Abstract
Toyocamycin, a nucleoside antibiotic, is a fungicide with the potential to control plant pathogens. In this study, three rounds of genome shuffling screening were applied to enhance the toyocamycin production in Streptomyces diastatochromogenes 1628. After three rounds of genome-shuffling screening, the toyocamycin production increased by 10.8-fold that of wild-type, and 2.64-fold of its parental strain. By optimization of its nutrition condition in medium, the highest production of toyocamycin reached 1173.6 mg/L in TY-producing medium. In addition, the mechanism for the improvement of shuffled strains was investigated. Recombinants with increased toyocamycin production exhibited higher transcriptional level of the toy cluster and product resistance. Furthermore, the rise of ATP hydrolysis rate indicated that intracellular ATP exhibit a significant role in tuning the toy cluster by an ATP-binding pathway-specific regulator. In all, we obtained S. diastatochromogenes mutants with enhanced toyocamycin production, and provided a valuable clue for the activation of secondary metabolites.
1. Introduction
The Streptomyces produces various kinds of antibiotics, including polyketides, nucleosides, pyrrolidines, and so on, which have attracted interest for their antitumor, antiparasitic, and immunosuppressant functions [1,2,3,4]. The metabolic pathways of these antibiotics have been comprehensively studied [5]. There are complex regulation networks in Streptomyces, and many intracellular compounds involve the switch from primary to secondary metabolisms during fermentation, such as S-adenosylmethionine (SAM), ppGpp, and triacylglycerols [6,7,8]. Therefore, the strategies to change the metabolism extensively often result in higher secondary metabolites production and more efficient fermentation productivity [9,10].
Toyocamycin belongs to the nucleoside’s antibiotic division and exhibits various biological activities, such as antitumor, antibacterial, and antifungal activities [11,12]. Nowadays, the toyocamycin could be regarded as a promising agricultural antibiotic in controlling plant disease. Toyocamycin can be generated by Streptomyces diastatochromogenes 1628, isolated from the soil. The production of wild-type S. diastatochromogenes is just 59.64 mg/L, which is lower than the demand of large-scale agricultural applications [11]. Therefore, a series of modification by genetic engineering strategies have been applied to improve toyocamycin production. The structural genes toyG and toyF were overexpressed to enhance the synthesis pathway intensity [13]. The transcriptional level and translate rate were improved by optimized promoters and overexpressing the ribosome recycling factor, respectively [14,15]. The oxygen uptake rate was increased by overexpression of hemoglobin [14]. However, the highest production by metabolic engineering technology was 489.7 mg/L, which still faces difficulty in industrial application. Therefore, more efficient methods are necessary to enhance the production of toyocamycin.
Ribosome engineering has been proven to be an effective way to activate the secondary metabolic pathway and silent genes by spontaneous mutation [16]. The beneficial mutants were screened out by drug resistance (streptomycin, rifampin, paromomycin, and so on), leading to the common mutant in ribosome S12 protein (encoded by rpsL gene, high streptomycin resistant), 16S rRNA Methyltransferase (encoded by rmsG, low streptomycin resistant) and RNA polymerase β-subunit (encoded by rpoB gene, rifampin resistant) [6,17]. In order to enhance the toyocamycin production, an effective genome shuffling method was applied. The genome shuffling was a powerful method for the rapid evolution of strains toward desirable phenotypes [18]. It accelerates directed evolution through recursive recombination of improved progenies. By genome shuffling, interstrain or interspecific hybrids could be generated. Therefore, genome shuffling could be an ideal method to obtain the optimal phenotype after ribosome engineering, especially when several rounds of antibiotic screening have been employed and mutants with high production of secondary metabolites have been obtained. Furthermore, the genome shuffling could conveniently combine with other genetic engineering methods, such as screening beneficial traits from genome shuffling and UV irradiation or chemical agents [19]. However, the genome shuffling has not been used in combination with ribosome engineering to enhance toyocamycin production.
Therefore, in this study, genome shuffling and an antibiotic screening strategy were combined to improve toyocamycin production. The difference in genotype, growth rate, toyocamycin production, and resistance were determined between the wild type, parental strains, and recombinant strains. The comparison of genetic mutants in the structural gene expression level was analyzed to explain the reason for higher production. Furthermore, intracellular ATP and ATP hydrolysis rates were detected to reveal the complex energy metabolic change in mutants.
2. Method
2.1. Strains
The wild-type S. diastatochromogenes 1628 were stored in the China general microbiological culture collection (CGMCC No. 2060) and its mutants (SD10, SD88, SD99, SD143) were screened out by ribosome engineering methods [20]. The ribosome engineering method was developed to increase the expression of genes and antibiotic production in bacteria. The SD10, SD 88, SD99, and SD143 were obtained from colonies that grew within 5 to 10 days after spore suspensions were spread on GYM agar containing various concentrations of antibiotic (Table 1). The strains used in this study were listed in the Table 1. All the strains were stored as spore suspensions at −80 °C (Table 1).

Table 1.
The strains used in this study.
2.2. Media and Growth Conditions
The medium GYM, 2×GYM, was described previously [21]. TY-producing medium (TPM) contained (per liter): soybean meal 4%, bran 1%, soluble starch 2%, FeCl2 0.1 %, CaCO3 0.5%, NH4NO3 0.3%, and KHSO4 0.3% [20]. The LRM medium was prepared according to a reported recipe [22]. CP medium contained 1% sucrose, 0.2% peptone, 0.4 yeast extraction, 0.05% MgSO4·7H2O, 0.05% K2HPO4, 0.05% NaCl, pH 7.3. M3G medium was prepared with 5% glucose, 1% (NH4)2SO4, 0.5% yeast extraction, 0.05% MgSO4·7H2O, 0.08% KH2PO4, 1.4% K2HPO4, 0.03% FeSO4·7H2O, 0.05% ZnSO4·7H2O, pH 7.3. The agar 2% was added to make solid culture.
For screening purposes, the candidates were fermented in small-scale bottles containing 50 mL GYM medium. A spore suspension volume of 0.5 mL (approximately 1 × 107 spores per mL) was inoculated into 50 mL of the above medium and was incubated at 28 °C on a rotary shaker set to 200 rpm. The supernatant was obtained by centrifuging at 8000 rpm for 5 min to test the production of toyocamycin by HPLC.
To test the phenotype stability, a spore suspension R-33 volume of 0.5 mL (approximately 1 × 107 spores per mL) was cultured in the GYM medium in the shakes containing 50 mL GYM medium for 4 days at 28 °C. 1 mL of supernatant was obtained by centrifuging at 8000 rpm for 5 min for toyocamycin production analysis. Then, 0.5 mL of the former culture was added to a new 50 mL GYM medium and cultured for 4 days. The transfer was repeated four times to test the stability of toyocamycin production.
The fermentation of wild-type and R3-13 was carried out in 250 mL shakes with 50 mL medium. The seed was cultivated in GYM medium at 28 °C, 200 rpm for 2 days. Then, the fermentation cultures (GYM, 2×GYM, TPM, CP, M3G), inoculated with spore suspension volume of 0.5 mL (approximately 1 × 107 spores per mL), were cultivated at 28 °C, 200 rpm for 96 h. Samples were taken during the fermentation, and the toyocamycin concentration and cell dry weight were measured.
2.3. Genome Shuffling
The spore suspension was grown in the GYM culture at 28 °C for 2 days. Then, 5% of the suspension was added to GYM culture and cultivated to a mid-exponential growth phase (about 2 days). The cells were centrifuged at 8000× g for 5 min, and washed by 10.3% sucrose and P-buffer twice. The P-buffer was a hyperosmotic solution, composed of the following, per liter: 103 g sucrose, 0.25 g K2SO4, 2.02 g MgCl2·6H2O, 0.368% CaCl2, 0.05% KH2PO4, 10% TES solution, pH 7.2. The TES solution (pH 7.2) contained microelement, per liter: 40 mg ZnCl2, 10 mg CuCl2·2H2O, 10 mg NaB4O7·10H2O, 200 mg FeCl2·6H2O, 10 mg MnCl2·4H2O, 10 mg (NH4)6Mo7O24·4H2O. The cells were treated with 2 mg/mL lysozyme at 37 °C for 2 h to prepare protoplasts. During the enzymolysis, the cells were shaken for 10 min. Then, the protoplasts were harvested by centrifuge at 2000 rpm for 5 min and suspended in the P-buffer. Finally, the protoplasts, washed twice by P-buffer, were filtered by gauze 200. The protoplasts from different starting strains were mixed in the same proportion and centrifuged at 1000 rpm for 5 min. PEG was added into the protoplasts to fuse the cells for 10 min. Then, the P-buffer was added and the fused protoplasts were obtained by centrifuging for 5 min at 1000 rpm. The fused protoplasts were spread out in regeneration medium LRM [22] and cultivated at 28 °C for 6 days. Then the strains were screened by drug-resistant plates (containing 200 μg/mL streptomycin, 200 μg/mL rifampin, and 30 μg/mL paromomycin) to identify the highly drug-resistant strains [23]. These strains were fermented on a small scale and the toyocamycin production was measured. The selected strains with higher yielding mutants were used as the starting strains for subsequent rounds of genome shuffling. Three successive rounds of genome shuffling were performed by the same methods, and in the second round of genome shuffling, the concentrations of antibiotic in drug-resistant plates were increased to 250 μg/mL streptomycin, 250 μg/mL rifampin, and 35 μg/mL paromomycin. In the third round of genome shuffling, the screening antibiotic concentrations were increased to 275 μg/mL streptomycin, 275 μg/mL rifampin, and 40 μg/mL paromomycin. The production of toyocamycin was measured by high-performance liquid chromatograph (HPLC).
2.4. The Toyocamycin Resistance of the Mutants and Parent Strains
The product toyocamycin was used to determine the change in product resistance of mutants and parent strains. These strains were cultured in the GYM medium containing the increased toyocamycin concentrations (0, 100, 300, 500 μg/mL). After 96 h of shaking culture at 28 °C, the dry cell weight (DCW) was measured [22].
2.5. Transcriptional Analysis by Real-Time qPCR
The RNA was isolated from the mycelia of S. diastatochromogenes 1628 and its mutants were grown in GYM medium for 72 h at 28 °C. The mycelium was collected by centrifuging at 10,000 rpm for 10 min, and the collected mycelium was immediately frozen in liquid nitrogen. The cells were lysed and homogenized by the liquid nitrogen grinding method. The total RNA was purified using the RNA mini extract kit (Takara, Kusatsu, Japan) according to the instructions of the kit. The quality of RNA was determined by an agarose gel. Then, the RNA was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Kusatsu, Japan). The concentrations of cDNA were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Illkirch, France).
qPCR was conducted in a 15 μL volume using the SYBR Premix ExTaq GC kit (Takara, Kusatsu, Japan); the sequence of primers used in this study is shown in the Supplementary Table S1. Each reaction mixture contained 7.5 μL of SYBR Premix Ex TaqTM (2×), 1 μL of template, 0.3 μL of forward primer, 0.3 μL of reverse primer, 0.3 μL of ROX reference dye (50×), and 5.6 μL of RNase-free H2O. The thermocycling conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 30 s. qPCR results were conducted in the Step One Plus Real time PCR system (Applied Biosystem, Waltham, MA, USA), and data was analyzed by the Step One software with the hrdB gene used as a reference gene for normalization. The STDEV indicates the standard deviation from three independent experiment replicates. The quantification of relative gene expression was analyzed using the 2−ΔΔCt method [24].
2.6. Intercellular ATP and ATP Hydrolysis Rate
50 mg of the mycelium were harvested from GYM medium at 48 h by centrifugation for 10 min (12,000 rpm, 4 °C) and washed twice with phosphate buffer (pH 7.4). Then the samples were disrupted for 5 min using an ultrasonic oscillation (Sonics VCX750, amplitude 25%). All the procedures were conducted at 4 °C. The supernatant was used to measure the intercellular ATP after removing the cellular debris by centrifugation. The ATP luminescence kit (TianGen, China) and a GLOMAXTM luminometer were used to determine intercellular ATP concentration [25]. The protein concentration was detected by a BCA protein assay kit (TianGen, China).
The ATP hydrolysis rate by the total cell supernatant was measured by an enzyme-coupled assay according to the previous study [26] but with several differences. The assay solution contained 10 μM ATP substrate, 100 μg protein of total cell supernatant, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.5), 10 mM Mg2+, 400 μM Zn2+, 100 mM KCl, and 15% glycerol. After reaction for 10 min at 37 °C, the reaction was stopped by a final concentration of 0.4 M trichloroacetic acid. Then the reaction solution was centrifuged at 12,000 rpm for 10 min to remove the protein. The supernatant was used to measure the ATP concentration using the ATP luminescence kit using a GLOMAXTM luminometer. The specific ATP hydrolysis activity was defined as the micromoles of ATP usage per minute per milligram of protein.
2.7. The Genotype Sequencing of Strains after Genome Shuffling
The rpsL, rmsG, and rpoB genes were amplified by PCR from the wild-type S. diastatochromogenes and its mutants’ genome by using the primers in the Table S1. The amplification protocol was carried out using the LA-taq enzyme with GC buffer and the purified PCR product was sequenced by Shanghai Sangon BioTech Co. (Shanghai, China). The mutant site was obtained by aligning the sequencing results with the wild-type sequence.
2.8. Analytical Methods
The toyocamycin concentration was taken from the clear supernatant determined by HPLC. The condition of HPLC was described previously [27]. The toyocamycin concentration was confirmed with the HPLC equipped with a column of Agilent SB-C18 (4.6 mm × 250 mm, 5 μm); the flow phase contained 90% methanol and 10% H2O (v/v). The UV detection of toyocamycin was at 279 nm. The injection volume was 10 μL and the whole process was performed at 30 °C. The data of the production were analyzed by one-way ANOVA in Excel and the significant level was obtained by unpaired Student’s t-test.
3. Results
3.1. Screening Hybrid Strains by Improving the Mutant rate of Genome Shuffling
After several rounds of antibiotic screening, further enhanced production of secondary metabolites was hard to obtain [20]. Genome shuffling could assemble the genetic diversity from different strains into a selected mutant by extensive homologous recombination. Thus, the beneficial traits from the starting strains with the desired phenotype could be inherited. In this study, the selected mutants SD10, SD88, SD99, and SD143 were used as the parental strains and the colonies were cultured on antibiotic resistance plates (Table 1). Three successive rounds of genome shuffling were applied and the candidate high producers were screened out by comparing their toyocamycin production.
As shown in the Table 1, the toyocamycin production and drug tolerance in S. diastatochromogenes 1628 were increased gradually after each round of genome shuffling, and the production of toyocamycin was related to increased drug-screening concentration. For the first round of genome shuffling, 2 strains (R1-31, R1-41) were screened out from 65 clones with the highest toyocamycin production. The production of toyocamycin increased by 6.4-fold and 6.2-fold compared to the wild type S. diastatochromogenes 1628, and reached 443.9 mg/L and 434.4 mg/L, respectively. Afterwards, the second round of genome shuffling was carried out by screening the drug-resistant mutants with a higher drug concentration (Str 250 mg/L, Rif 200 mg/L, Par 35 mg/L). R2-8 were obtained with 558.5 mg/L toyocamycin production, and were 9.3-fold that of the wild type strain. The percentage of positive mutants in the second round of genome shuffling was 13.6% (Table 2). After the third round of genome shuffling, R3-33 were obtained with a higher drug resistance (Str 275 mg/L, Rif 275 mg/L, and Par 40 mg/L), and the production of toyocamycin reached 644.5 mg/L, which was 10.8-fold that of the wild-type production. The toyocamycin production of R1-31, R1-41, R2-8, and R3-33 were increased significantly, compared with the wild-type by one-way analysis of variance (p < 0.01). At the same time, the positive mutant rate decreased (Table 2) among the colonies appearing on the agar plates with the increases in drug concentration.

Table 2.
The screening and toyocamycin production of drug-resistant mutants.
There were several genes commonly mutant during the ribosome engineering, including rpsL, rsmG, and rpoB, but not all mutations occured in these three genes [20,28]. In order to identify the change of genome occurring, we sequenced and analyzed the rpsL, rmsG, and rpoB genes between the mutants and these parental strains. As shown in Table 3, for the first-round shuffling, R1-31 had a deficiency at the 40 bp of rmsG, which was obtained from the strain SD143. R-41 had a mutation at Cys59Ter, which could affect the activity of RsmG. While in the second round of shuffling, R2-8 retained the mutation Cys59Ter of R1-41. The third round of shuffling also maintained the mutation Cys59Ter in the rmsG of R3-33. Interestingly, a new mutation Gly122Val was introduced in the rpsL of R3-33, which may occur in the screening using higher streptomycin. There was no mutation in the rpoB gene, but all the mutants showed higher rifampin-resistance during the three rounds of shuffling. This phenomenon may imply the presence of other unknown mutations in the genome which were responsible for the rifampin tolerance. Therefore, these mutations in these three genes are not the only reason for the increased antibiotic-resistance and improved toyocamycin titers.

Table 3.
The relative genotype of stains after genome shuffling.
3.2. Fermentation of Shuffled Recombinant Strains
Biomass and toyocamycin production are shown in Figure 1A,B. The biomass (mycelial dry weight) increased from 0 to 90 h for both 1628 and R3-33 in the GYM. The mycelial dry weight was highest (4.18 mg/mL) for 1628 at 96 h, while the highest mycelial dry weight was 4.61 mg/mL at 90 h for the R3-33, indicating the cell growth rate was similar for wild type strain and R3-33 from 12 to 96 h. At the same time, a dramatic increase of toyocamycin production occurred in this period for the mutant R3-33. At 0~36 h, the secondary metabolic toyocamycin could not be generated in both wild type and R3-33, because it is the primary phase for strain growth. Then, the toyocamycin production increased sharply from 36 h to 84 h for R3-33, and the highest production reached 644.5 mg/L at 84 h, which was 10.8-fold of the production in wild-type S. diastatochromogenes 1628 (59.67 mg/L) at 84 h. The productivity of R-33 reached 139.80 mg/g DCW at 96 h, which is significantly higher than that of wild-type 14.27 mg/g DCW. Therefore, the mutant R3-33 has a significant difference in cell growth and toyocamycin production.
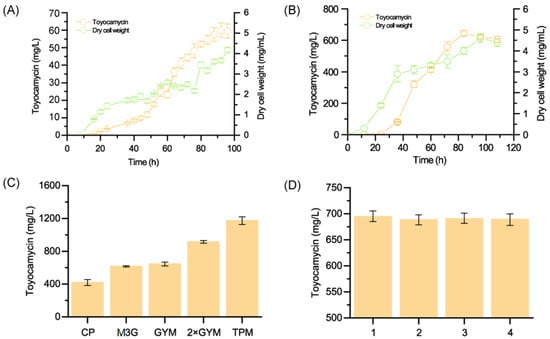
Figure 1.
The comparison of toyocamycin production by wild-type and R3-33. The antibiotic production and dry cell weight in wild-type (A) and R3-33 (B) from 0 h to 96 h in GYM medium at 28 °C; (C) the toyocamycin production in different medium; (D) the production stability of R3-33 in four rounds of generation.
The different culture nutritional conditions could affect the biomass and the secondary metabolites. Therefore, five kinds of medium were used to obtain higher toyocamycin production, including GYM, 2×GYM, TPM, CP, and M3G. The highest toyocamycin production in different culture mediums is shown in Figure 1C. The lowest toyocamycin production was 410 mg/mL in CP culture, while the highest production reached 1173.6 mg/L in TPM, which was twice that of the production in GYM (p < 0.01). No difference in production was shown in M3G and GYM (p > 0.1). The significant difference between CP, GYM, 2×GYM, and TPM indicate that the nutrition condition was crucial for toyocamycin production. The most suitable culture medium was TPM.
The stability of positive phenotype R-33 is critical for the industrial fermentation. Therefore, the selected colonies R3-33 were cultured for four generations and measured the toyocamycin production. All the generations showed similar biomass and toyocamycin production (688.4~694.3 mg/L), which suggested that the mutant R3-33 is genetically stable and suitable for the practical use (Figure 1D).
3.3. The Product Resistance of the Mutantions and Parent Strains
The antibiotic resistance screening to streptomycin, rifampin, and paromomycin made the mutants show diversity of product resistance. Figure 2 shows the toyocamycin resistance of wild-type, parental strains, and genome shuffling mutants R1-31, R1-41, R2-8, and R3-33. The wild-type showed nearly no growth (DCW value 0.22 g/L) in the GYM medium containing toyocamycin over 100 μg/mL, while the parental strains with higher toyocamycin production could not grow in the presence of toyocamycin above 300 μg/mL (DCW values 0.13 g/L~0.24 g/L). The improved recombinants R1-31, R1-41, R2-8, and R3-33, derived from genome shuffling, could grow under 500 μg/mL, which indicated that the recombinants from genome shuffling have more resistance to both the antibiotics and products. The results suggest that the resistance to the antibiotic was correlated to the secondary metabiotic production.
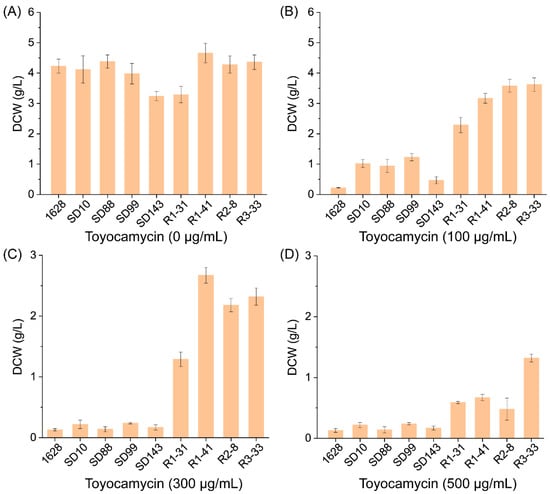
Figure 2.
The product resistance of the mutants and parent strains. The DCWs were monitored after inoculating for 96 h at 28 °C in the GYM medium with increasing concentrations of toyocamycin (A) 0 μg/mL; (B) 100 μg/mL; (C) 300 μg/mL; (D) 500 μg/mL.
3.4. Transcriptional Level of Toy Cluster Genes in Wild-Type and Mutant Strains
The transcriptional level of toyocamycin biosynthesis cluster was determined by RT-qPCR analysis with RNA extracted from S. diastatohromogenes 1628 and its mutants (Figure 3). Because the toyocamycin was the secondary metabolite for Streptomyces, the expression of the toy cluster was active at the late-exponential phase (72 h). Results of RT-qPCR indicated that the transcriptional level of the pathway-specific regulator ToyA and the structural genes in the all the four mutants (R1-31, R1-41, R2-8, and R3-33) increased obviously compared with the wild-type strains. The relative expression levels of toyA in R3-33 were markedly higher than that of R1-31, R1-41, and R2-8, which were 1.4-times of that in wild-type. At the same time, other structural genes toyB, toyF, toyG, and toyM of R3-33 were over twice that of wild-type. Furthermore, the expression level of structural genes had a positive correlation with toyocamycin production. In 72 h of fermentation, the mRNA level in R3-33 was the highest, and the R2-8 was the second highest. Therefore, the improvement of TM production could be due to the increased transcriptional level of toy genes.
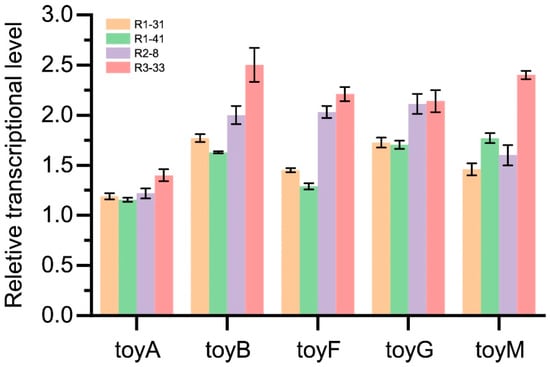
Figure 3.
Relative expression levels of pathway-specific regulator gene toyA, and structural genes toyB, toyF, toyG, and toyM in R1-31, R1-41, R2-8 and R3-33. The mRNA was extracted from strains grown in GYM medium for 72 h.
3.5. The Intracellular ATP Level and ATP Hydrolysis Rate in the Mutants
The pathway-specific regulator ToyA could sense the change of intracellular ATP (inATP) to affect the production of TM. However, the inATP level in the S. diastatohromogenes and its mutants, as well as the change of ATP hydrolysis rate, was not clear. The SD1628 has the highest inATP level of 21 μmol/g DCW, while the mutants screened from the first round of genome shuffling decreased to around 10 μmol/g DCW. The inATP level of R2-8 and R3-33 was lower than R1-31 and R1-41, and were 4.39 μmol/g DCW and 2.95 μmol/g DCW, respectively (Figure 4A). In addition, the ATP hydrolysis rate increased from 1.84 μmol/mg protein/min to 2.72 μmol/mg protein/min, which indicated that the intracellular pathway has a higher ratio to utilize ATP (Figure 4B). The detailed mechanism of increased ATP hydrolysis rate needs more precise metabolome analysis in the future.
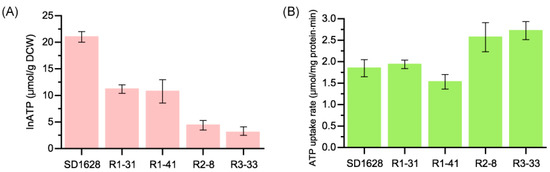
Figure 4.
The intracellular ATP level and ATP hydrolysis rate in vitro in wild-type and its mutants. (A) The amount of inATP measured at 48 h; (B) the ATP hydrolysis rate of wild-type SD1628 and its mutants.
4. Discussion
The improvement of antibiotic production is critical procedure in industrial strain modification. In order to increase toyocamycin production, genome shuffling was applied to achieve the target phenotypes. Several published articles have indicated that genome shuffling could combine other genetic engineering methods to effectively improve product yield, including tylosin in Streptomyces fradiae [29], fructosyltransferase in Aspergillus oryae [30], and succinic acid in Actinobacillus succinogenes [31]. For butanol fermentation, after chemical mutagenesis and genome shuffling, the new strain increased productivity and yield by 6.8% under high gravity conditions [32]. In our case, the solo antibiotic mutants of S. diastatochromogenes were used as the parental strains and multiple rounds of genome shuffling were applied to generate mutants with diverse genotypes. The toyocamycin production increased each round by 15.4% to 57.9% by testing the toyocamycin production of several strains in comparison with the WT.
Antibiotic resistance was a promising method to improve secondary metabolic pathways, which is called ribosome engineering. The antibiotics commonly used to screen mutants are kanamycin, gentamicin, rifampin, streptomycin, fusidic acid, and so on. In addition, the product was also used for improving the product yield by increasing product-resistance. For example, enhancing pristinamycin-resistance in S. pristinaespiralis would relieve feedback regulation of pristinamycin on its own biosynthesis [23]. However, in this study, we found the antibiotic-screening mutants could also increase product resistance, although the product was not used to screen mutants. A similar phenomenon was also observed in the S. viridochromogenes mutants using streptomycin-screening but resulting in increasing avilamycin resistance after genome shuffling and ribosome engineering [22]. Although the mechanism of how the antibiotic resistance was correlated with product productivity was unclear, it is an interesting phenomenon to explain.
The energy metabolic could be seen as the bridge to connect the primary metabolic and secondary metabolic pathway, which was at a high level to maintain the rapid growth rate at primary metabolism and a low level to stimulate the silent biosynthesis pathway when the nutrients were limited. A previous study showed that intracellular adenosine triphosphate (inATP) level is negatively correlated with tautomycetin biosynthesis, which is regulated by a pathway-specific transcriptional regulator TmcN belonging to the large ATP-binding LuxR (LAL) family [33]. The ATPase domain of TmcN can sense the endogenous pool of ATP, then activate the silent pathway of tautomycetin [34]. In addition, a similar phenomenon existed in other strains. For example, the generation of an artificial ATP deficit triggers antibiotic production in S. lividans [35]. The antibiotic production is correlated with highly active oxidative metabolism in S. coelicolor M145 [25]. Although extensive research has been conducted to characterize the transcriptional level of the biosynthesis pathway, the shifted energy metabolic could play a significant role in secondary metabolites, especially for the pathway controlled by an ATP-binding LuxR transcriptional regulator. In this study, ToyA is the specific regulation factor in toyocamycin synthesis clusters, which was found to activate the transcription of toyocamycin biosynthetic genes. ToyA is an ATP dependent enzyme, belonging to the large ATP-binding regulators of the luxR family (LAL) family, due to the well-conserved motifs of Walker A and Walker B and high homology to the ATPase domain. In addition, the ToyA has a C-terminal DNA binding domain which may bind to the toyocamycin cluster to regulate its transcription [36]. The results in this study provide a primary insight into the energy change in different mutants, indicating that the mutants with higher ATP hydrolysis rate generated higher production of toyocamycin. We will conduct further research to reveal the energetic shift between primary and secondary metabolism in the future.
Despite the ATP signal factor, other factors have included intracellular signals such as SAM for the stimulation of silent genes; the translation efficiency exerts a positive impact on the secondary metabolites production. There was a rsmG mutation harbored in all four strains after genome shuffling, which make the SAM-dependent 16S rRNA methyltransferase inactive. The mutations result in the absence of m7G modification in 16S rRNA, stimulating the expression of SAM synthetase markedly by perception of the change in 16S rRNA and catalyzing the synthesis of SAM from methionine and ATP [37]. It has been proven that SAM is an effective intracellular signal in Streptomyces and Bacillus [38]. For example, the rsmG mutants in S. coelicolor improve the antibiotic actinorhodin production by improving the SAM level [39,40]. In addition, the overexpression of SAM synthetase via multicopy plasmid or addition of SAM to the medium display leads to enhanced antibiotics in S. lividans, S. griseus, S. griseoflaveus, and S. coelicolor [40,41,42]. In B. subtilis, the intracellular SAM activates the bacilysin pathway by mutants in the enzyme involved in te SAM-recycling pathway [43]. However, although R1-31, R1-41, R2-8 and R3-33 have a similar mutant in the rsmG, the inactivation of RsmG was not the only factor that improved the toyocamycin production, especially with the genome shuffling method applied. For example, genome shuffling was applied using the ribosome engineering mutant as a starting strain for the enhanced production of tiancimycin-A, which lead to the deficiency of rsmG in the strains after genome shuffling. However, the knockouts of rsmG in the wild-type could not lead to the improvement of production of tiancimycin-A [44]. Therefore, the rsmG mutant was not the only reason explaining why the secondary metabolites production were enhanced. There may exist other mutants which were not detected in the genome of potential strains and mutants. The highest toyocamycin production was obtained by R3-33, which contains another mutant in rpsL (encoding the 16S ribosomal RNA), improving the translation efficiency by enhancing the ribosome stability over that of wild-type. The mutants in rpsL contribute to higher protein synthesis during the stationary phases and higher secondary metabolites production. This phenomenon has been proven in several strains, including Streptomyces spp., B. subtilis, Erwinia carotovora, and E. coli [45,46].
In conclusion, this study achieved a higher toyocamycin production by genome shuffling and antibiotic screening. Apart from the solo mutants in ribosome, genome shuffling could lead to a comprehensive shift in energy metabolic and intracellular signal synthesis. The mutants saw an incredible increase in ATP-hydrolysis rate, which primarily reveals that intercellular energy metabolic flux change could effectively stimulate the transcription of silent genes by the ATP-related pathway regulator.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8100535/s1, Table S1: The primers used in this study.
Author Contributions
Conceptualization, Y.S. and X.S.; methodology, Z.Z.; software, X.Z.; validation, J.Y., Y.S. and Z.Z.; formal analysis, X.Z.; investigation, Y.S.; resources, X.S.; data curation, Y.S.; writing—original draft preparation, Y.S.; writing—review and editing, X.S. and X.Y.; supervision, X.Y.; project administration, X.Y.; funding acquisition, Y.S and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31901930), Zhejiang Provincial Natural Science Foundation of China (LGN22C140006) and Zhejiang Provincial Programs for Science and Technology Development (2019C02015, 2017C32006). And The APC was funded by Zhejiang Provincial Programs for Science and Technology Development (2019C02015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are within the manuscript and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bolourian, A.; Mojtahedi, Z. Immunosuppressants produced by Streptomyces: Evolution, hygiene hypothesis, tumour rapalog resistance and probiotics. Environ. Microbiol. Rep. 2018, 10, 123–126. [Google Scholar] [CrossRef]
- Novakova, R.; Núñez, L.E.; Homerova, D.; Knirschova, R.; Feckova, L.; Rezuchova, B.; Sevcikova, B.; Menéndez, N.; Morís, F.; Cortés, J. Increased heterologous production of the antitumoral polyketide mithramycin A by engineered Streptomyces lividans TK24 strains. Appl. Microbiol. Biotechnol. 2018, 102, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T.-H.; Chan, K.-G.; Lee, L.-H.; Goh, B.-H. Streptomyces bacteria as potential probiotics in aquaculture. Front. Microbiol. 2016, 7, 79. [Google Scholar] [CrossRef]
- Yao, J.Y.; Guo, J.L.; Zang, C.S.; Mi, G.Q.; Jia, Y.Y.; Yin, W.L.; Cao, Z.; Xia, Y.C.; Pan, X.Y.; Ling, L.Y. Antiparasitic efficacy of natamycin isolated from Streptomyces gilvosporeu AXY-25 against Ichthyophthirius multifiliis. Aquaculture 2019, 506, 465–469. [Google Scholar] [CrossRef]
- Baltz, R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 2016, 43, 343–370. [Google Scholar] [CrossRef]
- Funane, K.; Tanaka, Y.; Hosaka, T.; Murakami, K.; Miyazaki, T.; Shiwa, Y.; Gibu, S.; Inaoka, T.; Kasahara, K.; Fujita, N. Combined drug resistance mutations substantially enhance enzyme production in Paenibacillus agaridevorans. J. Bacteriol. 2018, 200, e00188-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Inaoka, T.; Okamoto, S.; Ochi, K. A novel insertion mutation in Streptomyces coelicolor ribosomal S12 protein results in paromomycin resistance and antibiotic overproduction. Antimicrob. Agents Chemother. 2009, 53, 1019–1026. [Google Scholar] [CrossRef]
- Wang, W.; Li, S.; Li, Z.; Zhang, J.; Fan, K.; Tan, G.; Ai, G.; Lam, S.M.; Shui, G.; Yang, Z. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat. Biotechnol. 2019, 38, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Magocha, T.A.; Zabed, H.; Yang, M.; Yun, J.; Zhang, H.; Qi, X. Improvement of industrially important microbial strains by genome shuffling: Current status and future prospects. Bioresour. Technol. 2018, 257, 281–289. [Google Scholar] [CrossRef]
- Ochi, K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017, 70, 25. [Google Scholar] [CrossRef] [PubMed]
- Bing, Y.U.; Xuping, S.; Xiaoping, Y.U. Antifungal Activity of Toyocamycin on Rhizoctonia solani Kühn. Chin. J. Biol. Control 2011, 27, 373. [Google Scholar]
- McCarty, R.M.; Bandarian, V. Deciphering deazapurine biosynthesis: Pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem. Biol. 2008, 15, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Ma, Z.; Xu, X.; Bechthold, A.; Bian, Y.; Shentu, X.; Yu, X. Engineering Streptomyces diastatochromogenes 1628 to increase the production of toyocamycin. Eng. Life Sci. 2015, 15, 779–787. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, J.; Bechthold, A.; Tao, L.; Shentu, X.; Bian, Y.; Yu, X. Development of intergeneric conjugal gene transfer system in Streptomyces diastatochromogenes 1628 and its application for improvement of toyocamycin production. Curr. Microbiol. 2014, 68, 180–185. [Google Scholar] [CrossRef]
- Xu, X.; Wang, J.; Bechthold, A.; Ma, Z.; Yu, X. Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J. Microbiol. Biotechnol. 2017, 33, 30. [Google Scholar] [CrossRef]
- Hosaka, T.; Ohnishi-Kameyama, M.; Muramatsu, H.; Murakami, K.; Tsurumi, Y.; Kodani, S.; Yoshida, M.; Fujie, A.; Ochi, K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009, 27, 462. [Google Scholar] [CrossRef]
- Wang, G.; Hosaka, T.; Ochi, K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl. Environ. Microbiol. 2008, 74, 2834–2840. [Google Scholar] [CrossRef]
- Hospet, R.; Thangadurai, D.; Cruz-Martins, N.; Sangeetha, J.; Anu Appaiah, K.A.; Chowdhury, Z.Z.; Bedi, N.; Soytong, K.; Al Tawahaj, A.R.M.; Jabeen, S. Genome shuffling for phenotypic improvement of industrial strains through recursive protoplast fusion technology. Crit. Rev. Food Sci. Nutr. 2021, 1–10. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, J.; Ji, X.; Fang, Z.; Wu, Z.; Chen, J.; Du, G. Evolutionary engineering of industrial microorganisms-strategies and applications. Appl. Microbiol. Biotechnol. 2018, 102, 4615–4627. [Google Scholar] [CrossRef]
- Shentu, X.; Liu, N.; Tang, G.; Tanaka, Y.; Ochi, K.; Xu, J.; Yu, X. Improved antibiotic production and silent gene activation in Streptomyces diastatochromogenes by ribosome engineering. J. Antibiot. 2016, 69, 406. [Google Scholar] [CrossRef]
- Ochi, K. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: Significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 1987, 169, 3608–3616. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.Q.; Zhou, Y.H.; Chen, X.S.; Wu, J.Y.; Wei, P.; Yuan, L.X.; Yao, J.M. Genome shuffling and ribosome engineering of Streptomyces virginiae for improved virginiamycin production. Bioprocess Biosyst. Eng. 2018, 41, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Jin, Z.; Wang, H.; Jin, Q.; Jin, X.; Cen, P. Evolution of Streptomyces pristinaespiralis for resistance and production of pristinamycin by genome shuffling. Appl. Microbiol. Biotechnol. 2008, 80, 261–267. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, A.; Holland, I.; Virolle, M. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci. Rep. 2017, 7, 200. [Google Scholar] [CrossRef]
- Yang, Y.; Gunasekara, M.; Muhammednazaar, S.; Li, Z.; Hong, H. Proteolysis mediated by the membrane-integrated ATP-dependent protease FtsH has a unique nonlinear dependence on ATP hydrolysis rates. Protein Sci. 2019, 28, 1262–1275. [Google Scholar] [CrossRef]
- Ma, Z.; Tao, L.; Bechthold, A.; Shentu, X.; Bian, Y.; Yu, X. Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 2014, 98, 5051–5058. [Google Scholar] [CrossRef]
- Shentu, X.-P.; Cao, Z.-Y.; Xiao, Y.; Tang, G.; Ochi, K.; Yu, X.-P. Substantial improvement of toyocamycin production in Streptomyces diastatochromogenes by cumulative drug-resistance mutations. PLoS ONE 2018, 13, e0203006. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Perry, K.; Vinci, V.A.; Powell, K.; Stemmer, W.P.C.; Cardayré, S.B.D. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 2002, 415, 644–646. [Google Scholar] [CrossRef]
- Wang, S.; Duan, M.; Liu, Y.; Fan, S.; Lin, X.; Zhang, Y. Enhanced production of fructosyltransferase in Aspergillus oryzae by genome shuffling. Biotechnol. Lett. 2017, 39, 391–396. [Google Scholar] [CrossRef]
- Zheng, P.; Zhang, K.; Yan, Q.; Xu, Y.; Sun, Z. Enhanced succinic acid production by Actinobacillus succinogenes after genome shuffling. J. Ind. Microbiol. Biotechnol. J. 2013, 40, 831–840. [Google Scholar] [CrossRef]
- Wang, P.M.; Zheng, D.Q.; Liu, T.Z.; Tao, X.L.; Feng, M.G.; Min, H.; Jiang, X.H.; Wu, X.C. The combination of glycerol metabolic engineering and drug resistance marker-aided genome shuffling to improve very-high-gravity fermentation performances of industrial Saccharomyces cerevisiae. Bioresour. Technol. 2012, 108, 203–210. [Google Scholar] [CrossRef]
- Li, M.; Chen, Y.; Wu, S.; Tang, Y.; Deng, Y.; Yuan, J.; Dong, J.; Li, H.; Tang, L. TmcN is involved in ATP regulation of tautomycetin biosynthesis in Streptomyces griseochromogenes. Biochem. Biophys. Res. Commun. 2016, 478, 221–226. [Google Scholar] [CrossRef]
- Meng, L.; Li, M.; Yang, S.H.; Kim, T.J.; Suh, J.W. Intracellular ATP levels affect secondary metabolite production in Streptomyces spp. Biosci. Biotechnol. Biochem. 2011, 75, 1576–1581. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seghezzi, N.; Darbon, E.; Martel, C.; David, M.; Lejeune, C.; Esnault, C.; Virolle, M.J. The Generation of an artificial ATP deficit triggers antibiotic production in Streptomyces lividans. Antibiotics 2022, 11, 1157. [Google Scholar] [CrossRef]
- Xu, J.; Song, Z.; Xu, X.; Ma, Z.; Yu, X. ToyA, a positive pathway-specific regulator for toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 2019, 103, 7071–7084. [Google Scholar] [CrossRef]
- Okamoto, S.; Tamaru, A.; Nakajima, C.; Nishimura, K.; Tanaka, Y.; Tokuyama, S.; Suzuki, Y.; Ochi, K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007, 63, 1096–1106. [Google Scholar] [CrossRef]
- Tanaka, Y.; Komatsu, M.; Okamoto, S.; Tokuyama, S.; Kaji, A.; Ikeda, H.; Ochi, K. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl. Environ. Microbiol. 2009, 75, 4919–4922. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Hosaka, T.; Tokuyama, S.; Okamoto, S.; Ochi, K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3 (2). J. Bacteriol. 2007, 189, 3876–3883. [Google Scholar] [CrossRef]
- Okamoto, S.; Lezhava, A.; Hosaka, T.; Okamoto-Hosoya, Y.; Ochi, K. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3 (2). J. Bacteriol. 2003, 185, 601–609. [Google Scholar] [CrossRef]
- Hosaka, T.; Xu, J.; Ochi, K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol. Microbiol. 2006, 61, 883–897. [Google Scholar] [CrossRef]
- Saito, N.; Kurosawa, K.; Xu, J.; Okamoto, S.; Ochi, K. Effect of S-adenosylmethionine on antibiotic production in Streptomyces griseus and Streptomyces griseoflavus. Actinomycetologica 2003, 17, 47–49. [Google Scholar] [CrossRef]
- Tojo, S.; Kim, J.-Y.; Tanaka, Y.; Inaoka, T.; Hiraga, Y.; Ochi, K. The mthA mutation conferring low-level resistance to streptomycin enhances antibiotic production in Bacillus subtilis by increasing the S-adenosylmethionine pool size. J. Bacteriol. 2014, 196, 1514–1524. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, C.; Lin, J.; Zhuang, Z.; Kong, W.; Liu, L.; Huang, Y.; Duan, Y.; Zhu, X. Genome shuffling based on different types of ribosome engineering mutants for enhanced production of 10-membered enediyne tiancimycin-A. Appl. Microbiol. Biotechnol. 2020, 104, 4359–4369. [Google Scholar] [CrossRef]
- Barnard, A.M.; Simpson, N.J.; Lilley, K.S.; Salmond, G.P. Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology 2010, 156, 1030–1039. [Google Scholar] [CrossRef]
- Hosoya, Y.; Okamoto, S.; Muramatsu, H.; Ochi, K. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 1998, 42, 2041–2047. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).