Bacillus velezensis Identification and Recombinant Expression, Purification, and Characterization of Its Alpha-Amylase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Extraction of Genomic DNA from WangLB Bacterium
2.2. Identification of WangLB Bacterium by 16S rDNA
2.3. Morphological and Biochemical Identification of the WangLB Bacterium
2.4. Alpha-Amylase Activity Assay
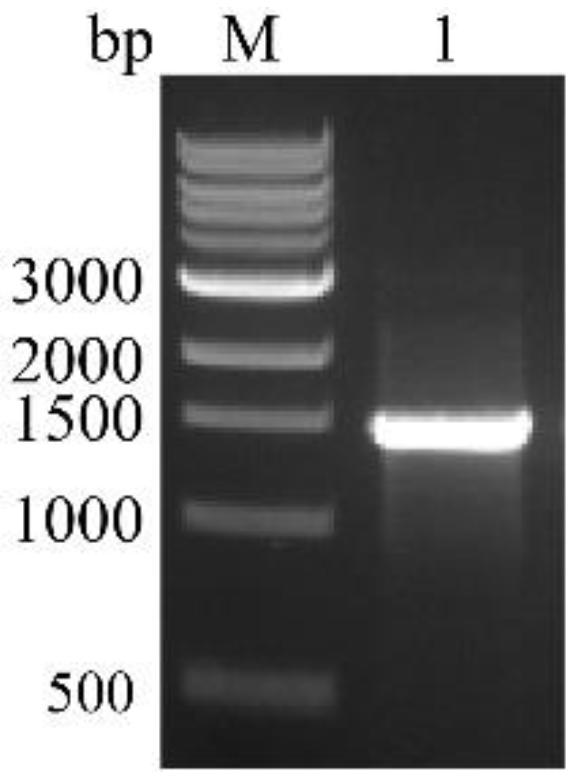
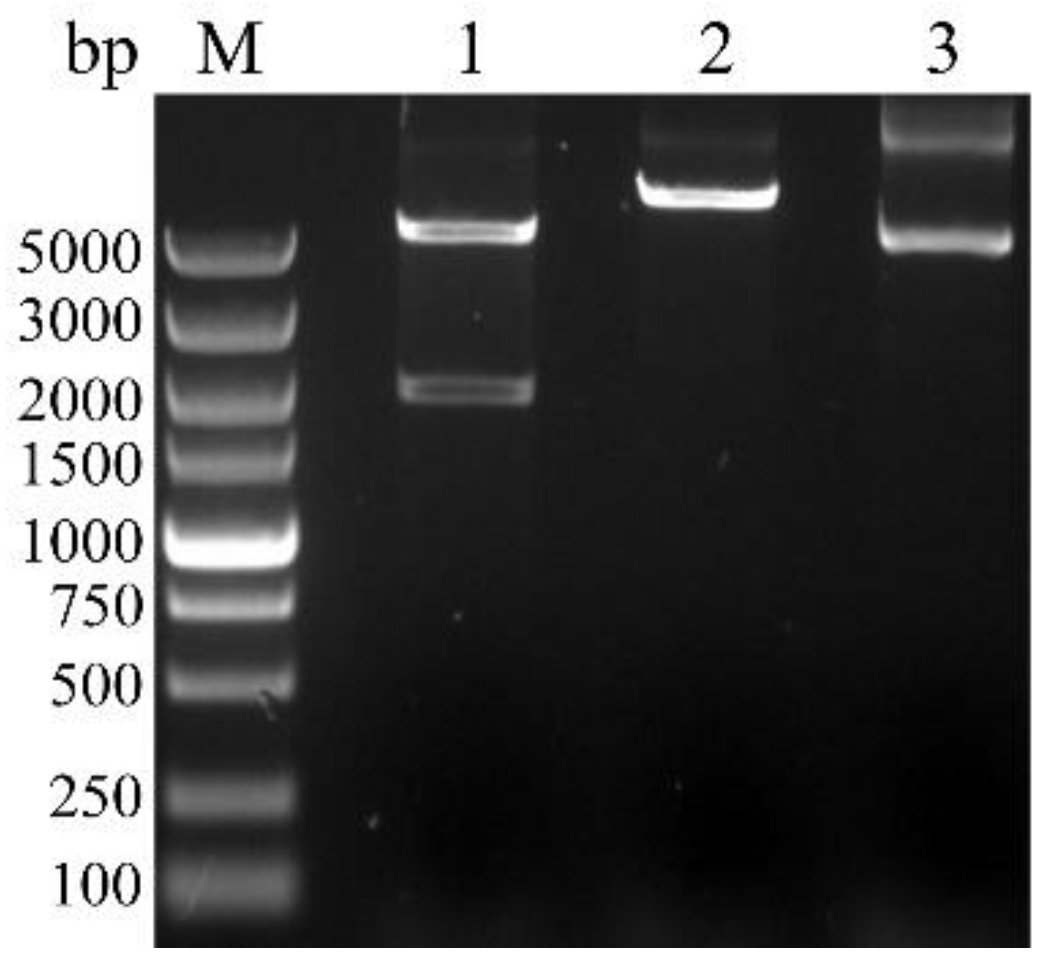
2.5. Cloning and Prokaryotic Expression Vector Construction of BvAmylase Gene
2.6. Prokaryotic Expression and Purification of the BvAmylase
2.7. Characterization of BvAmylase Enzyme
2.7.1. Effect of Temperature on BvAmylase Activity and Stability
2.7.2. Effect of pH on BvAmylase Activity and Stability
2.7.3. Effect of Some Metal Ions on BvAmylase Activity
2.7.4. Effect of Some Detergents and Organic Solvents on BvAmylase Activity
2.7.5. Determination of Km and Vmax for BvAmylase
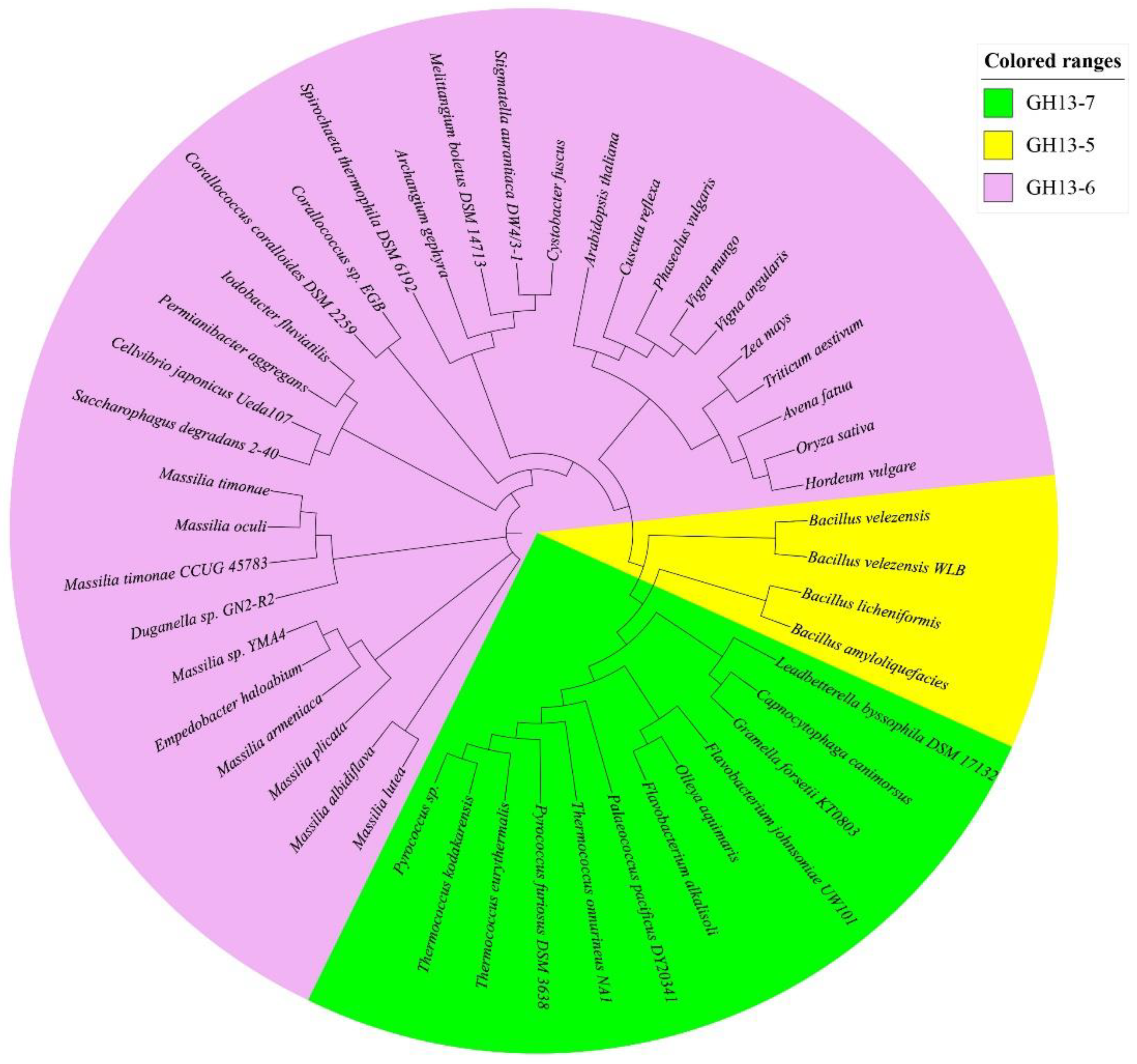
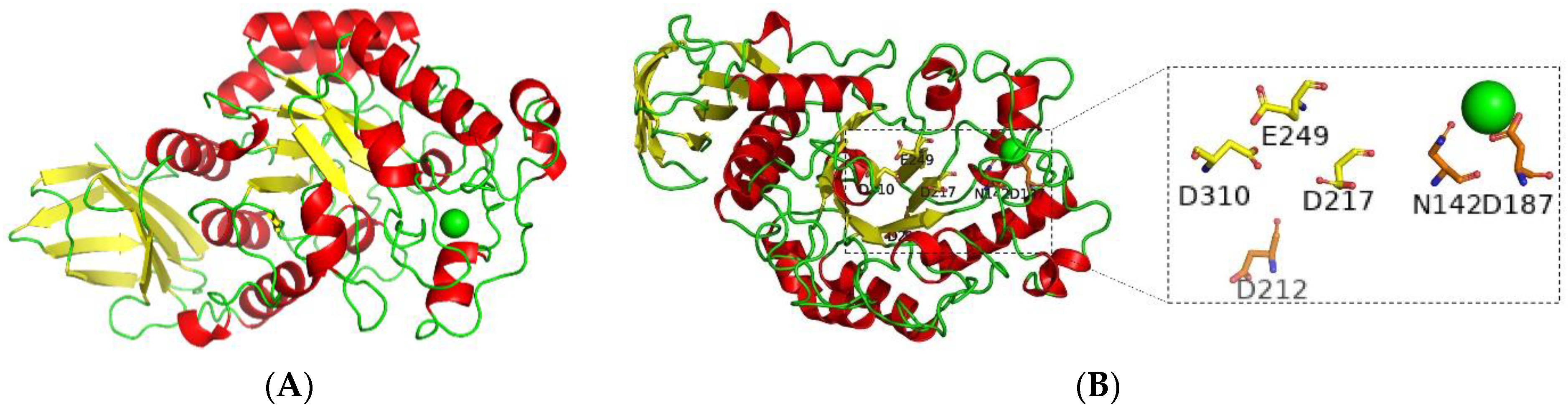
2.7.6. Bioinformatics of BvAmylase Gene
2.7.7. Statistical Analysis
3. Results
3.1. Identification of WangLB Bacterium by 16S rDNA
3.2. Morphological and Biochemical Identification of WangLB
3.3. Cloning, Sequence Analysis of the BvAmylase Gene, and Construction of the Prokaryotic Expression Plasmid
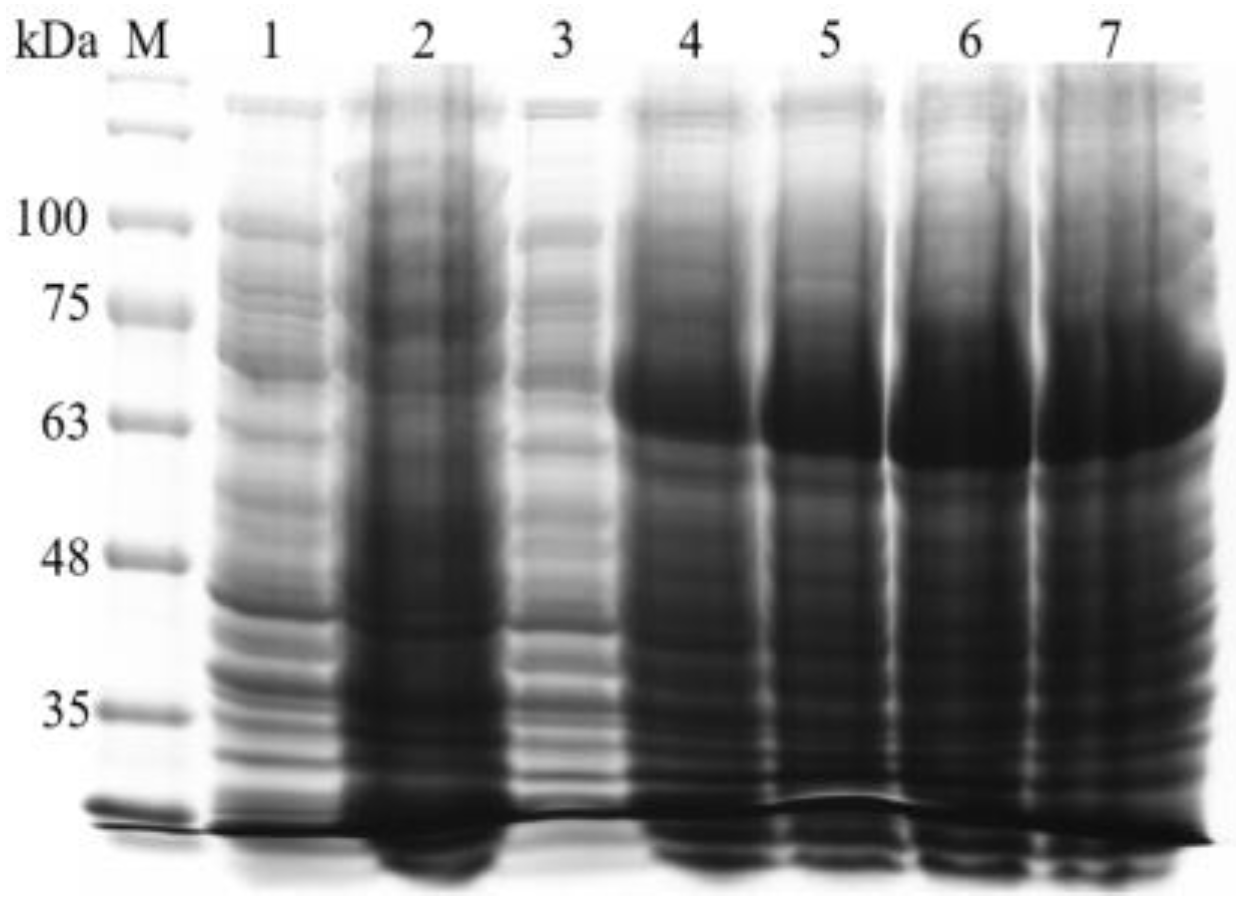
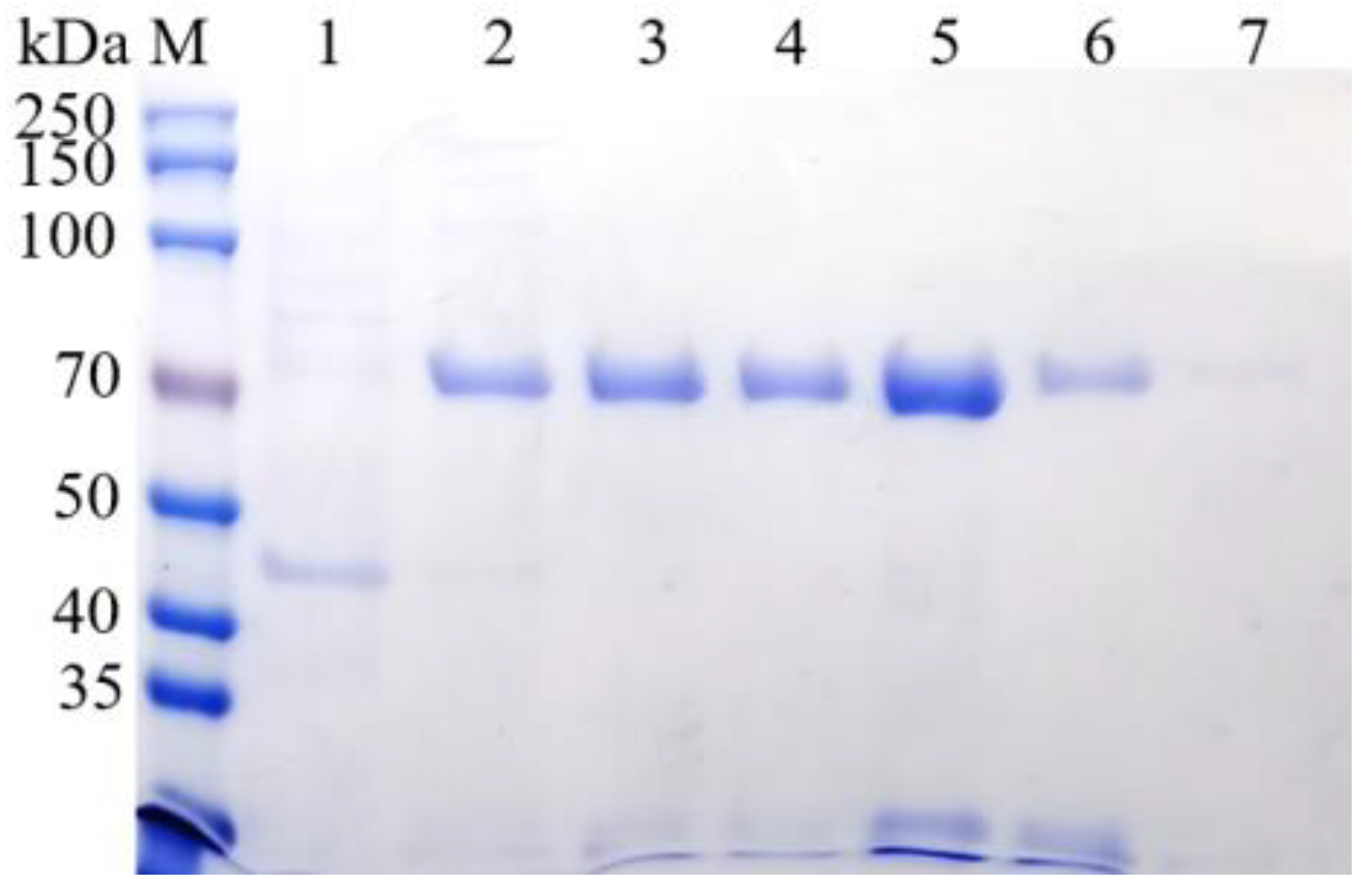
3.4. Prokaryotic Expression and Purification of BvAmylase Gene of WangLB
3.5. Characterization of BvAmylase of WangLB
3.5.1. Effect of Temperature on the α-Amylase Activity and Stability
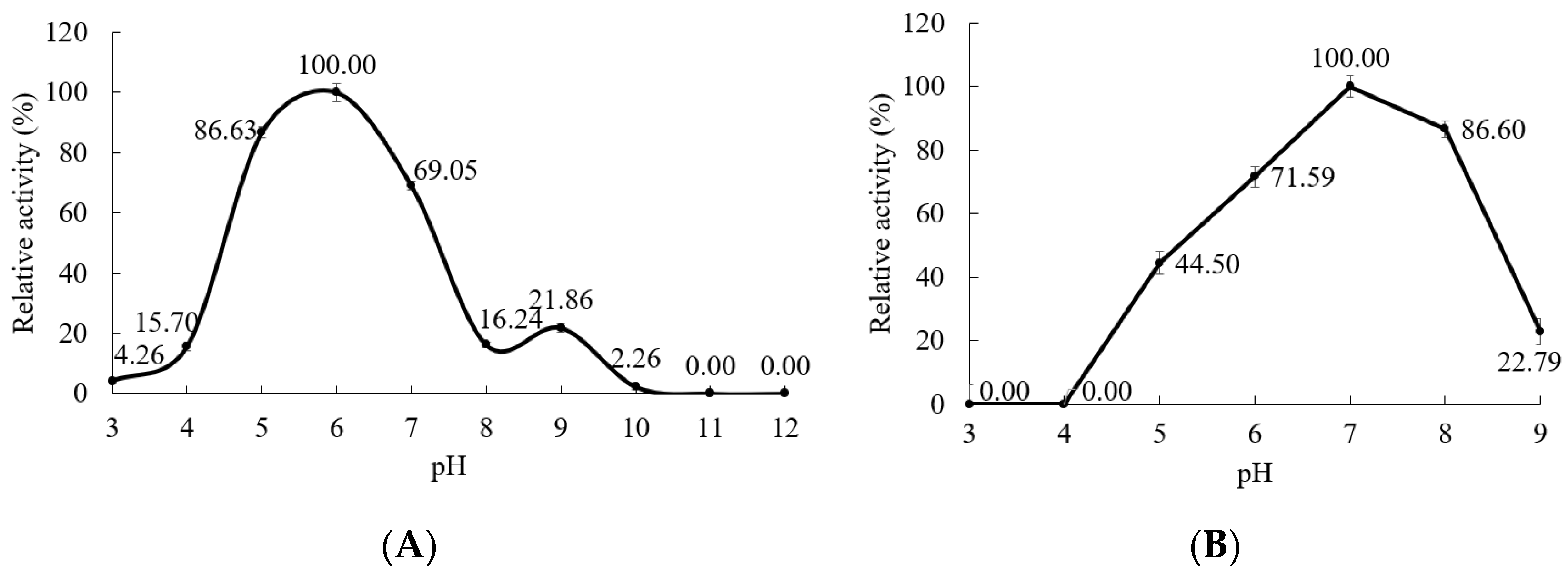
3.5.2. Effect of pH on the α-Amylase Activity and Stability
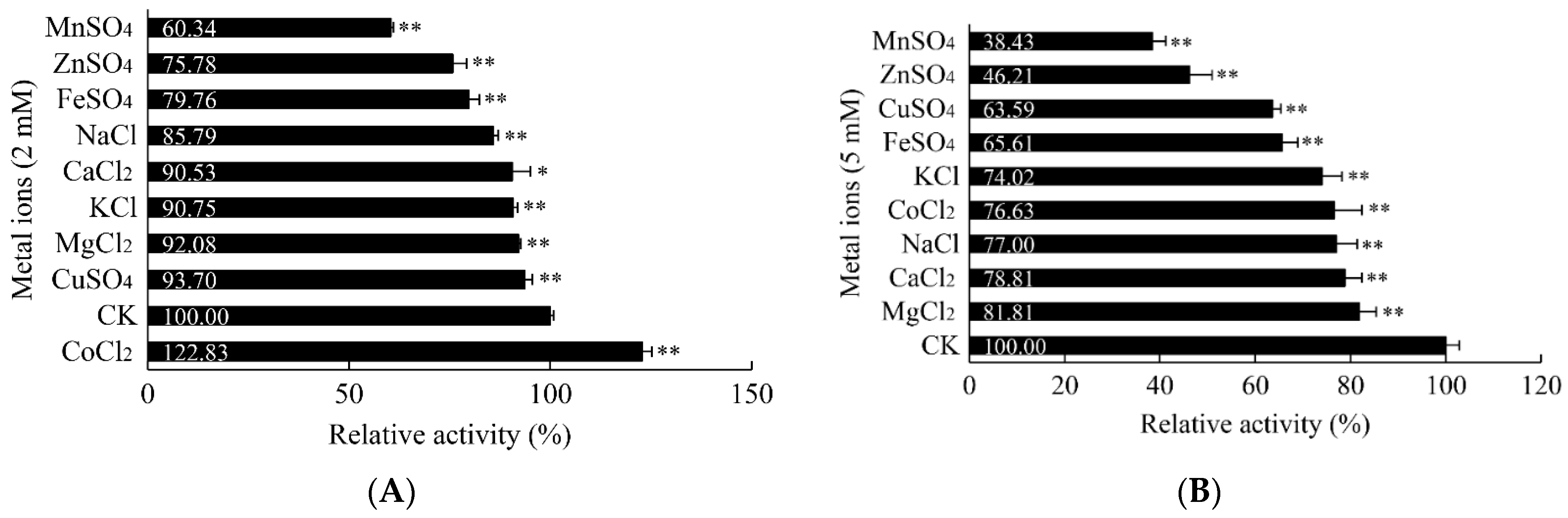
3.5.3. Influence of Metal Ions on the α-Amylase Activity
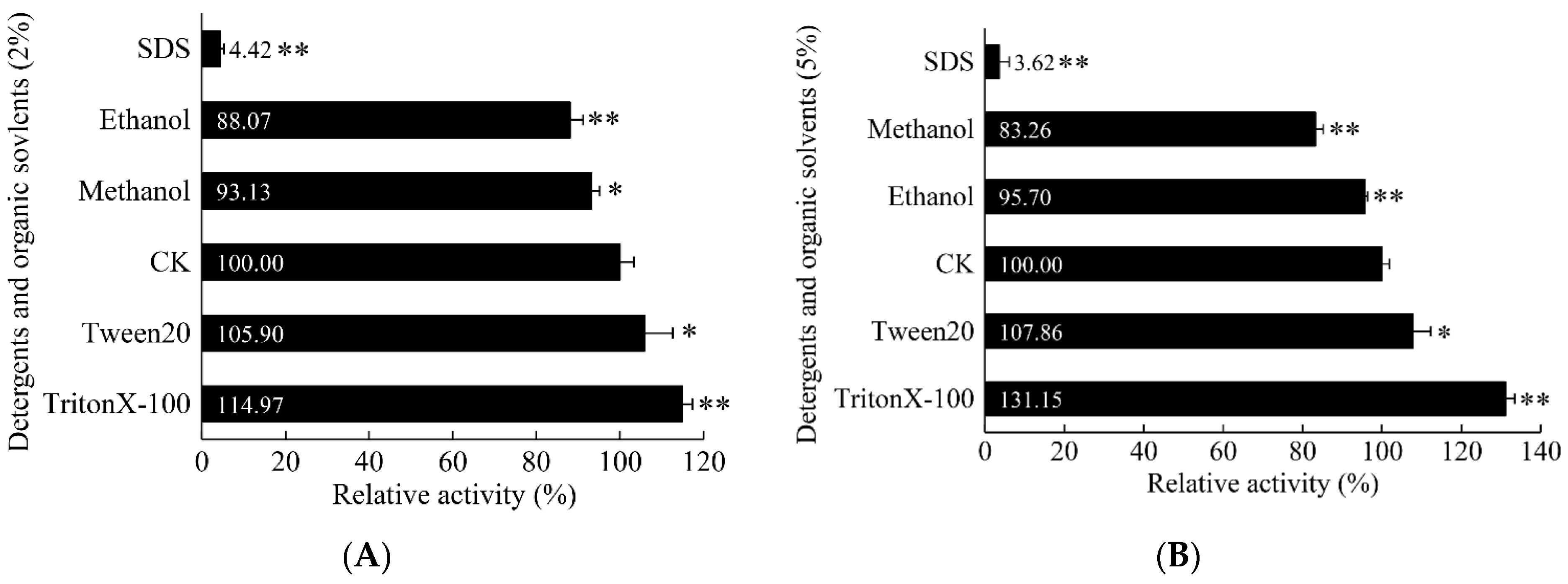
3.5.4. Influence of Detergents and Organic Solvents on α-Amylase Activity
3.5.5. Determination of the Kinetic Parameters of BvAmylase
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gupta, R.; Gigras, P.; Mohapatra, H.; Goswami, V.K.; Chauhan, B. Microbial α-amylases: A biotechnological perspective. Process Biochem. 2003, 38, 1599–1616. [Google Scholar] [CrossRef]
- van der Maarel, M.J.E.C.; van der Veen, B.; Uitdehaag, J.C.M.; Leemhuis, H.; Dijkhuizen, L. Properties and applications of starch-converting enzymes of the α-amylase family. J. Biotechnol. 2002, 94, 137–155. [Google Scholar] [CrossRef] [Green Version]
- Omemu, A.M.; Akpan; Bankole, M.D. Hydrolysis of raw tuber starches by amylase of Aspergillus niger AM07 isolated from the soil. Afr. J. Biotechnol. 2005, 4, 19–25. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Park, J.; Gu, L.; Li, D. An extremely thermostable maltogenic amylase from Staphylothermus marinus: Bacillus expression of the gene and its application in genistin glycosylation. Int. J. Biol. Macromol. 2018, 107, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Matpan Bekler, F.; Güven, K.; Gül Güven, R. Purification and characterization of novel α-amylase from Anoxybacillus ayderensis FMB1. Biocatal. Biotransfor. 2020, 39, 322–332. [Google Scholar] [CrossRef]
- Tagomori, B.Y.; dos Santos, F.C.; Barbosa-Tessmann, I.P. Recombinant expression, purification, and characterization of an α-amylase from Massilia timonae. 3 Biotech 2021, 11, 13. [Google Scholar] [CrossRef]
- Farooq, M.A.; Ali, S.; Hassan, A.; Tahir, H.M.; Mumtaz, S.; Mumtaz, S. Biosynthesis and industrial applications of α-amylase: A review. Arch. Microbiol. 2021, 203, 1281–1292. [Google Scholar] [CrossRef]
- Pinto, E.S.M.; Dorn, M.; Feltes, B.C. The tale of a versatile enzyme: Alpha-amylase evolution, structure, and potential biotechnological applications for the bioremediation of n-alkanes. Chemosphere 2020, 250, 126202. [Google Scholar] [CrossRef]
- Janecek, S.; Svensson, B.; Henrissat, B. Domain evolution in the alpha-amylase family. J. Mol. Evol. 1997, 45, 322–331. [Google Scholar] [CrossRef]
- Sahnoun, M.; Jemli, S.; Trabelsi, S.; Ayadi, L.; Bejar, S. Aspergillus Oryzae S2 alpha-Amylase domain C involvement in activity and specificity: In vivo proteolysis, molecular and docking studies. PLoS ONE 2016, 11, e0153868. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Jeyaseelan, J.; Liu, Y.; Qin, W. Characterization and optimization of amylase production in WangLB, a high amylase-producing strain of Bacillus. Appl. Biochem. Biotechnol. 2016, 180, 136–151. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, S.; Singh, A.; Batra, N.; Singh, J. Microbial production and biotechnological applications of α-galactosidase. Int. J. Biol. Macromol. 2020, 150, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.S.; Gupta, N.; Beliya, E.; Tiwari, S.; Jadhav, S.K. Aspects and recent trends in microbial alpha-amylase: A review. Appl. Biochem. Biotechnol. 2021, 193, 2649–2698. [Google Scholar] [CrossRef]
- Liu, X.; Kokare, C. Chapter 11 Microbial Enzymes of Use in Industry. In Biotechnology of Microbial Enzymes; Brahmachari, G., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 267–298. [Google Scholar]
- van den Burg, B. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 2003, 6, 213–218. [Google Scholar] [CrossRef]
- Rabbee, M.; Ali, M.; Choi, J.; Hwang, B.; Jeong, S.; Baek, K.H. Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Lee, Y.S.; Fang, S.J.; Park, I.H.; Choi, Y.L. Recombinant expression and characterization of an organic-solvent-tolerant alpha-amylase from Exiguobacterium sp. DAU5. Appl. Biochem. Biotechnol. 2013, 169, 1870–1883. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, X.; Liang, J.; Li, X.; Du, L.; Huang, R. Identification of a halophilic alpha-amylase gene from Escherichia coli JM109 and characterization of the recombinant enzyme. Biotechnol. Lett. 2013, 35, 1061–1065. [Google Scholar] [CrossRef]
- Zcan, D.; Spaholu, H.M. Simultaneous production of alpha and beta amylase enzymes using separate gene bearing recombinant vectors in the same Escherichia coli cells. Turk. J. Biol. 2020, 44, 201–207. [Google Scholar] [CrossRef]
- Solat, N.; Shafiei, M. A novel pH and thermo-tolerant halophilic alpha-amylase from moderate halophile Nesterenkonia sp. strain F: Gene analysis, molecular cloning, heterologous expression and biochemical characterization. Arch. Microbiol. 2021, 203, 3641–3655. [Google Scholar] [CrossRef]
- Montor-Antonio, J.J.; Hernandez-Heredia, S.; Avila-Fernandez, A.; Olvera, C.; Sachman-Ruiz, B.; Del Moral, S. Effect of differential processing of the native and recombinant alpha-amylase from Bacillus amyloliquefaciens JJC33M on specificity and enzyme properties. 3 Biotech 2017, 7, 336. [Google Scholar] [CrossRef]
- Gupta, N.; Beliya, E.; Paul, J.S.; Tiwari, S.; Kunjam, S.; Jadhav, S.K. Molecular strategies to enhance stability and catalysis of extremophile-derived α-amylase using computational biology. Extremophiles 2021, 25, 221–233. [Google Scholar] [CrossRef]
- Demirkan, E.S.; Mikami, B.; Adachi, M.; Higasa, T.; Utsumi, S. α-Amylase from B. amyloliquefaciens: Purification, characterization, raw starch degradation and expression in E. coli. Process Biochem. 2005, 40, 2629–2636. [Google Scholar] [CrossRef]
- Liu, X.D.; Xu, Y. A novel raw starch digesting α-amylase from a newly isolated Bacillus sp. YX-1: Purification and characterization. Bioresour. Technol. 2008, 99, 4315–4320. [Google Scholar] [CrossRef] [PubMed]
- Salem, K.; Elgharbi, F.; Ben Hlima, H.; Perduca, M.; Sayari, A.; Hmida-Sayari, A. Biochemical characterization and structural insights into the high substrate affinity of a dimeric and Ca2+ independent Bacillus subtilis alpha-amylase. Biotechnol. Prog. 2020, 36, e2964. [Google Scholar] [CrossRef]
- Afrisham, S.; Badoei-Dalfard, A.; Namaki-Shoushtari, A.; Karami, Z. Characterization of a thermostable, CaCl2-activated and raw-starch hydrolyzing alpha-amylase from Bacillus licheniformis AT70: Production under solid state fermentation by utilizing agricultural wastes. J. Mol. Catal. B Enzym. 2016, 132, 98–106. [Google Scholar] [CrossRef]
- Stamford, T.L.; Stamford, N.P.; Coelho, L.C.; Araujo, J.M. Production and characterization of a thermostable alpha-amylase from Nocardiopsis sp. endophyte of yam bean. Bioresour. Technol. 2001, 76, 137–141. [Google Scholar] [CrossRef]
- Xie, F.; Quan, S.; Liu, D.; Ma, H.; Li, F.; Zhou, F.; Chen, G. Purification and characterization of a novel α-amylase from a newly isolated Bacillus methylotrophicus strain P11-2. Process Biochem. 2014, 49, 47–53. [Google Scholar] [CrossRef]
- Aggarwal, R.; Dutta, T.; Sheikh, J. Extraction of amylase from the microorganism isolated from textile mill effluent vis a vis desizing of cotton. Sustain. Chem. Pharm. 2019, 14, 100178. [Google Scholar] [CrossRef]
- Simair, A.A.; Khushk, I.; Qureshi, A.S.; Bhutto, M.A.; Chaudhry, H.A.; Ansari, K.A.; Lu, C. Amylase production from thermophilic Bacillus sp. BCC 021-50 isolated from a marine environment. Fermentation 2017, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Wang, Y.; Tong, B.; Chen, X.; Chen, J. Purification and biochemical characterization of a thermostable and acid-stable alpha-amylase from Bacillus licheniformis B4-423. Int. J. Biol. Macromol. 2018, 109, 329–337. [Google Scholar] [CrossRef]
- Deljou, A.; Arezi, I. Production of thermostable extracellular α-amylase by a moderate thermophilic Bacillus licheniformis isolated from Qinarje Hot Spring (Ardebil prov. of Iran). Period. Biol. 2017, 118, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Burhan, A.; Nisa, U.; Gökhan, C.; Ömer, C.; Ashabil, A.; Osman, G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochem. 2003, 38, 1397–1403. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Malekzadeh, F.; Malik, K.A. Production of amylase by newly isolated moderate halophile, Halobacillus sp. strain MA-2. J. Microbiol. Methods 2003, 52, 353–359. [Google Scholar] [CrossRef]
- Raul, D.; Biswas, T.; Mukhopadhyay, S.; Das, S.K.; Gupta, S. Production and partial purification of alpha amylase from Bacillus subtilis (MTCC 121) using solid state fermentation. Biochem. Res. Int. 2014, 2014, 568141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kannan, T.R.; Kanagaraj, C. Molecular characteristic of α-AMYLASE enzymes producing from Bacillus lichenformis (JQ946317) using solid state fermentation. Biocatal. Agri. Biotechnol. 2019, 20, 101240. [Google Scholar] [CrossRef]
- Khusro, A.; Barathikannan, K.; Aarti, C.; Agastian, P. Optimization of thermo-alkali stable amylase production and biomass yield from Bacillus sp. under submerged cultivation. Fermentation 2017, 3, 7. [Google Scholar] [CrossRef]
- John Ravindar, D.; Elangovan, N. Molecular identification of amylase producing Bacillus subtilis and detection of optimal conditions. J. Pharm. Res. 2013, 6, 426–430. [Google Scholar] [CrossRef]
- Haki, G.D.; Anceno, A.J.; Rakshit, S.K. Atypical Ca2+-independent, raw-starch hydrolysing α-amylase from Bacillus sp. GRE1: Characterization and gene isolation. World J. Microbiol. Biotechnol. 2008, 24, 2517–2524. [Google Scholar] [CrossRef]
- Jabbour, D.; Sorger, A.; Sahm, K.; Antranikian, G. A highly thermoactive and salt-tolerant α-amylase isolated from a pilot-plant biogas reactor. Appl. Microbiol. Biotechnol. 2013, 97, 2971–2978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Yi, Z.; Fang, Y.; Jin, Y.; Zhao, H. Biochemical and synergistic properties of a novel alpha-mylase from Chinese nong-flavor Daqu. Microb. Cell Fact. 2021, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Satyanarayana, T. Characteristics of a high maltose-forming, acid-stable, and Ca2+-independent α-amylase of the acidophilic Bacillus acidicola. Appl. Biochem. Biotech. 2013, 171, 2053–2064. [Google Scholar] [CrossRef] [PubMed]
- Alikhajeh, J.; Khajeh, K.; Ranjbar, B.; Naderi-Manesh, H.; Lin, Y.H.; Liu, E.; Guan, H.H.; Hsieh, Y.C.; Chuankhayan, P.; Huang, Y.C.; et al. Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: Implications for thermal stability. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Satyanarayana, T. Microbial acid-stable α-amylases: Characteristics, genetic engineering and applications. Process Biochem. 2013, 48, 201–211. [Google Scholar] [CrossRef]
- Sajedi, R.H.; Naderi-Manesh, H.; Khajeh, K.; Ahmadvand, R.; Ranjbar, B.; Asoodeh, A.; Moradian, F. A Ca-independent α-amylase that is active and stable at low pH from the Bacillus sp. KR-8104. Enzyme Microb. Tech. 2005, 36, 666–671. [Google Scholar] [CrossRef]
- Liang, X.; Wang, F.; Luo, X.; Feng, Y.L.; Feng, J.X. Purification and characterization of a highly efficient calcium-independent α-Amylase from Talaromyces pinophilus 1-95. PLoS ONE 2015, 10, e0121531. [Google Scholar] [CrossRef] [Green Version]
- Burhanoğlu, T.; Sürmeli, Y.; Şanlı-Mohamed, G. Identification and characterization of novel thermostable α-amylase from Geobacillus sp. GS33. Int. J. Biol. Macromol. 2020, 164, 578–585. [Google Scholar] [CrossRef]
- Li, X.; Yu, H.Y. Characterization of an organic solvent-tolerant alpha-amylase from a halophilic isolate, Thalassobacillus sp. LY18. Folia Microbiol. 2012, 57, 447–453. [Google Scholar] [CrossRef]
- Zafar, A.; Aftab, M.N.; Din, Z.; Aftab, S.; Iqbal, I.; Haq, I. Cloning, purification and characterization of a highly thermostable amylase gene of Thermotoga petrophila into Escherichia coli. Appl. Biochem. Biotechnol. 2016, 178, 831–848. [Google Scholar] [CrossRef]






| Reaction Pore | Experiments | Abbreviation | Result |
|---|---|---|---|
| 1 | β-Xylosidase | BXYL | − |
| 3 | L-lysine arylaminase | LysA | − |
| 4 | L-aspartate arylaminase | AspA | + |
| 5 | Leucine arylaminase | LeuA | + |
| 7 | Phenylalanine arylaminase | PheA | + |
| 8 | L-proline arylaminase | ProA | + |
| 9 | β-galactosidase | BGAL | − |
| 10 | L-pyrrolidone arylaminase | PyrA | − |
| 11 | α-galactosidase | AGAL | + |
| 12 | Alanine arylaminase | AlaA | + |
| 13 | Tyrosine arylaminase | TyrA | + |
| 14 | β-N-acetylglucosaminidase | BNAG | − |
| 15 | Alanine phenylalanine proline arylaminase | APPA | + |
| 18 | Cyclodextrin | CDEX | − |
| 19 | D-galactose | dGAL | − |
| 21 | Glycogen | GLYG | − |
| 22 | Inositol | INO | − |
| 24 | Methyl-a-D-glucopyranoside acidification | MdG | − |
| 25 | Ellman | ELLM | − |
| 26 | Methyl-d-xyloside | MdX | − |
| 27 | α-mannosidase | AMAN | − |
| 29 | Maltotriose | MTE | − |
| 30 | Glycine arylaminase | GlyA | − |
| 31 | D-mannitol | dMAN | − |
| 32 | D-mannose | dMNE | − |
| 34 | D-melezitose | dMLZ | − |
| 36 | N-acetyl-D-glucosamine | NAG | − |
| 37 | Palatinose | PLE | − |
| 39 | L-rhamnose | IRHA | − |
| 41 | β-glucosidase | BGLU | + |
| 43 | β-mannosidase | BMAN | − |
| 44 | Phosphorylcholine | PHC | − |
| 45 | Pyruvate | PVATE | + |
| 46 | α-glucosidase | AGLU | − |
| 47 | D-tagatose | dTAG | − |
| 48 | D-trehalose | dTRE | − |
| 50 | Inulin | INU | − |
| 53 | D-glucose | dGLU | − |
| 54 | D-ribose | dRIB | − |
| 56 | Putrescine assimilation | PSCNa | − |
| 58 | Growth in 6.5% NaCI | NaCl 6.5% | − |
| 59 | Kanamycin resistance | KAN | − |
| 60 | Oleandomycin resistance | OLD | − |
| 61 | Esculin hydrolysis | ESC | + |
| 62 | Tetrazolium Red | TTZ | + |
| 63 | Resistance to polymyxin B | POLYB R | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Li, C.; Chen, X.; Chio, C.; Shrestha, S.; Qin, W. Bacillus velezensis Identification and Recombinant Expression, Purification, and Characterization of Its Alpha-Amylase. Fermentation 2021, 7, 227. https://doi.org/10.3390/fermentation7040227
Zhang X, Li C, Chen X, Chio C, Shrestha S, Qin W. Bacillus velezensis Identification and Recombinant Expression, Purification, and Characterization of Its Alpha-Amylase. Fermentation. 2021; 7(4):227. https://doi.org/10.3390/fermentation7040227
Chicago/Turabian StyleZhang, Xiaodong, Caixia Li, Xuantong Chen, Chonlong Chio, Sarita Shrestha, and Wensheng Qin. 2021. "Bacillus velezensis Identification and Recombinant Expression, Purification, and Characterization of Its Alpha-Amylase" Fermentation 7, no. 4: 227. https://doi.org/10.3390/fermentation7040227
APA StyleZhang, X., Li, C., Chen, X., Chio, C., Shrestha, S., & Qin, W. (2021). Bacillus velezensis Identification and Recombinant Expression, Purification, and Characterization of Its Alpha-Amylase. Fermentation, 7(4), 227. https://doi.org/10.3390/fermentation7040227








