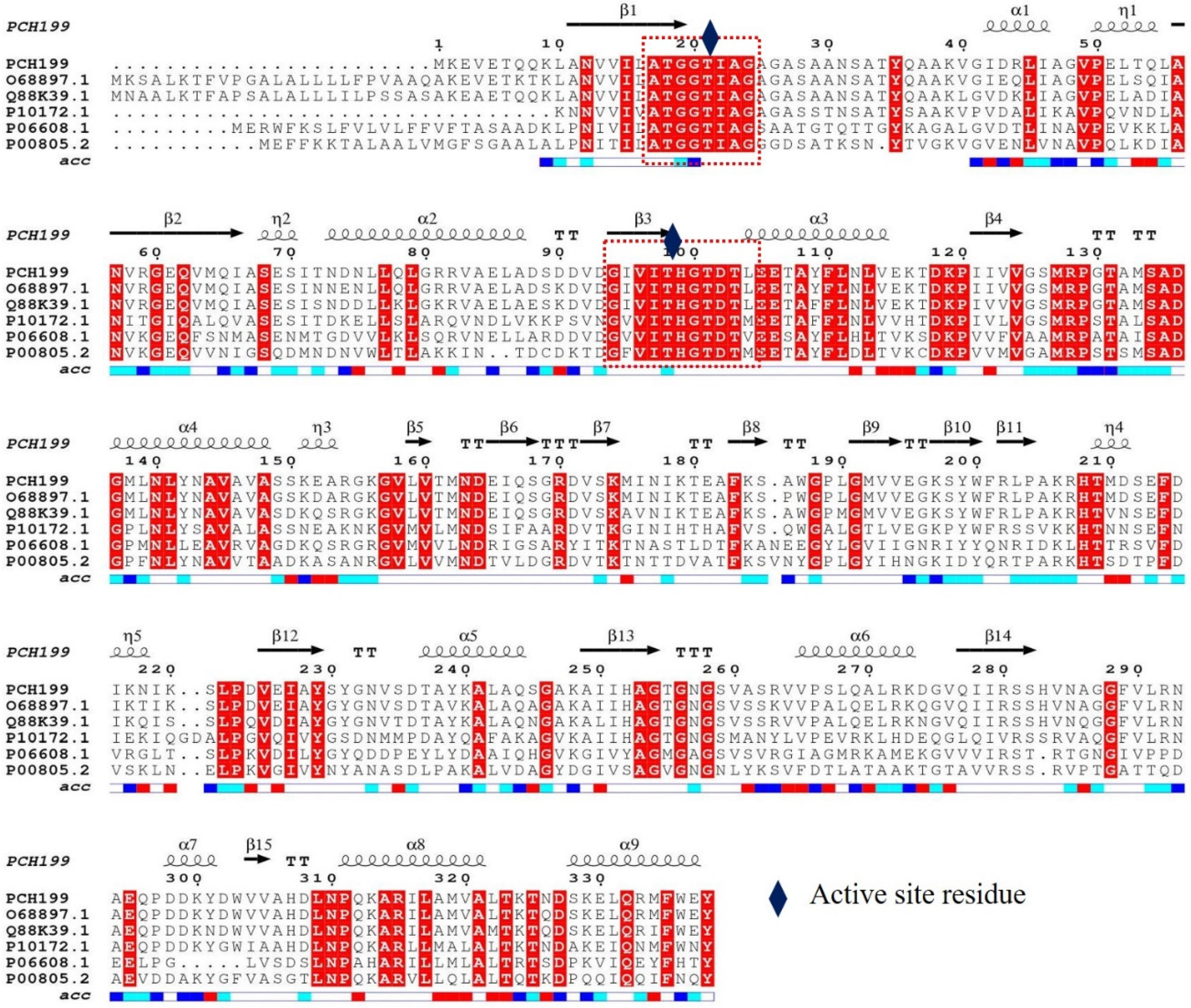
2.2. Sequence Analysis and Cloning of Pg-asn II
2.3. Expression of Recombinant Pg-ASNase II
2.4. Production Parameter Optimization
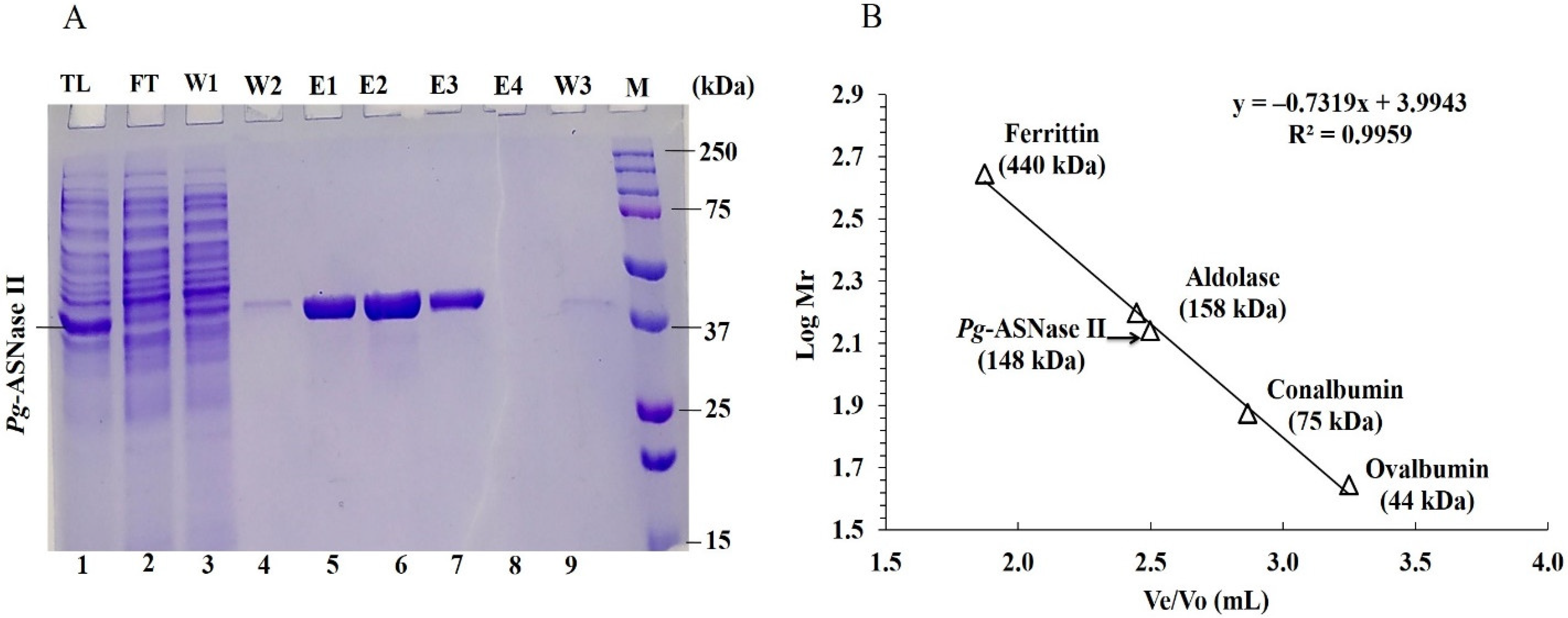
2.5. Purification of Recombinant Pg-ASNase II
2.6. Pg-ASNase II Activity
2.7. Biochemical Characteristics of Pg-ASNase II
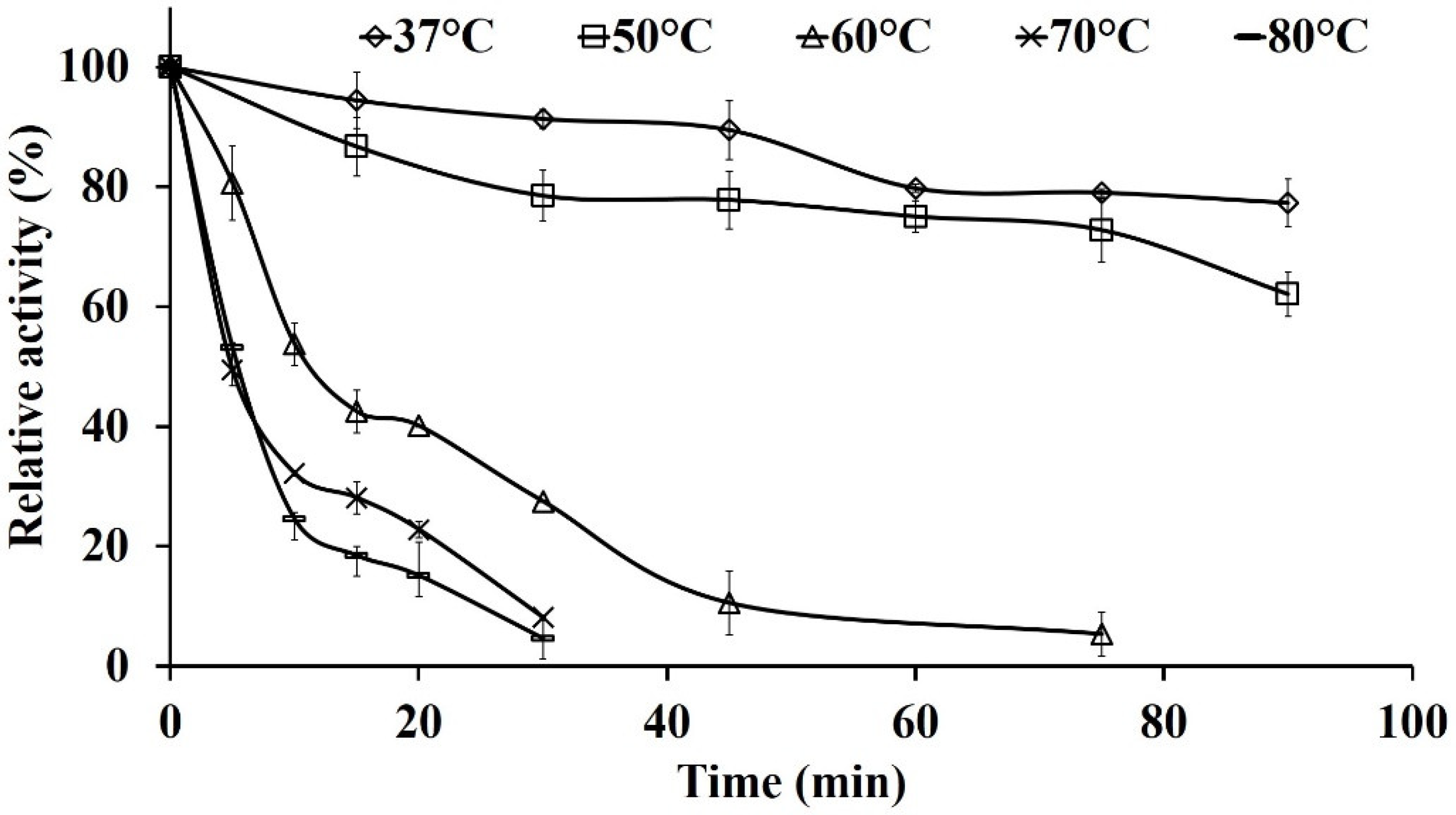
2.7.1. Thermal Stability of Pg-ASNase II
Evaluation of Deactivation Constant (kd) and Half-life Time (t1/2)
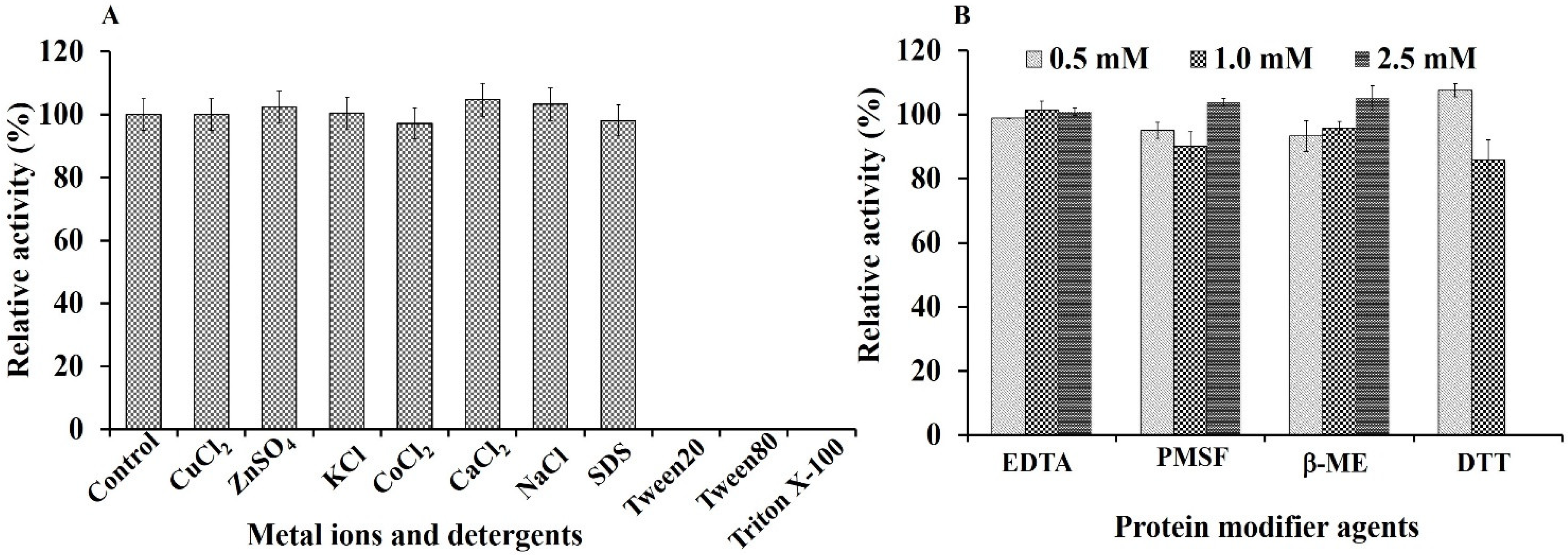
2.7.2. Effect of Metal Ions, Detergents, and Protein Modifying Agents
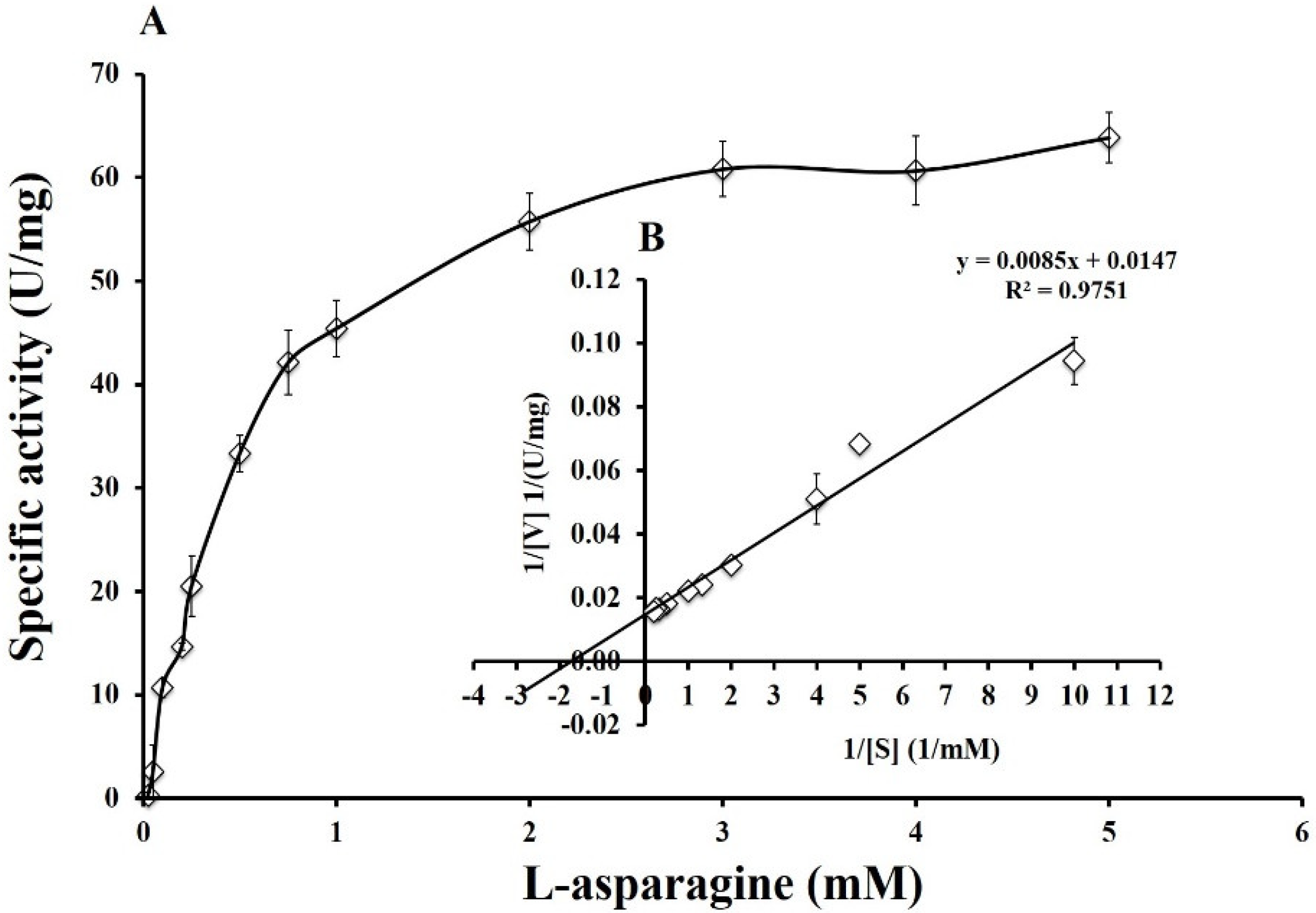
2.7.3. Determination of Kinetic Parameters: Km, Vmax, kcat, and kcat/Km
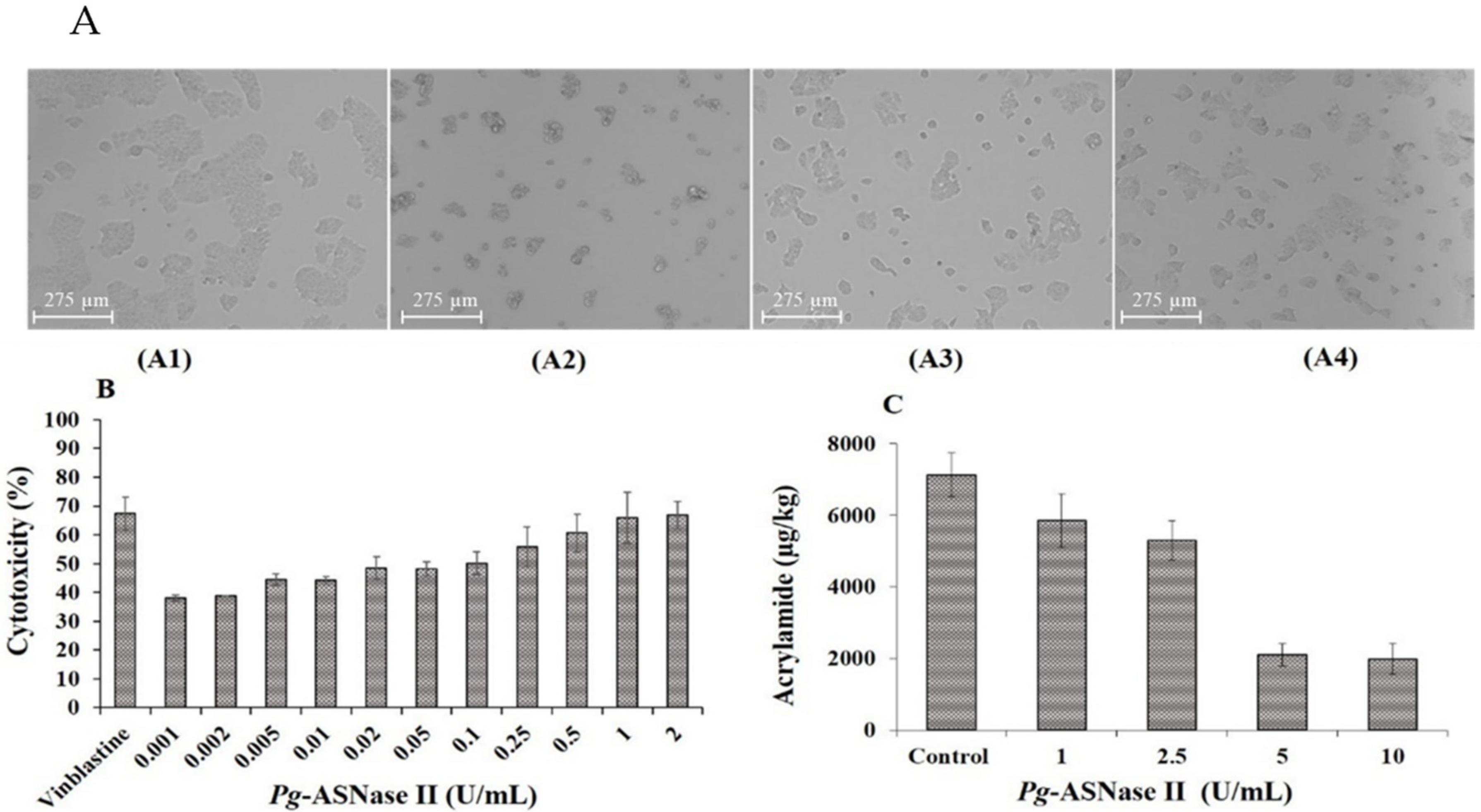
2.8. Cytotoxicity Effect of Pg-ASNase II on Cancer Cell Line
2.9. Estimation of Acrylamide Mitigation in the Potato Chips
3. Results
3.1. Sequence Analysis, Cloning, and Expression of Pg-ASNase II
3.2. Production Parameter Optimization
3.3. Purification of Pg-ASNase II
3.4. Effect of Different Reaction Parameters on Enzyme Activity
3.5. Effect of Metal Ions, Detergents, and Protein Modifier Agents
3.6. Enzyme Kinetics of Pg-ASNase II
3.7. Anti-Proliferation Studies
3.8. Acrylamide Reduction
4. Discussion
4.1. In-Silico Analysis, Expression, and Purification of Pg-ASNase II
4.2. Effect of Reaction Conditions and Kinetic Behavior of Pg-ASNase II
4.3. Potential Applications of Pg-ASNase II
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Zhang, X.; Xu, S.; Xu, M.; Yang, T.; Wang, L.; Zhang, H.; Fang, H.; Osire, T.; Rao, Z. Insight into the thermostability of thermophilic L-asparaginase and non-thermophilic L-asparaginase II through bioinformatics and structural analysis. Appl. Microbiol. Biotechnol. 2019, 103, 7055–7070. [Google Scholar] [CrossRef] [PubMed]
- Kelo, E.; Noronkoski, T.; Mononen, I. Depletion of L-asparagine supply and apoptosis of leukemia cells induced by human glycosylasparaginase. Leukemia 2009, 23, 1167–1171. [Google Scholar] [CrossRef]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Kilberg, M.S.; Bussolati, O. Asparagine synthetase in cancer: Beyond acute lymphoblastic leukemia. Front. Oncol. 2020, 9, 1480. [Google Scholar] [CrossRef]
- Michalska, K.; Jaskolski, M. Structural aspects of L-asparaginases, their friends and relations. Acta Biochim. Pol. 2006, 53, 627–640. [Google Scholar] [CrossRef]
- Yun, M.K.; Nourse, A.; White, S.W.; Rock, C.O.; Heath, R.J. Crystal structure and allosteric regulation of the cytoplasmic Escherichia coli L-asparaginase I. J. Mol. Biol. 2007, 369, 794–811. [Google Scholar] [CrossRef]
- Schalk, A.M.; Nguyen, H.A.; Rigouin, C.; Lavie, A. Identification and structural analysis of an L-asparaginase enzyme from guinea pig with putative tumor cell killing properties. J. Biol. Chem. 2014, 289, 33175–33186. [Google Scholar] [CrossRef]
- Covini, D.; Tardito, S.; Bussolati, O.; Chiarelli, L.R.; Pasquetto, M.V.; Digilio, R.; Valentini, G.; Scotti, C. Expanding targets for a metabolic therapy of cancer: L-asparaginase. Recent Pat. Anticancer Drug Discov. 2012, 7, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Baskar, G.; Lalitha, K.; Aiswarya, R.; Naveenkumar, R. Synthesis, characterization and synergistic activity of cerium-selenium nanobiocomposite of fungal L-asparaginase against lung cancer. Mater. Sci. Eng. C 2018, 93, 809–815. [Google Scholar] [CrossRef]
- Knott, S.R.; Wagenblast, E.; Khan, S.; Kim, S.Y.; Soto, M.; Wagner, M.; Turgeon, M.O.; Fish, L.; Erard, N.; Gable, A.L.; et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 2018, 554, 378–381. [Google Scholar] [CrossRef]
- Kotzia, G.A.; Labrou, N.E. L-Asparaginase from Erwinia chrysanthemi 3937: Cloning, expression and characterization. J. Biotechnol. 2007, 127, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Horvat, T.Z.; Pecoraro, J.J.; Daley, R.J.; Buie, L.W.; King, A.C.; Rampal, R.K.; Tallman, M.S.; Park, J.H.; Douer, D. The use of Erwinia asparaginase for adult patients with acute lymphoblastic leukemia after pegaspargase intolerance. Leuk. Res. 2016, 50, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.H.; da Silva Fiúza, T.; de Morais, S.B.; Trevizani, R. Circumventing the side effects of L-asparaginase. Biomed. Pharmacother. 2021, 139, 111616. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Wan, X.; Geng, X.; Xue, D.; Xie, Z.; Chen, C. Microbial L-asparaginase for application in acrylamide mitigation from food: Current research status and future perspectives. Microorganisms 2021, 9, 1659. [Google Scholar] [CrossRef]
- Mottram, D.S.; Wedzicha, B.L.; Dodson, A.T. Acrylamide is formed in the Maillard reaction. Nature 2002, 419, 448–449. [Google Scholar] [CrossRef] [PubMed]
- Rifai, L.; Saleh, F.A. A review on acrylamide in food: Occurrence, toxicity, and mitigation strategies. Int. J. Toxicol. 2020, 39, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Da Cunha, M.C.; dos Santos Aguilar, J.G.; de Melo, R.R.; Nagamatsu, S.T.; Ali, F.; de Castro, R.J.S.; Sato, H.H. Fungal L-asparaginase: Strategies for production and food applications. Food Res. Int. 2019, 126, 108658. [Google Scholar] [CrossRef]
- Kumar, V.; Thakur, V.; Ambika, K.S.; Singh, D. Bioplastic reservoir of diverse bacterial communities revealed along altitude gradient of Pangi-Chamba trans-Himalayan region. FEMS Microbiol. Lett. 2018, 365, 144. [Google Scholar] [CrossRef]
- Kumar, V.; Thakur, V.; Kumar, V.; Kumar, R.; Singh, D. Genomic insights revealed physiological diversity and industrial potential for Glaciimonas sp. PCH181 isolated from Satrundi glacier in Pangi-Chamba Himalaya. Genomics 2020, 112, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, S.; Singh, D. Metagenomic insights into Himalayan glacial and kettle lake sediments revealed microbial community structure, function, and stress adaptation strategies. Extremophiles 2022, 26, 3. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, S.; Darnal, S.; Patial, V.; Singh, A.; Thakur, V.; Kumar, S.; Singh, D. Optimized chromogenic dyes-based identification and quantitative evaluation of bacterial L-asparaginase with low/no glutaminase activity bioprospected from pristine niches in Indian trans-Himalaya. 3 Biotech 2019, 9, 275. [Google Scholar] [CrossRef]
- Kumar, S.; Darnal, S.; Patial, V.; Kumar, V.; Kumar, V.; Kumar, S.; Singh, D. Molecular cloning, characterization, and in-silico analysis of l-asparaginase from Himalayan Pseudomonas sp. PCH44. 3 Biotech 2022, 12, 162. [Google Scholar] [CrossRef]
- Patial, V.; Kumar, V.; Joshi, R.; Gupta, M.; Singh, D. Acrylamide mitigation in foods using recombinant L-asparaginase: An extremozyme from Himalayan Pseudomonas sp. PCH182. Food Res. Int. 2022, 162, 111936. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Bergmans, H.E.N.; van Die, I.M.; Hoekstra, W.P.M. Transformation in Escherichia coli: Stages in the process. J. Bacteriol. 1981, 146, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Imada, A.; Igarasi, S.; Nakahama, K.; Isono, M. Asparaginase and glutaminase activities of microorganisms. J. Gen. Microbiol. 1973, 76, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Paleologos, E.K.; Kontominas, M.G. Determination of acrylamide and methacrylamide by normal phase high performance liquid chromatography and UV detection. J. Chromatogr. A 2005, 1077, 128–135. [Google Scholar] [CrossRef]
- Farahat, M.G.; Amr, D.; Galal, A. Molecular cloning, structural modeling and characterization of a novel glutaminase-free L-asparaginase from Cobetia amphilecti AMI6. Int. J. Biol. Macromol. 2020, 143, 685–695. [Google Scholar] [CrossRef]
- Saeed, H.; Hemida, A.; El-Nikhely, N.; Abdel-Fattah, M.; Shalaby, M.; Hussein, A.; Eldoksh, A.; Ataya, F.; Aly, N.; Labrou, N.; et al. Highly efficient Pyrococcus furiosus recombinant L-asparaginase with no glutaminase activity: Expression, purification, functional characterization, and cytotoxicity on THP-1, A549 and Caco-2 cell lines. Int. J. Biol. Macromol. 2020, 156, 812–828. [Google Scholar] [CrossRef]
- Lubkowski, J.; Wlodawer, A. Structural and biochemical properties of L-asparaginase. FEBS J. 2021, 288, 4183–4209. [Google Scholar] [CrossRef]
- Silaban, S.; Gaffar, S.; Simorangkir, M.; Maksum, I.P.; Subroto, T. Effect of IPTG concentration on recombinant human prethrombin-2 expression in Escherichia coli BL21(DE3) ArcticExpress. IOP Conf. Ser. Earth Environ. Sci. 2018, 217, 012039. [Google Scholar] [CrossRef]
- Radha, R.; Arumugam, N.; Gummadi, S.N. Glutaminase free L-asparaginase from Vibrio cholerae: Heterologous expression, purification and biochemical characterization. Int. J. Biol. Macromol. 2018, 111, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, R.; Manonmani, H.K. Expression and characterization of recombinant l-asparaginase from Pseudomonas fluorescens. Protein Expr. Purif. 2018, 143, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Bhargavi, M.; Jayamadhuri, R. Isolation and screening of marine bacteria producing anti-cancer enzyme L-asparaginase. Am. J. Mar. Sci. 2016, 4, 1–3. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Singh, A.; Mukherjee, K.J.; Panda, A.K. Refolding and purification of recombinant L-asparaginase from inclusion bodies of E. coli into active tetrameric protein. Front. Microbiol. 2014, 5, 486. [Google Scholar] [CrossRef] [PubMed]
- Bento, H.; Paiva, G.B.; Almeida, M.R.; Silva, C.G.; Carvalho, P.J.; Tavares, A.P.; Pedrolli, D.B.; Santos-Ebinuma, V.C. Aliivibrio fischeri L-Asparaginase production by engineered Bacillus subtilis: A potential new biopharmaceutical. Bioprocess. Biosyst. Eng. 2022, 45, 1635–1644. [Google Scholar] [CrossRef]
- Saeed, H.; Elsawy, E.; Shalaby, M.; Abdel-Fattah, M.; Hemida, A.; Eldoksh, A.; Ataya, F.S.; Nematalla, H.; Elkewedi, M.; Labrou, N.N.; et al. L-asparaginase from Dickeya chrysanthemi: Expression, purification and cytotoxicity assessment. Prep. Biochem. Biotechnol. 2022, 52, 668–680. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, Y.; Park, G.H.; Umasuthan, N.; Heo, S.J.; de Zoysa, M.; Jung, W.K.; Lee, D.W.; Kim, H.; Kang, D.H.; et al. A newly identified glutaminase-free L-asparaginase (L-ASPG86) from the marine bacterium Mesoflavibacter zeaxanthinifaciens. J. Microbiol. Biotechnol. 2016, 26, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.A.; Deraz, S.F.; Soliman, H.M.; El-Deeb, N.M.; El-Ewasy, S.M. Purification, characterization, cytotoxicity and anticancer activities of L-asparaginase, anti-colon cancer protein, from the newly isolated alkaliphilic Streptomyces fradiae NEAE-82. Sci. Rep. 2016, 6, 32926. [Google Scholar] [CrossRef]
- Husain, I.; Sharma, A.; Kumar, S.; Malik, F. Purification and characterization of glutaminase free asparaginase from Enterobacter cloacae: In-vitro evaluation of cytotoxic potential against human myeloid leukemia HL-60 cells. PLoS ONE 2016, 11, e0148877. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Liu, Y.; Mu, Q.; Jiang, Z.; Yang, S. Biochemical characterization of a novel L-asparaginase from Paenibacillus barengoltzii being suitable for acrylamide reduction in potato chips and mooncakes. Int. J. Biol. Macromol. 2017, 96, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, B.; Basit, A.; Khurshid, M.; Bashir, Q. Characterization of a thermostable, allosteric L-asparaginase from Anoxybacillus flavithermus. Int. J. Biol. Macromol. 2020, 152, 584–592. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, S.; Jiao, Y.; Wang, Y.; Wang, M.; Du, G. Gene cloning and expression of the l-asparaginase from Bacillus cereus BDRD-ST26 in Bacillus subtilis WB600. J. Biosci. Bioeng. 2019, 127, 418–424. [Google Scholar] [CrossRef]
- Özdemir, F.İ.; Orhan, M.D.; Atasavum, Z.; Tülek, A. Biochemical characterization and detection of antitumor activity of l-asparaginase from thermophilic Geobacillus kaustophilus DSM 7263T. Protein Expr. Purif. 2022, 199, 106146. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, D.; Chiarelli, L.R.; Pasquetto, M.V.; Stivala, S.; Valentini, G.; Scotti, C. Helicobacter pylori L-asparaginase: A promising chemotherapeutic agent. Biochem. Biophys. Res. Commun. 2008, 377, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Xu, M.; He, B.; Rao, Z. Cloning, expression, and characterization of L-asparaginase from a newly isolated Bacillus subtilis B11–06. J. Agric. Food Chem. 2013, 61, 9428–9434. [Google Scholar] [CrossRef]
- Darwesh, D.B.; Al-Awthan, Y.S.; Elfaki, I.; Habib, S.A.; Alnour, T.M.; Darwish, A.B.; Youssef, M.M. Anticancer Activity of Extremely Effective Recombinant L-Asparaginase from Burkholderia pseudomallei. J. Microbiol. Biotechnol. 2022, 32, 551–563. [Google Scholar] [CrossRef]
- Aly, N.; El-Ahwany, A.; Ataya, F.S.; Saeed, H. Bacillus sonorensis L-asparaginase: Cloning, expression in E. coli and characterization. Protein J. 2020, 39, 717–729. [Google Scholar] [CrossRef]
- Chohan, S.M.; Sajed, M.; un Naeem, S.; Rashid, N. Heterologous gene expression and characterization of TK2246, a highly active and thermostable plant type L-asparaginase from Thermococcus kodakarensis. Int. J. Biol. Macromol. 2020, 147, 131–137. [Google Scholar] [CrossRef]
- Zhang, J.F.; Shi, L.Y.; Wei, D.Z. Chemical modification of L-asparaginase from Escherichia coli with a modified polyethyleneglycol under substrate protection conditions. Biotechnol. Lett. 2004, 26, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Egler, R.A.; Ahuja, S.P.; Matloub, Y. L-asparaginase in the treatment of patients with acute lymphoblastic leukemia. J. Pharmacol. Pharmacother. 2016, 7, 62–71. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.A.; Deraz, S.F.; El-Ewasy, S.M.; Suddek, G.M. Purification, characterization and immunogenicity assessment of glutaminase free L-asparaginase from Streptomyces brollosae NEAE-115. BMC Pharmacol. Toxicol. 2018, 19, 51. [Google Scholar] [CrossRef] [PubMed]
- Warangkar, S.C.; Khobragade, C.N. Purification, characterization, and effect of thiol compounds on activity of the Erwinia carotovora L-asparaginase. Enzyme Res. 2010, 2010, 165878. [Google Scholar] [CrossRef]
- Sun, Z.; Qin, R.; Li, D.; Ji, K.; Wang, T.; Cui, Z.; Huang, Y. A novel bacterial type II l-asparaginase and evaluation of its enzymatic acrylamide reduction in french fries. Int. J. Biol. Macromol. 2016, 92, 232–239. [Google Scholar] [CrossRef]
- Shakambari, G.; Birendranarayan, A.K.; Angelaa Lincy, M.J.; Rai, S.K.; Ahamed, Q.T.; Ashokkumar, B.; Saravanan, M.; Mahesh, A.; Varalakshmi, P. Hemocompatible glutaminase free L-asparaginase from marine Bacillus tequilensis PV9W with anticancer potential modulating p53 expression. RSC Adv. 2016, 6, 25943–25951. [Google Scholar] [CrossRef]
- Narta, U.K.; Kanwar, S.S.; Azmi, W. Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Crit. Rev. Oncol. Hematol. 2007, 61, 208–221. [Google Scholar] [CrossRef]
- El-Naggar, N.E.A.; El-Shweihy, N.M. Bioprocess development for L-asparaginase production by Streptomyces rochei, purification and in-vitro efficacy against various human carcinoma cell lines. Sci. Rep. 2020, 10, 7942. [Google Scholar] [CrossRef]
- Mostafa, Y.; Alrumman, S.; Alamri, S.; Hashem, M.; Al-izran, K.; Alfaifi, M.; Elbehairi, S.E.; Taha, T. Enhanced production of glutaminase-free l-asparaginase by marine Bacillus velezensis and cytotoxic activity against breast cancer cell lines. Electron. J. Biotechnol. 2019, 42, 6–15. [Google Scholar] [CrossRef]
- Onishi, Y.; Prihanto, A.A.; Yano, S.; Takagi, K.; Umekawa, M.; Wakayama, M. Effective treatment for suppression of acrylamide formation in fried potato chips using L-asparaginase from Bacillus subtilis. 3 Biotech 2015, 5, 783–789. [Google Scholar] [CrossRef]
- Baskar, G.; Aiswarya, R. Overview on mitigation of acrylamide in starchy fried and baked foods. J. Sci. Food Agric. 2018, 98, 4385–4394. [Google Scholar] [CrossRef] [PubMed]

| A | ||||||
|---|---|---|---|---|---|---|
| Volume (mL) | Total Protein (mg) | Specific Activity (U/mg) | Total Activity (U) | Purification Fold | Yield (%) | |
| Crude | 10.0 | 10.0 | 3.00 | 30.0 | 1.0 | 100 |
| His-tag affinity | 1.2 | 0.42 | 59.22 | 25.0 | 20.0 | 83.0 |
| B | ||||||
| Volume (mL) | Total Protein (mg) | Specific Activity (U/mg) | Total Activity (U) | Purification Fold | Yield (%) | |
| Crude | 140 | 495.6 | 3.28 | 1625.56 | 1.0 | 100 |
| Q-Sepharose | 2.0 | 29.3 | 48.12 | 1409.91 | 14.67 | 86.73 |
| Temperature (°C) | Half-Life (min) | kd (min) | R2 Value |
|---|---|---|---|
| 37 | 625.15 | 1.10 × 10−3 | 0.98 |
| 50 | 316.89 | 2.1 × 10−3 | 0.96 |
| 60 | 46.77 | 14.8 × 10−3 | 0.98 |
| 70 | 18.85 | 36.7 × 10−3 | 0.95 |
| 80 | 17.86 | 38.8 × 10−3 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Darnal, S.; Patial, V.; Kumar, V.; Singh, D. Molecular Characterization of a Stable and Robust L-Asparaginase from Pseudomonas sp. PCH199: Evaluation of Cytotoxicity and Acrylamide Mitigation Potential. Fermentation 2022, 8, 568. https://doi.org/10.3390/fermentation8100568
Kumar S, Darnal S, Patial V, Kumar V, Singh D. Molecular Characterization of a Stable and Robust L-Asparaginase from Pseudomonas sp. PCH199: Evaluation of Cytotoxicity and Acrylamide Mitigation Potential. Fermentation. 2022; 8(10):568. https://doi.org/10.3390/fermentation8100568
Chicago/Turabian StyleKumar, Subhash, Sanyukta Darnal, Vijeta Patial, Virender Kumar, and Dharam Singh. 2022. "Molecular Characterization of a Stable and Robust L-Asparaginase from Pseudomonas sp. PCH199: Evaluation of Cytotoxicity and Acrylamide Mitigation Potential" Fermentation 8, no. 10: 568. https://doi.org/10.3390/fermentation8100568
APA StyleKumar, S., Darnal, S., Patial, V., Kumar, V., & Singh, D. (2022). Molecular Characterization of a Stable and Robust L-Asparaginase from Pseudomonas sp. PCH199: Evaluation of Cytotoxicity and Acrylamide Mitigation Potential. Fermentation, 8(10), 568. https://doi.org/10.3390/fermentation8100568






