Bacterial and Fungal Microbiota of Guinea Grass Silage Shows Various Levels of Acetic Acid Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Silage Preparation
2.2. Chemical Component and Fermentation Characteristic Analyses
2.3. MiSeq Preparation
2.4. Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
3.1. Fermentation Products
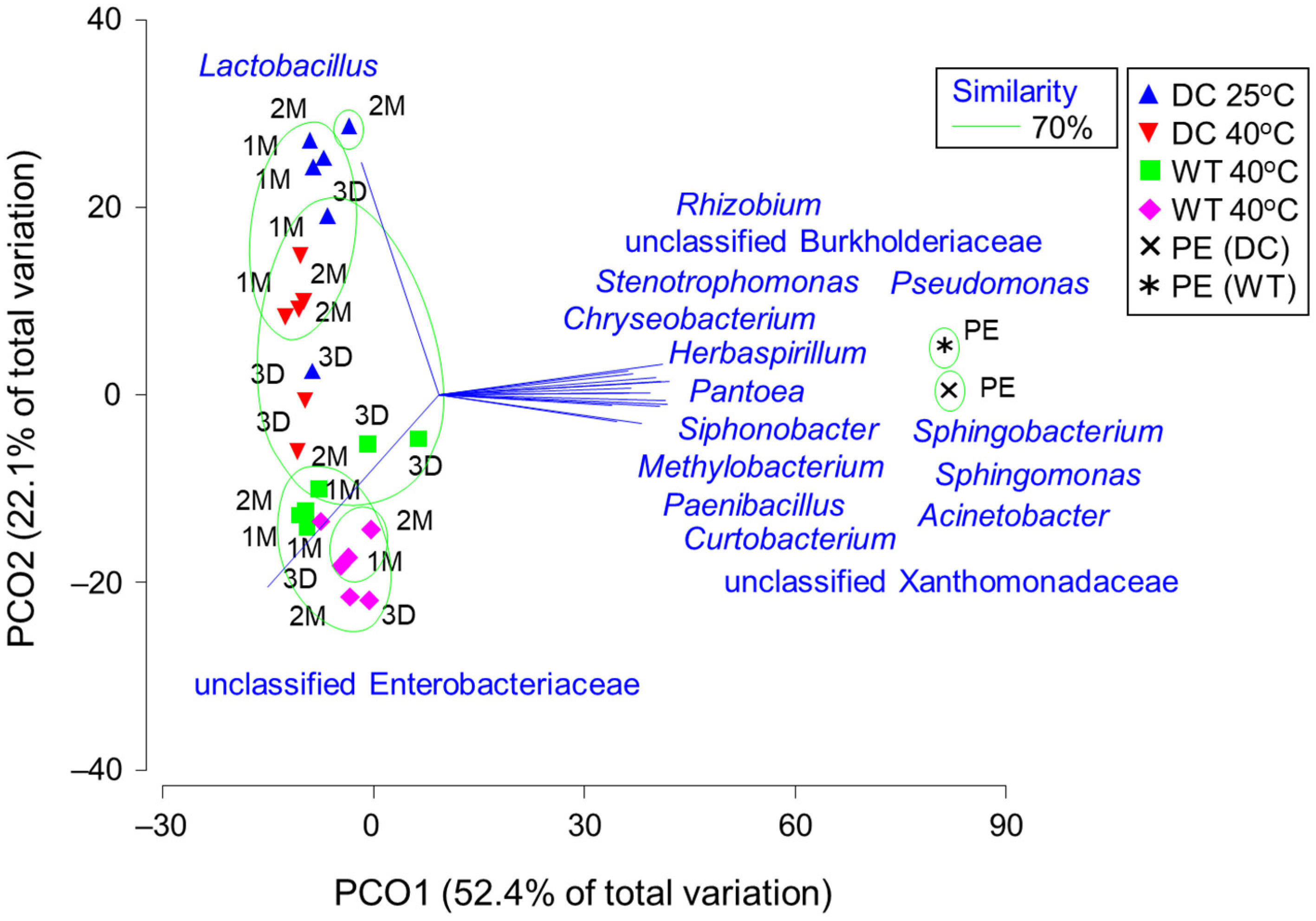
3.2. Bacterial Microbiota
3.3. Fungal Microbiota
3.4. Relationships between the Fermentation Products and Core Genera of Bacteria and Fungi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- McAllister, T.A.; Dunière, L.; Drouin, P.; Xu, S.; Wang, Y.; Munns, K.; Zaheer, R. Silage review: Using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 2018, 101, 4060–4074. [Google Scholar] [CrossRef] [Green Version]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Nishino, N.; Li, Y.; Wang, C.; Parvin, S. Effects of wilting and molasses addition on fermentation and bacterial community in guinea grass silage. Lett. Appl. Microbiol. 2012, 54, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.A.; Poppe, S.; Meier, H. The influence of wilting on the quality of tropical grass silage in Cuba. Arch. Anim. Nutr. 1989, 39, 785–792. [Google Scholar] [CrossRef]
- Parvin, S.; Nishino, N. Bacterial community associated with ensilage process of wilted guinea grass. J. Appl. Microbiol. 2009, 107, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Oude Elferink, S.J.W.H.; Krooneman, E.J.; Gottschal, J.C.; Spoelstra, S.F.; Faber, F.; Driehuis, F. Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 2001, 67, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartowsky, E.J.; Henschke, P.A. Acetic acid bacteria spoilage of bottled red wine–a review. Int. J. Food Microbiol. 2008, 125, 60–70. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.; Henderson, A.; Heron, S. The Biochemistry of Silage, 2nd ed.; Chalcombe: Marlow, UK, 1991. [Google Scholar]
- Tran, T.M.T.; Nguyen, H.V.; Nishino, N. A pilot examination of the fermentation products, aerobic stability and bacterial community of total mixed ration silage produced in Vietnam. Grassl. Sci. 2014, 60, 63–68. [Google Scholar] [CrossRef]
- Wali, A.; Nishino, N. Bacterial and fungal microbiota associated with the ensiling of wet soybean curd residue under prompt and delayed sealing conditions. Microorganisms 2020, 8, 1334. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004, 36, 808–812. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Ben Mahfoudh, K.; Wagner, M.; Loy, A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 2011, 77, 7846–7849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G.; Liu, S.; Armstrong, S.D. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Ni, K.; Zhang, Y.; Lin, Y.; Yang, F. Fermentation characteristics, chemical composition and microbial community of tropical forage silage under different temperatures. Asian-Australas. J. Anim. Sci. 2019, 32, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Ni, K.; Minh, T.T.; Tu, T.T.M.; Tsuruta, T.; Pang, H.; Nishino, N. Comparative microbiota assessment of wilted Italian ryegrass, whole crop corn, and wilted alfalfa silage using denaturing gradient gel electrophoresis and next-generation sequencing. Appl. Microbiol. Biotechnol. 2017, 101, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Zalar, P.; Sybren de Hoog, G.; Schroers, H.-J.; Frank, J.M.; Gunde-Cimerman, N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie Van Leeuwenhoek 2005, 87, 311–328. [Google Scholar] [CrossRef]
- Surussawadee, J.; Khunnamwong, P.; Srisuk, N.; Limtong, S. Papiliotrema siamense f.a., sp. nov., a yeast species isolated from plant leaves. Int. J. Syst. Evol. Microbiol. 2014, 64, 3058–3062. [Google Scholar] [CrossRef] [Green Version]
- Vujanovic, V. Tremellomycetes yeasts in kernel ecological niche: Early indicators of enhanced competitiveness of endophytic and mycoparasitic symbionts against wheat pathobiota. Plants 2021, 10, 905. [Google Scholar] [CrossRef]
- Kot, A.M.; Kieliszek, M.; Piwowarek, K.; Błażejak, S.; Mussagy, C.U. Sporobolomyces and Sporidiobolus–non-conventional yeasts for use in industries. Fungal Biol. Rev. 2021, 37, 41–58. [Google Scholar] [CrossRef]
- Muck, R.E.; Moser, L.E.; Pitt, R.E. Postharvest factors affecting ensiling. In Silage Science and Technology; Buxton, D.R., Muck, R.E., Harrison, J.H., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2003; pp. 251–304. [Google Scholar] [CrossRef]
- Bernardes, T.F.; Daniel, J.L.P.; Adesogan, A.T.; McAllister, T.A.; Drouin, P.; Nussio, L.G.; Huhtanen, P.; Tremblay, G.F.; Bélanger, G.; Cai, Y. Silage review: Unique challenges of silages made in hot and cold regions. J. Dairy Sci. 2018, 101, 4001–4019. [Google Scholar] [CrossRef]
- Weinberg, Z.G.; Szakacs, G.; Ashbell, G.; Hen, Y. The effect of temperature on the ensiling process of corn and wheat. J. Appl. Microbiol. 2001, 90, 561–566. [Google Scholar] [CrossRef] [Green Version]
- Gulfam, A.; Guo, G.; Tajebe, S.; Chen, L.; Liu, Q.; Yuan, X.; Bai, Y.; Shao, T. Characteristics of lactic acid bacteria isolates and their effect on the fermentation quality of Napier grass silage at three high temperatures. J. Sci. Food Agric. 2017, 97, 1931–1938. [Google Scholar] [CrossRef] [PubMed]
- Ohara, H.; Yahata, M. L-lactic acid production by Bacillus sp. in anaerobic and aerobic culture. J. Ferment. Bioeng. 1996, 81, 272–274. [Google Scholar] [CrossRef]
- Payot, T.; Chemaly, Z.; Fick, M. Lactic acid production by Bacillus coagulans—Kinetic studies and optimization of culture medium for batch and continuous fermentations. Enzyme Microb. Technol. 1999, 24, 191–199. [Google Scholar] [CrossRef]
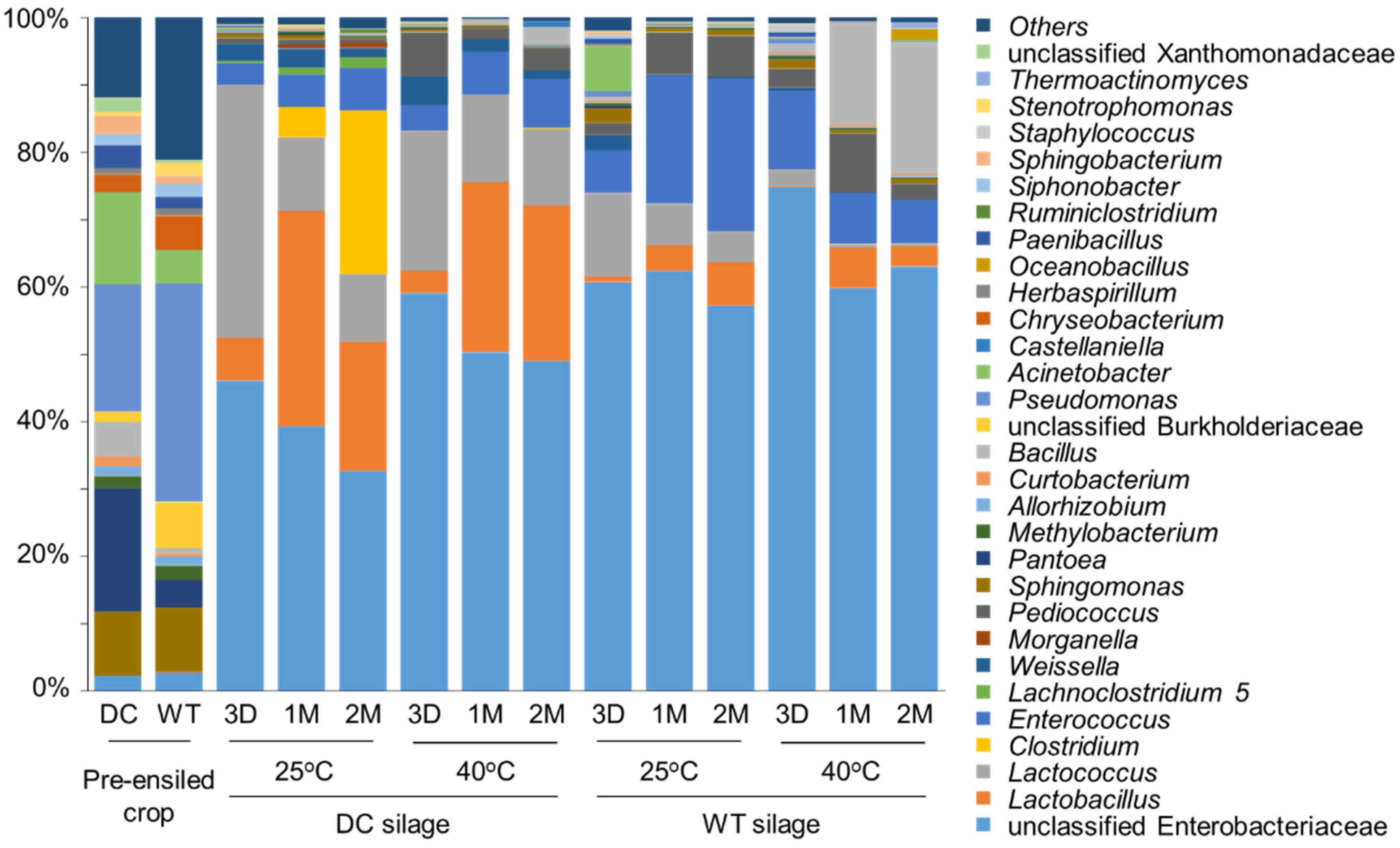



| DM (g/kg) | pH | Lactic Acid (g/kg DM) | Acetic Acid (g/kg DM) | Ethanol (g/kg DM) | L/A | NH3-N (g/kg DM) | Propionic Acid (g/kg DM) | Butyric Acid (g/kg DM) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 3 days | 164 | 5.51 | 15.0 a | 9.05 b | 3.19 b | 1.71 a | 1.33 b | ND | ND |
| 1 month | 162 | 5.51 | 9.44 ab | 25.1 b | 4.02 b | 0.34 b | 2.38 ab | 0.90 | 9.28 | |
| 2 months | 155 | 5.37 | 0.34 b | 49.5 a | 15.0 a | 0.01 b | 3.95 a | 3.47 | 21.6 | |
| SE | 3.41 | 0.16 | 3.56 | 5.16 | 2.16 | 0.20 | 0.54 | - | - | |
| 40 °C | 3 days | 168 z | 5.24 | 21.3 | 6.84 z | 1.63 y | 3.14 x | 1.30 y | ND | ND |
| 1 month | 176 y | 5.04 | 24.9 | 22.3 y | 2.25 y | 1.13 y | 1.99 x | ND | ND | |
| 2 months | 188 x | 4.95 | 25.2 | 27.4 x | 3.98 x | 0.93 y | 2.02 x | ND | ND | |
| SE | 1.16 | 0.10 | 2.95 | 1.14 | 0.32 | 0.39 | 0.08 | - | - | |
| Two-way ANOVA | ||||||||||
| T | <0.01 | <0.01 | <0.01 | <0.05 | <0.01 | <0.01 | <0.05 | - | - | |
| P | 0.126 | 0.319 | 0.253 | <0.01 | <0.01 | <0.01 | <0.01 | - | - | |
| T × P | <0.01 | 0.761 | <0.05 | <0.05 | <0.05 | 0.568 | 0.068 | - | - | |
| DM (g/kg) | pH | Lactic Acid (g/kg DM) | Acetic Acid (g/kg DM) | Ethanol (g/kg DM) | L/A | NH3-N (g/kg DM) | Propionic Acid (g/kg DM) | Butyric Acid (g/kg DM) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 3 days | 354 | 6.12 a | 0.96 b | 1.08 c | 0.77 c | 0.92 | 0.50 b | ND | ND |
| 1 month | 379 | 6.05 ab | 7.06 a | 7.43 b | 3.07 b | 1.01 | 1.51 a | ND | ND | |
| 2 months | 344 | 5.84 b | 9.36 a | 11.6 a | 6.98 a | 0.84 | 1.27 a | ND | ND | |
| SE | 17.5 | 0.06 | 0.98 | 1.00 | 0.49 | 0.20 | 0.16 | - | - | |
| 40 °C | 3 days | 376 | 6.16 x | 1.86 z | 2.33 y | 1.40 y | 0.81 | 0.68 y | ND | ND |
| 1 month | 419 | 5.77 y | 6.89 y | 5.48 y | 1.58 y | 1.30 | 1.23 x | ND | ND | |
| 2 months | 430 | 5.45 z | 13.1 x | 10.3 x | 5.25 x | 1.27 | 1.44 x | ND | ND | |
| SE | 20.0 | 0.05 | 1.45 | 1.01 | 0.56 | 0.22 | 0.09 | - | - | |
| Two-way ANOVA | ||||||||||
| T | <0.01 | <0.01 | 0.164 | 0.432 | 0.068 | 0.251 | 0.823 | - | - | |
| P | 0.217 | <0.01 | <0.01 | <0.01 | <0.01 | 0.404 | <0.01 | - | - | |
| T × P | 0.254 | <0.01 | 0.296 | 0.280 | 0.085 | 0.424 | 0.159 | - | - | |
| Unclassified Enterobacteriaceae | Lactobacillus | Lactococcus | Pediococcus | Enterococcus | Weissella | Clostridium | Bacillus | ||
|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 3 days | 46.1 | 6.32 | 37.6 a | 0.81 | 3.24 | 2.58 | ND | 0.01 |
| 1 month | 39.3 | 32.0 | 10.9 b | 0.83 | 4.79 | 2.79 | 4.54 | 0.15 | |
| 2 months | 32.7 | 19.2 | 10.1 b | 0.57 | 6.34 | 1.31 | 24.3 | 0.02 | |
| SE | 5.31 | 6.69 | 2.10 | 0.36 | 0.83 | 1.29 | - | 0.03 | |
| 40 °C | 3 days | 59.1 | 3.36 y | 20.8 x | 6.36 x | 3.82 z | 4.32 | ND | ND |
| 1 month | 50.3 | 25.3 x | 13.0 y | 1.59 y | 6.37 y | 1.82 | ND | 0.48 | |
| 2 months | 49.0 | 23.1 x | 11.4 y | 3.29 xy | 7.23 x | 1.38 | 0.18 | 2.32 | |
| SE | 4.19 | 3.10 | 0.87 | 0.86 | 0.19 | 1.33 | - | - | |
| Two-way ANOVA | |||||||||
| T | 0.118 | <0.05 | <0.01 | <0.05 | <0.01 | 0.342 | 0.132 | <0.01 | |
| P | <0.05 | 0.672 | <0.05 | <0.01 | 0.084 | 0.801 | 0.074 | <0.01 | |
| T × P | 0.858 | 0.612 | <0.01 | <0.05 | 0.712 | 0.605 | 0.139 | <0.01 | |
| Unclassified Enterobacteriaceae | Lactobacillus | Lactococcus | Pediococcus | Enterococcus | Weissella | Clostridium | Bacillus | ||
|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 3 days | 60.7 | 0.85 b | 12.4 a | 1.76 | 6.43 b | 2.25 a | ND | 0.52 |
| 1 month | 62.4 | 3.76 ab | 6.18 b | 6.22 | 19.0 a | 0.19 b | ND | ND | |
| 2 months | 57.2 | 6.47 a | 4.52 b | 6.12 | 22.8 a | 0.22 b | 0.01 | 0.06 | |
| SE | 3.40 | 1.23 | 0.75 | 1.39 | 1.98 | 0.09 | - | - | |
| 40 °C | 3 days | 74.8 | 0.22 | 2.35 | 2.77 | 11.8 | 0.44 | 0.01 | 1.25 |
| 1 month | 59.8 | 5.99 | 0.53 | 8.77 | 7.61 | 0.08 | 0.02 | 15.1 | |
| 2 months | 63.0 | 3.04 | 0.34 | 2.39 | 6.59 | 0.01 | 0.00 | 19.5 | |
| SE | 7.65 | 1.59 | 0.68 | 3.06 | 2.53 | 0.25 | 0.00 | 11.0 | |
| Two-way ANOVA | |||||||||
| T | 0.427 | <0.05 | <0.01 | 0.165 | 0.108 | <0.01 | 0.819 | 0.528 | |
| P | 0.277 | 0.618 | <0.01 | 0.977 | <0.01 | <0.01 | 0.286 | 0.113 | |
| T × P | 0.424 | 0.217 | <0.05 | 0.437 | <0.01 | <0.01 | 0.528 | 0.493 | |
| Wallemia | Mucor | Saitozyma | Hannaella | Sporobolomyces | Papiliotrema | Moesziomyces | Cladosporium | ||
|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 3 days | 6.58 c | 0.3 | 32.7 a | 6.90 | 14.6 a | 17.7 a | 6.67 | 1.74 |
| 1 month | 47.3 a | ND | 4.24 b | 1.61 | 1.97 b | 5.42 ab | ND | 2.28 | |
| 2 months | 32.1 b | ND | 7.70 b | 1.07 | 1.03 b | 0.02 b | 1.91 | 2.45 | |
| SE | 3.12 | - | 4.83 | 2.54 | 2.22 | 2.85 | - | 0.63 | |
| 40 °C | 3 days | 22.4 | ND | 20.8 | 5.42 | 8.98 | 16.1 | 9.90 x | 2.07 |
| 1 month | 50.1 | ND | 12.7 | ND | 6.44 | 2.73 | 0.68 y | 9.67 | |
| 2 months | 42.9 | ND | 13.0 | 1.13 | 2.86 | 0.40 | 1.45 y | 4.55 | |
| SE | 11.6 | - | 5.92 | - | 2.16 | 4.16 | 1.61 | 5.05 | |
| Two-way ANOVA | |||||||||
| T | <0.05 | - | <0.05 | 0.087 | <0.01 | <0.01 | <0.01 | 0.556 | |
| P | 0.207 | - | 0.900 | 0.593 | 0.910 | 0.671 | 0.323 | 0.308 | |
| T × P | 0.753 | - | 0.208 | 0.916 | 0.134 | 0.911 | 0.412 | 0.618 | |
| Wallemia | Mucor | Saitozyma | Hannaella | Sporobolomyces | Papiliotrema | Moesziomyces | Cladosporium | ||
|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 3 days | 1.82 | 0.26 | 33.9 a | 6.17 | 12.5 | 12.8 | 8.71 a | 3.87 |
| 1 month | 1.08 | 42.7 | 13.9 b | 0.71 | 12.5 | 7.42 | 1.09 b | 2.13 | |
| 2 months | 4.93 | 3.98 | 19.7 ab | 3.29 | 10.6 | 12.4 | 6.67 a | 4.41 | |
| SE | 2.92 | 22.6 | 3.41 | 2.50 | 5.79 | 3.25 | 0.86 | 2.64 | |
| 40 °C | 3 days | 6.36 | 1.16 | 18.6 | 2.14 | 9.95 | 26.7 | 4.82 x | 5.63 |
| 1 month | 8.08 | 0.58 | 14.9 | 7.91 | 16.6 | 17.0 | 1.75 y | 3.99 | |
| 2 months | 18.7 | 0.11 | 13.1 | 3.54 | 13.7 | 13.7 | 0.62 y | 3.00 | |
| SE | 6.78 | 0.75 | 3.27 | 1.85 | 5.01 | 8.10 | 0.52 | 1.61 | |
| Two-way ANOVA | |||||||||
| T | 0.321 | 0.401 | <0.05 | 0.910 | 0.826 | 0.458 | <0.01 | 0.747 | |
| P | 0.095 | 0.294 | <0.05 | 0.548 | 0.737 | 0.154 | <0.01 | 0.694 | |
| T × P | 0.673 | 0.397 | 0.127 | 0.107 | 0.806 | 0.609 | <0.01 | 0.709 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, J.; Nishino, N. Bacterial and Fungal Microbiota of Guinea Grass Silage Shows Various Levels of Acetic Acid Fermentation. Fermentation 2022, 8, 10. https://doi.org/10.3390/fermentation8010010
Hou J, Nishino N. Bacterial and Fungal Microbiota of Guinea Grass Silage Shows Various Levels of Acetic Acid Fermentation. Fermentation. 2022; 8(1):10. https://doi.org/10.3390/fermentation8010010
Chicago/Turabian StyleHou, Jianjian, and Naoki Nishino. 2022. "Bacterial and Fungal Microbiota of Guinea Grass Silage Shows Various Levels of Acetic Acid Fermentation" Fermentation 8, no. 1: 10. https://doi.org/10.3390/fermentation8010010
APA StyleHou, J., & Nishino, N. (2022). Bacterial and Fungal Microbiota of Guinea Grass Silage Shows Various Levels of Acetic Acid Fermentation. Fermentation, 8(1), 10. https://doi.org/10.3390/fermentation8010010






