Abstract
Succinic acid (SA) is one of the top candidate value-added chemicals that can be produced from biomass via microbial fermentation. A considerable number of cell factories have been proposed in the past two decades as native as well as non-native SA producers. Actinobacillus succinogenes is among the best and earliest known natural SA producers. However, its industrial application has not yet been realized due to various underlying challenges. Previous studies revealed that the optimization of environmental conditions alone could not entirely resolve these critical problems. On the other hand, microbial in silico metabolic modeling approaches have lately been the center of attention and have been applied for the efficient production of valuable commodities including SA. Then again, literature survey results indicated the absence of up-to-date reviews assessing this issue, specifically concerning SA production. Hence, this review was designed to discuss accomplishments and future perspectives of in silico studies on the metabolic capabilities of SA producers. Herein, research progress on SA and A. succinogenes, pathways involved in SA production, metabolic models of SA-producing microorganisms, and status, limitations and prospects on in silico studies of A. succinogenes were elaborated. All in all, this review is believed to provide insights to understand the current scenario and to develop efficient mathematical models for designing robust SA-producing microbial strains.
1. Introduction
In the past few years, tremendous attempts and successes have been witnessed in the development of the green economy through the production of chemicals, fuels, materials, etc., from bio-based sources [1]. To begin with, succinic acid (SA) is one of the top potential value-added chemicals that can be produced biotechnologically from biomass resources [2,3]. More interestingly, SA as a high-value platform chemical can be co-produced with high-yield products such as biofuels via integrated biorefinery approaches [4,5] that could offset the process cost [6] and also alleviate waste management issues. In fact, the application of SA ranges from being a specialty chemical in pharmaceutical, food and agricultural areas to being a precursor for industrially important bulk chemicals [7,8]. Among the well-known SA producers are natural host rumen bacteria, model microorganisms and non-conventional microbial cell factories. To this end, Actinobacillus succinogenes, being one of the best natural SA producers, has been given more emphasis in the current review.
From the pre-genomic era to date, the metabolic control and optimization of environmental conditions have been implemented in the microbial production of value-added commodities. However, obviously, there are other complicated genotypic traits that are beyond the appliance of these strategies. In addition, traditional mutagenesis and screening methods have also been employed in the development of improved strains to produce desired products [9]. The success stories of these traditional methods in strain improvement are still undeniable. However, despite being time- and resource-consuming, they could also pose some problems such as the possession of undesired mutations [10], uncontrolled multiple traits, and irreversible damage to the host cell [11]. Furthermore, Lee et al. explained that it could be challenging to recognize which genes have to be manipulated from many lists of genes of a given organism to generate the desired phenotype [12]. On the other hand, taking advantage of technological advancements in high-throughput (HT) techniques and the availability of vast genomic data provides a new paradigm for the rational design of cell factories via the development of predictive mathematical models [9,13]. Hence, these mathematical models would assist in predicting in silico outcomes of genetic and phenotypic traits rather than choosing desired strains resulting from tedious random mutagenesis [9]. Besides, in silico metabolic models combined with other computational, evolutionary and comparative genomic analyses could serve as proof-of-concept for the successful metabolic engineering of cell factories for the synthesis of desired products such as SA [12,14].
Despite the availability of numerous studies and huge potential applications, there is a lack of up-to-date comprehensive reviews covering the implications of in silico metabolic modeling in SA production. The only claimed review in this regard was first published online at the end of 2016 [14]. Therefore, the current study included topics that were not discussed in the previous review, viz., A. succinogenes, new advancements and prospective in this area. This review presented highlights on advancements of research and development (R&D), metabolic pathways and in silico studies generally on SA production and specifically regarding A. succinogenes. Finally, the status quo and future perspectives of in silico metabolic modeling in SA production were summarized. We believe this review can be taken as the first step (from the lists below) in the quest towards the development of robust SA-producing hosts by applying computational methods: (1) understand, summarize the current scenario, identify gaps and suggest possible prospects, (2) design and employ state-of-the-art models, (3) validate via experimentation and literature, and (4) implement an in silico model in vitro and in vivo.
2. Major R&D Advancements on SA and A. succinogenes
When we look back to timelines of SA, its discovery, application and production should be assessed. Figure 1 demonstrates major milestones in the history of SA and A. succinogenes. To begin with, documents revealed that SA was first purified in 1546 by distillation from amber (SA is also known as amber acid), and since then it has been used for various applications [15,16]. However, until recently, the majority of SA has been derived from petroleum-based sources. Encouragingly, SA was proposed twice as “top value-added chemicals from biomass” in 2004 [2] and in the revised 2010 [3] studies. Following this, the first commercial bio-based SA production plants were launched and it was reported that these companies contributed half of the annual global SA production [17,18]. As far as A. succinogenes is concerned, a patent for the production of SA using this strain was registered in 1996 [19], and the first strain isolated from bovine rumen was published in 1999 [20].
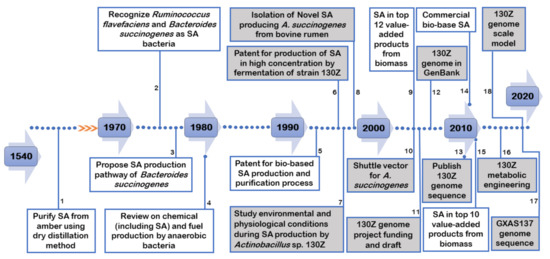
Figure 1.
Major timelines of succinic acid and A. succinogenes. The shaded boxes represent A. succinogenes. The numbers represent the sources of the studies: 1 [15,16], 2 [21], 3 [22], 4 [23], 5 [24], 6 [19], 7 [25], 8 [20], 9 [2], 10 [26], 11 [27], 12 [27], 13 [8], 14 [18], 15 [3], 16 [28], 17 [29], 18 [30].
A literature survey was conducted using the Web of Science database against the terms “succinic acid” and “Actinobacillus succinogenes” separately, employing the “Basic Search” option. According to the search results, there were 19,338 and 426 studies about SA and A. succinogenes, respectively. Figure 2 shows the number of these publications filtered from 1999 to 2020. The research trends of SA revealed that the number of publications has increased chronologically with slight exceptions. For instance, the number of studies in 2018 (the highest) was increased almost 5-fold compared with the 1999 counterpart. This is, in fact, one indication of the attraction of the subject matter to the scientific community. Unlike SA, the number of studies on A. succinogenes indicated yearly fluctuations, with only one published study in 2000 and the absence of publications in the next two consecutive years. Additionally, the number of publications has increased dramatically since 2007, which coincided with the release of A. succinogenes’ genome sequence to GenBank (Figure 1). As a matter of fact, from the total number of publications regarding A. succinogenes, more than 96% of them have been disseminated since the aforementioned year. This shows that the aftermath of the availability of the organism’s genomic data has led to a significant growth of interest and opened a new avenue in research. It is worth mentioning that genomic data are also a stepping stone for in silico metabolic modeling, which is the core theme of this review. Moreover, the highest number of publications for both SA and A. succinogenes was recorded in 2018 (Figure 2).
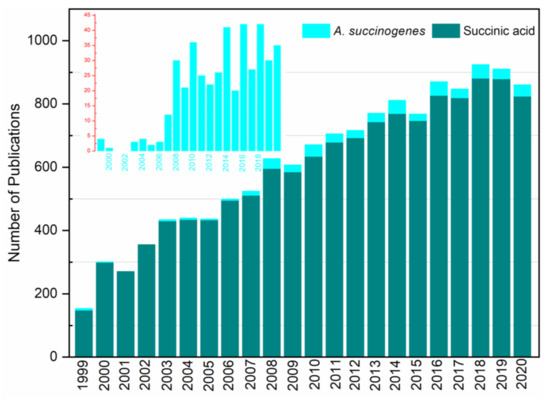
Figure 2.
Literature survey results on succinic acid and A. succinogenes. The inset graph represents A. succinogenes.
3. Succinic Acid Production Pathways
It is important to have an unequivocal understanding about SA production pathways before directly moving to the topic of metabolic modeling. This is partly because finding appropriate biochemical pathways from the metabolic network represents the most challenging task in computational metabolic modeling [31]. Therefore, the realization of the participating pathways in SA production is a crucial step to develop plausible metabolic models and implement the existing ones accordingly. In line with this, applying accumulated knowledge of metabolic pathways combined with the optimization of fermentation processes and other advanced techniques has enabled the development of efficient strains capable of producing target products with high efficiency [32,33].
The tricarboxylic acid (TCA) cycle is among the major biochemical hubs with inevitable functions in the cell mainly for energy generation and precursor synthesis [34]. Obviously, being an intermediate compound of the TCA cycle [35], it is possible to infer that almost all living things can produce SA, whereas the choice of host strain relies on SA being the major end product and, so far, rumen microbes such as A. succinogenes are potentiality considered as the best natural SA producers [36]. In general, based on the downstream metabolites of glycolysis and the TCA cycle, there are three pathways that lead to SA production, namely the reductive and oxidative branches of the TCA cycle and the glyoxylate shunt (GS) (Figure 3). Accordingly, various organisms follow one or more of these routes to produce SA. This diversity may arise due to the absence and/or inactivation of certain enzymes, environmental conditions (e.g., mode of fermentation) or any other reasons. However, interestingly, it is possible to manipulate biochemical pathways by adding novel routes, redirecting existing pathways or removing unnecessary ones through genetic engineering to construct efficient tailor-made cell factories. What is more fascinating is if the TCA cycle has to be exploited for chemical production, SA is the best candidate to be produced with maximum possible routes, whereby the two branches of the TCA cycle are linked via the GS [34] (Figure 3).
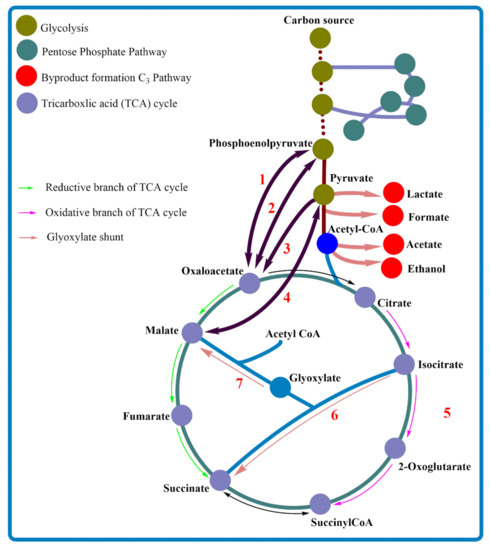
Figure 3.
Major metabolic pathways of succinic acid production. The numbers represent alternative routes towards succinic acid.
Phosphoenolpyruvate (PEP) and pyruvate are important branching nodes for SA production. As shown in Figure 3, the reductive branch of the TCA cycle possibly splits from these nodes and is directed towards SA predominantly by carboxylation reaction. In total, four routes are shown in Figure 3, with two routes from each node represented by 1 to 4. Route 1 and 2 depict the route from PEP to oxaloacetate (OAA), catalyzed by PEP carboxylase and PEP carboxykinase. Route 3 and 4 are directed from pyruvate to OAA and malate, catalyzed by pyruvate carboxylase and malic enzyme, respectively. All in all, this pathway is active under anaerobic conditions and covers TCA cycle intermediate metabolites of OAA, malate, fumarate and SA as an end product. The detailed reactions and mechanisms have been reported in previous publications [34,37,38,39]. Microbes may possess one or more of these alternative pathways, and their efficiencies in SA production have been evaluated previously (Table 1). According to Kim et al., the overexpression of PEP carboxykinase from A. succinogenes’ genome to Escherichia coli improved SA production compared with the native PEP carboxylase [40]. The common byproducts of SA production (formate, acetate, lactate and ethanol) occur at the C3 pathway diverted from pyruvate and acetyl-CoA and cause the competition of carbon flux between the product formation C4 pathway (reductive TCA pathway) and byproduct formation C3 pathway (Figure 3). Researchers attempted to overexpress one or more of the above-mentioned carboxylation enzymes and could enhance SA production and reduce byproduct formations through redirecting carbon flow towards SA-producing reductive branches of the TCA cycle [38,41,42,43].

Table 1.
A few examples of metabolic pathways employed for succinic acid production.
In the oxidative TCA cycle, pyruvate enters into the TCA cycle via acetyl-CoA and is converted to citrate, isocitrate and eventually to SA under aerobic fermentation, as shown in Figure 3 route 5. Likewise, SA production using GS, shown in Figure 3 route 6 and 7, is catalyzed by isocitrate lyase and malate synthase, respectively. This pathway also operates optimally under aerobic conditions. The central metabolic pathway of A. succinogenes lacks GS, and the TCA cycle is also incomplete due to the absence of oxidative TCA cycle enzymes of citrate synthase and isocitrate dehydrogenase [8] (Figure 4). Hence, the wild strain depends entirely on the reductive branch of the TCA cycle for SA production. Table 1 demonstrates SA production by various microorganisms using the aforementioned three major pathways and their combinations. It is also worth noting that each route has merits and demerits. In this regard, Raab et al. explained that the concurrent operation of oxidative and reductive routes appears to be more advantageous than exclusively oxidative or reductive, compromising the advantages and disadvantages of each pathway [58]. As a remark, the final goal of metabolic modifications should be to maximize SA production and at the same time minimize/eliminate byproduct formation to achieve a homo-SA production system.
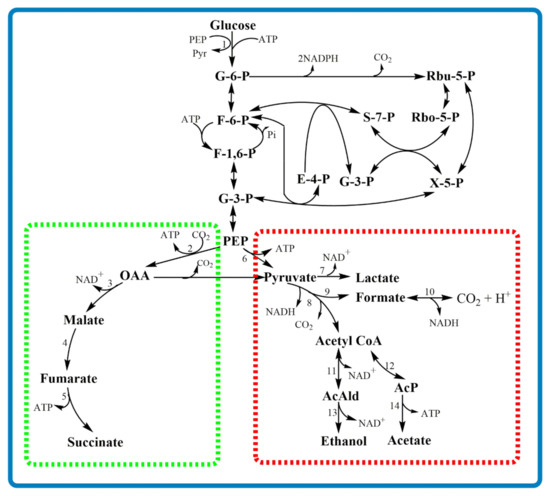
Figure 4.
Central metabolic pathway of A. succinogenes. The green dashed box represents the reductive TCA cycle (C4) for succinic acid production (incomplete TCA cycle) and the red dashed box represents the byproduct formation routes (C3). Numbers represent enzymes involved in the metabolic reactions: 1, PEP: glucose phosphotransferase or hexokinase; 2, PEP carboxykinase; 3, malate dehydrogenase; 4, fumarase; 5, fumarate reductase; 6, pyruvate kinase; 7, lactate dehydrogenase; 8, pyruvate dehydrogenase; 9, pyruvate-formate lyase; 10, formate dehydrogenase; 11, phosphotransacetylase; 12, alcohol dehydrogenase; 13, acetyl-kinase; 14, acetaldehyde dehydrogenase. See the abbreviations in the designated section “Abbreviations”.
4. Metabolic Models of SA Producers
This section describes the developments of in silico metabolic modeling in selected SA-producing strains. The detailed description of various computational methods of metabolic modeling is not within the scope of this review as these concepts have been exhaustively reviewed elsewhere [9,10,14,59,60,61,62,63,64]. Scholars have proposed several metabolic modeling approaches and each method comes with intrinsic advantages and disadvantages. Hence, the application of a specific modeling strategy should consider viable parameters accordingly. In fact, various resources are publicly available for users to engage with for such purposes. For the sake of convenience, Copeland et al. [65], Jing et al. [66] and O’Shea et al. [67] listed out databases, software tools, resources, etc., that could be employed for metabolic modeling studies. Combining accumulated biological knowledge, experimental studies, genomic information and appropriate aforementioned computational inputs, one can possibly execute a metabolic model of a particular biocatalyst for target product synthesis.
Metabolic modeling studies can be generally based on either of the two broad assumptions or their combinations: dynamic/kinetic and static/stoichiometric/steady-state [9,68]. The former describes the variation of metabolites with time by applying differential equations, while the latter assumes that the production and loss of metabolites are equal, resulting in net zero production [31,61]. Kinetic models require a myriad of experimental data for enzyme rate parameters. On the contrary, constraint-based metabolic models, under the assumption of steady-state conditions, involve fundamentally fewer parameters for construction [61]. Hence, this model is commonly employed for the metabolic synthesis of desired products including SA.
Flux balance analysis (FBA) based on constraints has become a universally applicable strategy for metabolic studies [68,69]. In FBA, the flow of metabolites in the metabolic network is analyzed using mathematical approaches. In principle, in silico approaches are executed at dry labs and these simulation results could provide theoretical backgrounds to move to wet labs for the actual experimental studies. This is because of the presence of a plethora of conditions that are practically challenging or even impossible to be tested experimentally in vitro or in vivo. Metabolic models of SA-producing microbes from the very earliest study by Lee et al. [70] to the latest ones are listed chronologically in Table 2. Most of the computational studies were basically focused on non-native strains such as E. coli to explore the metabolic capabilities of microbes and hunt for efficient ways to produce SA. These metabolic models led to the identification and construction of alternative SA pathways (as discussed in the above section) and finally the development of model-guided experimental strategies for enhanced SA production. To begin with, in probably the first in silico study for SA production, Lee et al. constructed a metabolic pathway of E. coli and further conducted metabolic flux analysis (MFA) to calculate flux distributions for the maximum possible SA yield [70]. MFA results of this study revealed that the theoretical SA yield could be improved through recruiting the pyruvate carboxylation pathway (see Figure 3, route 4) rather than the native PEP carboxylation. Interestingly, this theoretical assumption was ultimately validated by experimental set ups.

Table 2.
In silico metabolic studies on succinic acid-producing microorganisms.
Likewise, other potential microbes were also evaluated for their metabolic capability of SA production. The fundamental difference in these in silico studies, besides the algorithms employed, is the scope of metabolic network coverage. The scope could range from central carbon, intermediate, to genome-scale metabolic models. Genome-scale metabolic models (GEMs) are by far the most inclusive models that can help us to predict system-wide phenotypic and genotypic traits so as to facilitate the manipulation of the metabolic network of an organism [13]. Herewith, we have included GEMs of the most commonly employed SA-producing microorganisms (Table 3). As it can be seen in Table 3, each version (if any) of GEM of a given microbe evolves with the incorporation of more metabolites, metabolic reactions, genes and so forth (in reference to the previous version) in the quest towards the construction of the most comprehensive model. In this regard, recent reviews pointed out that E. coli’s GEM appears to be the most complete [59] and the best validated [100] so far. On top of other advantages, one beauty of GEM construction is that it can be applied not only for specific target products like SA, but it can also be used for any aspects of studies of an organism. The GEMs shown in Table 3 may not be necessarily constructed for the purpose of SA production. Therefore, the public availability of the GEMs can be used as a reference (1) for the construction of their latest version, (2) to manipulate the organism’s metabolism for tailored studies and (3) to design GEMs for other related stains and purposes. Moreover, the availability of multiple GEMs of an organism may assist in picking the most suitable model based on specific interest. For instance, Agren et al. [78] considered the earliest GEM (iFF708) [101] to metabolically engineer S. cerevisiae for SA production, despite the availability of other latest models. The authors explained that they chose this model because it focuses on central carbon metabolism and includes relatively small subcellular compartments, which favors it for SA production studies. The power of in silico metabolic modeling has also been seen in a recent SA production study using M. succiniciproducens as a biocatalyst. In this research, GEM [75] was analyzed to characterize malate dehydrogenase (MDH) and finally the overexpression of genes encoding MDH led to the production of the highest overall SA reported to date [99]. This is one of the latest tangible pieces of evidence of applications of model-guided in silico studies in the journey of developing industrially compatible SA-producing strains.

Table 3.
Genome-scale metabolic models of A. succinogenes and other selected succinic acid producers.
5. Attempts at Metabolic Modeling of A. succinogenes
Despite the fact that excess studies have been published on the optimization of upstream, midstream and downstream processing steps of SA production using A. succinogenes, the application of in silico study on this microorganism has not yet been covered as expected. Additionally, one of the core objectives of this study is to expose the current status of computational studies on the development of this biocatalyst and inspire scholars to engage with the topic straightaway. In our previous review [11], we identified six major bottlenecks for industrial application of this strain: (1) byproduct formation, (2) auxotrophy, (3) pH sensitivity, (4) dearth of metabolic engineering tools, (5) redox imbalance (NADH limitations) and (6) product inhibition. Several attempts have been assessed to overcome these challenges. However, almost all of these efforts were fragmented and tedious in a way to solve these grand challenges with commonly employed traditional methods. Therefore, developing well-organized systematic strategies though computational approaches is crucial to alleviate these situations.
As shown in Table 2 and Table 3, there are very few in silico studies in general and only one GEM specifically regarding A. succinogenes. The majority of modeling studies were based on glucose metabolism focusing on the central carbon (specifically glycolysis and TCA cycle) pathways. This is most probably because glucose is the most preferable carbon source of the strain and the two pathways harbor the most important steps in the SA production process. Examples of non-glucose-based metabolic modeling studies include xylose [112,113], sugar mixture [113] and glycerol [88,114]. In general, the metabolic modeling study timeline of A. succinogenes could be seen by dividing it into two major separate eras as pre- and post-genomic. The fundamental research during the pre-genomic [115,116,117] and genomic [8] metabolic modeling studies of A. succinogenes was led by McKinlay et al. In the pre-genomic era, metabolic studies were essentially based on isotope labeling experiments. In the first study, a chemically defined medium was created to evaluate the intracellular metabolic flux and predict the SA production metabolic map of A. succinogenes using 13C labeling experiments [115]. Later on, the strain’s metabolic pathways and fluxes were determined by spectrometry (gas chromatography–mass spectrometry and nuclear magnetic resonance) through C-labeled product isotopes [116]. In the subsequent study, MFA was performed to estimate the influence of carbon dioxide and reductant concentrations on the strain’s metabolism [117]. All the above three experiments were based on C-labeled glucose substrate, in which the first one provided insights on the general SA production metabolism, the second focused on the product/byproduct metabolism and the last one emphasized the intermediate metabolites. Obviously, the results of these in vitro experiments could be viable assets for the construction and validation of the upcoming in silico metabolic studies after them.
The release of the complete genome sequence of A. succinogenes 130Z [8] was a phenomenal motivation for researchers to undergo progressive experiments and attempt a few in silico analysis studies as well. Remarkably, the output has been further stretched beyond this strain in a way that powerful genes of A. succinogenes were transferred into other microorganisms to develop efficient SA-producing cell factories. Besides, these sequence data could be applied in comparative genomic studies of related microbes. The post-genomic metabolic modeling studies are shown in Table 2. Rafieenia developed the metabolic model of A. succinogenes composed of 27 reactions and 28 metabolites with a sugar mixture of glucose and xylose by applying MFA via a computational technique [81]. Furthermore, constrained [88] and dynamic [90] MFA methods were employed to determine flux ranges and maximum productivity in A. succinogenes metabolism, respectively. Clearly, previously discussed modeling studies were all limited at the central carbon metabolism boundaries. Hence, the development of models with more inclusive and higher predictive power was needed. Nag et al. developed an extended intermediate model that contained nucleic acid, amino acid, lipid and glycogen metabolisms in addition to the central carbon metabolism [95]. However, this model still did not explicitly incorporate all the known metabolic pathways of A. succinogenes to have a comprehensive understanding of the strain’s metabolism. At last, the first and only GEM (iBP722) of A. succinogenes was published [30] two decades after the first GEM (i.e., Haemophilus influenzae) was released [118]. To date, to the best of our knowledge, there is no experimental study based on this GEM. All in all, advancements of in silico metabolic modeling of A. succinogenes upgraded from the first 27 reactions to 375 and finally a GEM with 1072 reactions (Table 2 and Table 3). The next model is expected to consider these achievements as frameworks and will be designed thereof.
6. Perspectives and Conclusions
Beholding the magnificent characteristics of computing such as the speed, versatility, accuracy and more, in silico study of metabolic modeling is generally priceless. We have listed some of the advancements, existing gaps and future outlooks in the development of computational methods for effective SA production (Table 4). The accumulated experimental data can be principally helpful to construct and validate in silico metabolic models. In the case of A. succinogenes, the majority of the modeling study relied on isotope labeling on a glucose substrate. The limitation of this approach is its restriction to only core metabolic networks so that it is unable to move forward to a GEM [119]. Besides, in such studies, non-glucose substrates were overlooked and this could impact the notion to exploit all available bioresources for SA production. Bear in mind that one of the fascinating advantages of A. succinogenes as an SA-producing biocatalyst is its utilization of a wide range of substrates.

Table 4.
Opportunities, gaps/challenges and perspectives for in silico metabolic modeling of succinic acid production.
Progress in in silico modeling is related to advancements in biotechnological HT techniques and access to omics data, bioinformatics tools and resources and literatures integrated with high-performance computing (HPC) systems. This is an interdisciplinary approach that needs collaboration from various experts from life, chemical and physical sciences and computational, mathematical and other backgrounds. In case technical gaps in HT and HPC and other issues are encountered, this could apparently be addressed by these collaborations and lessons learned from previous models. It is worth mentioning that the final goal of in silico modeling is to develop metabolically capable organisms for the large-scale production of target products. Nonetheless, most of the previous computational studies, particularly on SA production, remained at the theoretical stage. Evidently, a review by Valderrama-Gomez et al. [14] verified that only approximately 38% of the reported studies in the temporal years between 2002 and 2016 could be experimentally tested from the total computational studies. Unfortunately, none of them were the exact applications of their corresponding in silico predictions. It is recommended to construct highly predictive models, and this model should guide metabolic engineering strategy to achieve the outline goal: in silico → in vitro → in vivo.
As far as A. succinogenes is concerned, taking overwhelming advantages of this strain over other SA-producing strains and research advancements discussed in our previous review [11] and points raised here, it is crucial at this point to design an industrially robust biocatalyst of its kind.
Author Contributions
W.D. and Z.Q. conceived and designed the research. W.D. analyzed the data, generated figures and tables and wrote the manuscript. Z.W., X.L. and M.W. took part in editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Innovation Program of Hunan Province (2018RS3101), Furong Scholars Award Program of Hunan Province (Xiang Jiao Tong (2020) No. 58), High-tech Industry Technology Innovation Leading Plan (2020GK4106).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interests.
Abbreviations
| AcAld | acetaldehyde |
| ATP | adenosine triphosphate |
| C4 | 4 carbon |
| e.g., | example |
| F-6-P | fructose-6-phosphate |
| G-3-P | glyceraldehyde-3-phosphate |
| GEM | genome-scale metabolic model |
| HPC | high-performance computing |
| IDH | isocitrate dehydrogenase |
| NADH | nicotinamide adenine dinucleotide |
| OAA | oxaloacetate |
| PEP | phosphoenolpyruvate |
| PPP | pentose phosphate pathway |
| pyc | pyruvate carboxylase gene |
| R&D | research and development |
| Ru-5-P | ribulose-5-phosphate |
| SA | succinic acid |
| TCA | tricarboxylic acid |
| AcP | acetylphosphate |
| C3 | 3 carbon |
| CO2 | carbon dioxide |
| F-1,6-P | fructose-1,6-bisphosphate |
| FBA | flux balance analysis |
| G-6-P | glucose-6-phosphate |
| GS | glyoxylate shunt |
| HT | high throughput |
| MFA | metabolic flux analysis |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| OPR | open reading frame |
| Pi | inorganic phosphate |
| ptsG | PEP-dependent phosphotransferase system glucose-specific gene |
| Pyr | pyruvate |
| Rbo-5-P | ribose-5-phosphate |
| S-7-P | sedoheptulose-7-phosphate |
| SDH | succinate dehydrogenase |
| X-5-P | xylulose-5-phosphate |
References
- Dessie, W.; Luo, X.; Tang, J.; Tang, W.; Wang, M.; Qin, Z.; Tan, Y. Towards Full Utilization of Biomass Resources: A Case Study on Industrial Hemp Residue and Spent Mushroom Substrate. Processes 2021, 9, 1200. [Google Scholar] [CrossRef]
- Werpy, T.; Petersen, G. Top Value Added Chemicals from Biomass: Volume I—Results of Screening for Potential Candidates from Sugars and Synthesis Gas; National Renewable Energy Lab.: Golden, CO, USA, 2004. [Google Scholar]
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates—The US Department of Energy’s “Top 10” revisited. Green Chem. 2010, 12, 539–554. [Google Scholar] [CrossRef]
- Pateraki, C.; Patsalou, M.; Vlysidis, A.; Kopsahelis, N.; Webb, C.; Koutinas, A.A.; Koutinas, M. Actinobacillus succinogenes: Advances on succinic acid production and prospects for development of integrated biorefineries. Biochem. Eng. J. 2016, 112, 285–303. [Google Scholar] [CrossRef]
- Dessie, W.; Luo, X.; Wang, M.; Feng, L.; Liao, Y.; Wang, Z.; Yong, Z.; Qin, Z. Current advances on waste biomass transformation into value-added products. Appl. Microbiol. Biotechnol. 2020, 104, 4757–4770. [Google Scholar] [CrossRef]
- Bender, T.A.; Dabrowski, J.A.; Gagné, M.R. Homogeneous catalysis for the production of low-volume, high-value chemicals from biomass. Nat. Rev. Chem. 2018, 2, 35–46. [Google Scholar] [CrossRef]
- Zeikus, J.G.; Jain, M.K.; Elankovan, P. Biotechnology of succinic acid production and markets for derived industrial products. Appl. Microbiol. Biotechnol. 1999, 51, 545–552. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Laivenieks, M.; Schindler, B.D.; McKinlay, A.A.; Siddaramappa, S.; Challacombe, J.F.; Lowry, S.R.; Clum, A.; Lapidus, A.L.; Burkhart, K.B.; et al. A genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genom. 2010, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Cvijovic, M.; Bordel, S.; Nielsen, J. Mathematical models of cell factories: Moving towards the core of industrial biotechnology. Microb. Biotechnol. 2011, 4, 572–584. [Google Scholar] [CrossRef]
- Badri, A.; Srinivasan, A.; Raman, K. In Silico Approaches to Metabolic Engineering. In Current Developments in Biotechnology and Bioengineering; Gunasekaran, P., Noronha, S., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 161–200. [Google Scholar] [CrossRef]
- Dessie, W.; Xin, F.; Zhang, W.; Jiang, Y.; Wu, H.; Ma, J.; Jiang, M. Opportunities, challenges, and future perspectives of succinic acid production by Actinobacillus succinogenes. Appl. Microbiol. Biotechnol. 2018, 102, 9893–9910. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, J.M.; Song, H.; Lee, J.W.; Kim, T.Y.; Jang, Y.-S. From genome sequence to integrated bioprocess for succinic acid production by Mannheimia succiniciproducens. Appl. Microbiol. Biotechnol. 2008, 79, 11–22. [Google Scholar] [CrossRef]
- Milne, C.B.; Kim, P.-J.; Eddy, J.A.; Price, N.D. Accomplishments in genome-scale in silico modeling for industrial and medical biotechnology. Biotechnol. J. 2009, 4, 1653–1670. [Google Scholar] [CrossRef]
- Valderrama-Gomez, M.A.; Kreitmayer, D.; Wolf, S.; Marin-Sanguino, A.; Kremling, A. Application of theoretical methods to increase succinate production in engineered strains. Bioprocess Biosyst. Eng. 2017, 40, 479–497. [Google Scholar] [CrossRef]
- Smyth, H.F.; Carpenter, C.P.; Weil, C.S. Range-finding toxicity data: List IV. AMA Arch. Indust. Hyg. Occup. Med. 1951, 4, 119–122. [Google Scholar]
- Saxena, R.K.; Saran, S.; Isar, J.; Kaushik, R. Production and Applications of Succinic Acid. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Negi, S., Soccol, C.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 601–630. [Google Scholar] [CrossRef]
- Li, J.; Rong, L.; Zhao, Y.; Li, S.; Zhang, C.; Xiao, D.; Foo, J.L.; Yu, A. Next-generation metabolic engineering of non-conventional microbial cell factories for carboxylic acid platform chemicals. Biotechnol. Adv. 2020, 43, 107605. [Google Scholar] [CrossRef] [PubMed]
- Cok, B.; Tsiropoulos, I.; Roes, A.L.; Patel, M.K. Succinic acid production derived from carbohydrates: An energy and greenhouse gas assessment of a platform chemical toward a bio-based economy. Biofuels Bioprod. Biorefin. 2014, 8, 16–29. [Google Scholar] [CrossRef]
- Guettler, M.V.; Jain, M.K.; Rumler, D. Method for Making Succinic Acid, Bacterial Variants for Use in the Process, and Methods for Obtaining Variants. U.S. Patent 5,573,931, 12 November 1996. [Google Scholar]
- Guettler, M.V.; Rumler, D.; Jain, M.K. Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int. J. Syst. Evol. Microbiol. 1999, 49, 207–216. [Google Scholar] [CrossRef]
- Wolin, M.J. Metabolic interactions among intestinal microorganisms. Am. J. Clin. Nutr. 1974, 27, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.L. The pathway of formation of acetate and succinate from pyruvate by Bacteroides succinogenes. Arch. Microbiol. 1978, 117, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Zeikus, J.G. Chemical and fuel production by anaerobic bacteria. Annu. Rev. Microbiol. 1980, 34, 423–464. [Google Scholar] [CrossRef]
- Glassner, D.A.; Datta, R. Process for the Production and Purification of Succinic Acid. U.S. Patent 5,143,834, 1 September 1992. [Google Scholar]
- der Werf, M.J.V.; Guettler, M.V.; Jain, M.K.; Zeikus, J.G. Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch. Microbiol. 1997, 167, 332–342. [Google Scholar] [CrossRef]
- Kim, P.; Laivenieks, M.; McKinlay, J.; Vieille, C.; Gregory Zeikus, J. Construction of a shuttle vector for the overexpression of recombinant proteins in Actinobacillus succinogenes. Plasmid 2004, 51, 108–115. [Google Scholar] [CrossRef]
- JGI. Available online: https://genome.jgi.doe.gov/portal/actsu/actsu.info.html (accessed on 7 January 2021).
- Joshi, R.V.; Schindler, B.D.; McPherson, N.R.; Tiwari, K.; Vieille, C. Development of a markerless knockout method for Actinobacillus succinogenes. Appl. Environ. Microbiol. 2014, 80, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shen, N.; Qin, Y.; Zhu, J.; Li, Y.; Wu, J.; Jiang, M.-G. Complete Genome Sequence of Actinobacillus succinogenes GXAS137, a Highly Efficient Producer of Succinic Acid. Gen. Announc. 2018, 6, e01562-17. [Google Scholar] [CrossRef]
- Pereira, B.; Miguel, J.; Vilaça, P.; Soares, S.; Rocha, I.; Carneiro, S. Reconstruction of a genome-scale metabolic model for Actinobacillus succinogenes 130Z. BMC Syst. Biol. 2018, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Pitkänen, E.; Jouhten, P.; Rousu, J. Inferring branching pathways in genome-scale metabolic networks. BMC Syst. Biol. 2009, 3, 103. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, J.; Shang, X.; Wang, B.; Liu, S.; Chai, X.; Tan, T.; Zhang, Y.; Wen, T. A new genome-scale metabolic model of Corynebacterium glutamicum and its application. Biotechnol. Biofuels 2017, 10, 169. [Google Scholar] [CrossRef]
- Miklóssy, I.; Bodor, Z.; Sinkler, R.; Orbán, K.C.; Lányi, S.; Albert, B. In silico and in vivo stability analysis of a heterologous biosynthetic pathway for 1,4-butanediol production in metabolically engineered E. coli. J. Biomol. Str. Dyn. 2017, 35, 1874–1889. [Google Scholar] [CrossRef] [PubMed]
- Vuoristo, K.S.; Mars, A.E.; Sanders, J.P.M.; Eggink, G.; Weusthuis, R.A. Metabolic Engineering of TCA Cycle for Production of Chemicals. Trends Biotechnol. 2016, 34, 191–197. [Google Scholar] [CrossRef]
- Nghiem, N.P.; Kleff, S.; Schwegmann, S. Succinic Acid: Technology Development and Commercialization. Fermentation 2017, 3, 26. [Google Scholar] [CrossRef]
- Song, H.; Lee, S.Y. Production of succinic acid by bacterial fermentation. Enzym. Microb. Technol. 2006, 39, 352–361. [Google Scholar] [CrossRef]
- Cheng, K.-K.; Wang, G.-Y.; Zeng, J.; Zhang, J.-A. Improved Succinate Production by Metabolic Engineering. Biomed. Res. Int. 2013, 2013, 538790. [Google Scholar] [CrossRef]
- Vemuri, G.N.; Eiteman, M.A.; Altman, E. Effects of Growth Mode and Pyruvate Carboxylase on Succinic Acid Production by Metabolically Engineered Strains of Escherichia coli. Appl. Environ. Microbiol. 2002, 68, 1715–1727. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Z.; Zhu, N.; Wang, B.; Chen, T.; Zhao, X. Metabolic engineering of Escherichia coli and in silico comparing of carboxylation pathways for high succinate productivity under aerobic conditions. Microbiol. Res. 2014, 169, 432–440. [Google Scholar] [CrossRef]
- Kim, P.; Laivenieks, M.; Vieille, C.; Zeikus, J.G. Effect of Overexpression of Actinobacillus succinogenes Phosphoenolpyruvate Carboxykinase on Succinate Production in Escherichia coli. Appl. Environ. Microbiol. 2004, 70, 1238–1241. [Google Scholar] [CrossRef]
- Cui, Z.; Gao, C.; Li, J.; Hou, J.; Lin, C.S.K.; Qi, Q. Engineering of unconventional yeast Yarrowia lipolytica for efficient succinic acid production from glycerol at low pH. Metab. Eng. 2017, 42, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Vadali, R.V.; Bennett, G.N.; San, K.-Y. Increasing the Acetyl-CoA Pool in the Presence of Overexpressed Phosphoenolpyruvate Carboxylase or Pyruvate Carboxylase Enhances Succinate Production in Escherichia coli. Biotechnol. Prog. 2004, 20, 1599–1604. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Zhang, X.; Yang, P.; Liang, Q.; Qi, Q. A novel whole-phase succinate fermentation strategy with high volumetric productivity in engineered Escherichia coli. Bioresour. Technol. 2013, 149, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zheng, P.; Yu, F.; Yang, Q. A two-stage process for succinate production using genetically engineered Corynebacterium acetoacidophilum. Process Biochem. 2015, 50, 1692–1700. [Google Scholar] [CrossRef]
- Tsuji, A.; Okada, S.; Hols, P.; Satoh, E. Metabolic engineering of Lactobacillus plantarum for succinic acid production through activation of the reductive branch of the tricarboxylic acid cycle. Enzym. Microb. Technol. 2013, 53, 97–103. [Google Scholar] [CrossRef]
- Jojima, T.; Noburyu, R.; Suda, M.; Okino, S.; Yukawa, H.; Inui, M. Improving Process Yield in Succinic Acid Production by Cell Recycling of Recombinant Corynebacterium glutamicum. Fermentation 2016, 2, 5. [Google Scholar] [CrossRef]
- Yuzbashev, T.V.; Yuzbasheva, E.Y.; Sobolevskaya, T.I.; Laptev, I.A.; Vybornaya, T.V.; Larina, A.S.; Matsui, K.; Fukui, K.; Sineoky, S.P. Production of succinic acid at low pH by a recombinant strain of the aerobic yeast Yarrowia lipolytica. Biotechnol. Bioeng. 2010, 107, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, A.A.; Ledesma-Amaro, R.; Lin, C.S.K.; Coulon, F.; Thakur, V.K.; Kumar, V. Bioproduction of succinic acid from xylose by engineered Yarrowia lipolytica without pH control. Biotechnol. Biofuels 2020, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, B.; Chen, T.; Wang, Z.; Tang, Y.-J.; Zhao, X. Directed pathway evolution of the glyoxylate shunt in Escherichia coli for improved aerobic succinate production from glycerol. J. Ind. Microbiol. Biotechnol. 2013, 40, 1461–1475. [Google Scholar] [CrossRef]
- Zhu, L.-W.; Li, X.-H.; Zhang, L.; Li, H.-M.; Liu, J.-H.; Yuan, Z.-P.; Chen, T.; Tang, Y.-J. Activation of glyoxylate pathway without the activation of its related gene in succinate-producing engineered Escherichia coli. Metab. Eng. 2013, 20, 9–19. [Google Scholar] [CrossRef]
- Arikawa, Y.; Kobayashi, M.; Kodaira, R.; Shimosaka, M.; Muratsubaki, H.; Enomoto, K.; Okazaki, M. Isolation of sake yeast strains possessing various levels of succinate- and/or malate-producing abilities by gene disruption or mutation. J. Biosci. Bioeng. 1999, 87, 333–339. [Google Scholar] [CrossRef]
- Singh, A.; Cher Soh, K.; Hatzimanikatis, V.; Gill, R.T. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab. Eng. 2011, 13, 76–81. [Google Scholar] [CrossRef]
- Mao, Y.; Li, G.; Chang, Z.; Tao, R.; Cui, Z.; Wang, Z.; Tang, Y.-J.; Chen, T.; Zhao, X. Metabolic engineering of Corynebacterium glutamicum for efficient production of succinate from lignocellulosic hydrolysate. Biotechnol. Biofuels 2018, 11, 95. [Google Scholar] [CrossRef]
- Raab, A.M.; Gebhardt, G.; Bolotina, N.; Weuster-Botz, D.; Lang, C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab. Eng. 2010, 12, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Bennett, G.N.; San, K.-Y. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab. Eng. 2005, 7, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Litsanov, B.; Kabus, A.; Brocker, M.; Bott, M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 2012, 5, 116–128. [Google Scholar] [CrossRef]
- Khodayari, A.; Chowdhury, A.; Maranas, C.D. Succinate Overproduction: A Case Study of Computational Strain Design Using a Comprehensive Escherichia coli Kinetic Model. Front. Bioeng. Biotechnol. 2015, 2, 76. [Google Scholar] [CrossRef] [PubMed]
- Raab, A.M.; Lang, C. Oxidative versus reductive succinic acid production in the yeast Saccharomyces cerevisiae. Bioeng. Bugs 2011, 2, 120–123. [Google Scholar] [CrossRef]
- Fang, X.; Lloyd, C.J.; Palsson, B.O. Reconstructing organisms in silico: Genome-scale models and their emerging applications. Nat. Rev. Microbiol. 2020, 18, 731–743. [Google Scholar] [CrossRef]
- Sarkar, D.; Maranas, C.D. Engineering microbial chemical factories using metabolic models. BMC Chem. Eng. 2019, 1, 22. [Google Scholar] [CrossRef]
- Landon, S.; Rees-Garbutt, J.; Marucci, L.; Grierson, C. Genome-driven cell engineering review: In vivo and in silico metabolic and genome engineering. Essays Biochem. 2019, 63, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Jouhten, P. Metabolic modelling in the development of cell factories by synthetic biology. Comput. Struct. Biotechnol. J. 2012, 3, e201210009. [Google Scholar] [CrossRef]
- Bordbar, A.; Monk, J.M.; King, Z.A.; Palsson, B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014, 15, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Fleming, R.M.T.; Thiele, I.; Provan, G.; Nasheuer, H.P. Integrated stoichiometric, thermodynamic and kinetic modelling of steady state metabolism. J. Theor. Biol. 2010, 264, 683–692. [Google Scholar] [CrossRef]
- Copeland, W.B.; Bartley, B.A.; Chandran, D.; Galdzicki, M.; Kim, K.H.; Sleight, S.C.; Maranas, C.D.; Sauro, H.M. Computational tools for metabolic engineering. Metab. Eng. 2012, 14, 270–280. [Google Scholar] [CrossRef]
- Jing, L.S.; Shah, F.F.M.; Mohamad, M.S.; Hamran, N.L.; Salleh, A.H.M.; Deris, S.; Alashwal, H. Database and tools for metabolic network analysis. Biotechnol. Bioprocess Eng. 2014, 19, 568–585. [Google Scholar] [CrossRef]
- O’Shea, K.; Misra, B.B. Software tools, databases and resources in metabolomics: Updates from 2018 to 2019. Metabolomics 2020, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Saha, R. Computational Approaches on Stoichiometric and Kinetic Modeling for Efficient Strain Design. In Synthetic Metabolic Pathways: Methods and Protocols; Jensen, M.K., Keasling, J.D., Eds.; Springer: New York, NY, USA, 2018; pp. 63–82. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Hong, S.H.; Moon, S.Y. In silico metabolic pathway analysis and design: Succinic acid production by metabolically engineered Escherichia coli as an example. Genome Inform. 2002, 13, 214–223. [Google Scholar]
- Hong, S.H.; Kim, J.S.; Lee, S.Y.; In, Y.H.; Choi, S.S.; Rih, J.-K.; Kim, C.H.; Jeong, H.; Hur, C.G.; Kim, J.J. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat. Biotechnol. 2004, 22, 1275–1281. [Google Scholar] [CrossRef]
- Hong, S.H.; Lee, S.Y. Enhanced production of succinic acid by metabolically engineered Escherichia coli with amplified activities of malic enzyme and fumarase. Biotechnol. Bioprocess Eng. 2004, 9, 252. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, D.-Y.; Kim, T.Y.; Kim, B.H.; Lee, J.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Enhanced Production of Succinic Acid, Based on Genome Comparison and In Silico Gene Knockout Simulation. Appl. Environ. Microbiol. 2005, 71, 7880–7887. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, X.; Yang, Y.; Zhao, X. Genome-scale in silico aided metabolic analysis and flux comparisons of Escherichia coli to improve succinate production. Appl. Microbiol. Biotechnol. 2006, 73, 887–894. [Google Scholar] [CrossRef]
- Kim, T.Y.; Kim, H.U.; Park, J.M.; Song, H.; Kim, J.S.; Lee, S.Y. Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol. Bioeng. 2007, 97, 657–671. [Google Scholar] [CrossRef]
- Kim, T.Y.; Kim, H.U.; Song, H.; Lee, S.Y. In silico analysis of the effects of H2 and CO2 on the metabolism of a capnophilic bacterium Mannheimia succiniciproducens. J. Biotechnol. 2009, 144, 184–189. [Google Scholar] [CrossRef]
- Meijer, S.; Nielsen, M.L.; Olsson, L.; Nielsen, J. Gene deletion of cytosolic ATP: Citrate lyase leads to altered organic acid production in Aspergillus niger. J. Ind. Microbiol. Biotechnol. 2009, 36, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Agren, R.; Otero, J.M.; Nielsen, J. Genome-scale modeling enables metabolic engineering of Saccharomyces cerevisiae for succinic acid production. J. Ind. Microbiol. Biotechnol. 2013, 40, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Otero, J.M.; Cimini, D.; Patil, K.R.; Poulsen, S.G.; Olsson, L.; Nielsen, J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS ONE 2013, 8, e54144. [Google Scholar] [CrossRef]
- Becker, J.; Reinefeld, J.; Stellmacher, R.; Schäfer, R.; Lange, A.; Meyer, H.; Lalk, M.; Zelder, O.; von Abendroth, G.; Schröder, H.; et al. Systems-wide analysis and engineering of metabolic pathway fluxes in bio-succinate producing Basfia succiniciproducens. Biotechnol. Bioeng. 2013, 110, 3013–3023. [Google Scholar] [CrossRef] [PubMed]
- Rafieenia, R. Metabolic capabilities of Actinobacillus succinogenes for succinic acid production. Braz. J. Chem. Eng. 2014, 31, 859–865. [Google Scholar] [CrossRef]
- Chua, P.S.; Salleh, A.H.M.; Mohamad, M.S.; Deris, S.; Omatu, S.; Yoshioka, M. Identifying a gene knockout strategy using a hybrid of the bat algorithm and flux balance analysis to enhance the production of succinate and lactate in Escherichia coli. Biotechnol. Bioprocess Eng. 2015, 20, 349–357. [Google Scholar] [CrossRef]
- Jian, X.; Li, N.; Zhang, C.; Hua, Q. In silico profiling of cell growth and succinate production in Escherichia coli NZN111. Bioresour. Bioprocess 2016, 3, 48. [Google Scholar] [CrossRef][Green Version]
- Mienda, B.S.; Shamsir, M.S.; Illias, R.M. Model-aided atpE gene knockout strategy in Escherichia coli for enhanced succinic acid production from glycerol. J. Biomol. Str. Dyn. 2016, 34, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Mienda, B.S.; Shamsir, M.S.; Illias, R.M. Model-guided metabolic gene knockout of gnd for enhanced succinate production in Escherichia coli from glucose and glycerol substrates. Comput. Biol. Chem. 2016, 61, 130–137. [Google Scholar] [CrossRef]
- Choi, S.; Song, H.; Lim, S.W.; Kim, T.Y.; Ahn, J.H.; Lee, J.W.; Lee, M.-H.; Lee, S.Y. Highly selective production of succinic acid by metabolically engineered Mannheimia succiniciproducens and its efficient purification. Biotechnol. Bioeng. 2016, 113, 2168–2177. [Google Scholar] [CrossRef]
- Lee, J.W.; Yi, J.; Kim, T.Y.; Choi, S.; Ahn, J.H.; Song, H.; Lee, M.-H.; Lee, S.Y. Homo-succinic acid production by metabolically engineered Mannheimia succiniciproducens. Metab. Eng. 2016, 38, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Martinez, L.; Theodoropoulos, C. Estimation of flux distribution in metabolic networks accounting for thermodynamic constraints: The effect of equilibrium vs. blocked reactions. Biochem. Eng. J. 2016, 105, 347–357. [Google Scholar] [CrossRef]
- Jian, X.; Li, N.; Chen, Q.; Hua, Q. Model-guided identification of novel gene amplification targets for improving succinate production in Escherichia coli NZN111. Integr. Biol. 2017, 9, 830–835. [Google Scholar] [CrossRef]
- St. John, P.C.; Crowley, M.F.; Bomble, Y.J. Efficient estimation of the maximum metabolic productivity of batch systems. Biotechnol. Biofuels 2017, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Tafur Rangel, A.E.; Camelo Valera, L.C.; Gómez Ramírez, J.M.; González Barrios, A.F. Effects of metabolic engineering on downstream processing operational cost and energy consumption: The case of Escherichia coli’s glycerol conversion to succinic acid. J. Chem. Technol. Biotechnol. 2018, 93, 2011–2020. [Google Scholar] [CrossRef]
- Mohd Daud, K.; Zakaria, Z.; Shah, Z.A.; Mohamad, M.; Deris, S.; Omatu, S.; Corchado Rodríguez, J. A hybrid of differential search algorithm and flux balance analysis to: Identify knockout strategies for in silico optimization of metabolites production. Int. J. Adv. Soft Comput. Appl. 2018, 10, 84–107. [Google Scholar]
- Mienda, B.S. Escherichia coli genome-scale metabolic gene knockout of lactate dehydrogenase (ldhA), increases succinate production from glycerol. J. Biomol. Str. Dyn. 2018, 36, 3680–3686. [Google Scholar] [CrossRef]
- Arif, M.A.; Mohamad, M.S.; Abd Latif, M.S.; Deris, S.; Remli, M.A.; Mohd Daud, K.; Ibrahim, Z.; Omatu, S.; Corchado, J.M. A hybrid of Cuckoo Search and Minimization of Metabolic Adjustment to optimize metabolites production in genome-scale models. Comput. Biol. Med. 2018, 102, 112–119. [Google Scholar] [CrossRef]
- Nag, A.; St. John, P.C.; Crowley, M.F.; Bomble, Y.J. Prediction of reaction knockouts to maximize succinate production by Actinobacillus succinogenes. PLoS ONE 2018, 13, e0189144. [Google Scholar] [CrossRef]
- Widiastuti, H.; Lee, N.-R.; Karimi, I.A.; Lee, D.-Y. Genome-Scale In Silico Analysis for Enhanced Production of Succinic Acid in Zymomonas mobilis. Processes 2018, 6, 30. [Google Scholar] [CrossRef]
- Lee, M.K.; Mohamad, M.S.; Choon, Y.W.; Daud, K.M.; Nasarudin, N.A.; Ismail, M.A.; Ibrahim, Z.; Napis, S.; Sinnott, R.O. Comparison of Optimization-Modelling Methods for Metabolites Production in Escherichia coli. J. Integr. Bioinform. 2020, 17, 20190073. [Google Scholar] [CrossRef] [PubMed]
- Upton, D.J.; McQueen-Mason, S.J.; Wood, A.J. In silico evolution of Aspergillus niger organic acid production suggests strategies for switching acid output. Biotechnol. Biofuels 2020, 13, 27. [Google Scholar] [CrossRef]
- Ahn, J.H.; Seo, H.; Park, W.; Seok, J.; Lee, J.A.; Kim, W.J.; Kim, G.B.; Kim, K.-J.; Lee, S.Y. Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase. Nat. Commun. 2020, 11, 1970. [Google Scholar] [CrossRef]
- Mienda, B.S. Genome-scale metabolic models as platforms for strain design and biological discovery. J. Biomol. Str. Dyn. 2017, 35, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Famili, I.; Forster, J.; Nielsen, J.; Palsson, B.O. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. USA 2003, 100, 13134–13139. [Google Scholar] [CrossRef]
- Kjeldsen, K.R.; Nielsen, J. In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network. Biotechnol. Bioeng. 2009, 102, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Shinfuku, Y.; Sorpitiporn, N.; Sono, M.; Furusawa, C.; Hirasawa, T.; Shimizu, H. Development and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb. Cell Fact. 2009, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Xu, N.; Ye, C.; Liu, L.; Wu, J. Reconstruction and analysis of a genome-scale metabolic network of Corynebacterium glutamicum S9114. Gene 2016, 575, 615–622. [Google Scholar] [CrossRef]
- Reed, J.L.; Vo, T.D.; Schilling, C.H.; Palsson, B.O. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 2003, 4, R54. [Google Scholar] [CrossRef]
- Feist, A.M.; Henry, C.S.; Reed, J.L.; Krummenacker, M.; Joyce, A.R.; Karp, P.D.; Broadbelt, L.J.; Hatzimanikatis, V.; Palsson, B.Ø. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007, 3, 121. [Google Scholar] [CrossRef]
- Orth, J.D.; Conrad, T.M.; Na, J.; Lerman, J.A.; Nam, H.; Feist, A.M.; Palsson, B.Ø. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 2011, 7, 535. [Google Scholar] [CrossRef]
- O’Brien, E.J.; Lerman, J.A.; Chang, R.L.; Hyduke, D.R.; Palsson, B.Ø. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol. Syst. Biol. 2013, 9, 693. [Google Scholar] [CrossRef]
- Nookaew, I.; Jewett, M.C.; Meechai, A.; Thammarongtham, C.; Laoteng, K.; Cheevadhanarak, S.; Nielsen, J.; Bhumiratana, S. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: A scaffold to query lipid metabolism. BMC Syst. Biol. 2008, 2, 71. [Google Scholar] [CrossRef]
- Mo, M.L.; Palsson, B.Ø.; Herrgård, M.J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 2009, 3, 37. [Google Scholar] [CrossRef]
- Österlund, T.; Nookaew, I.; Bordel, S.; Nielsen, J. Mapping condition-dependent regulation of metabolism in yeast through genome-scale modeling. BMC Syst. Biol. 2013, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Bradfield, M.F.A.; Nicol, W. The pentose phosphate pathway leads to enhanced succinic acid flux in biofilms of wild-type Actinobacillus succinogenes. Appl. Microbiol. Biotechnol. 2016, 100, 9641–9652. [Google Scholar] [CrossRef]
- Pateraki, C.; Almqvist, H.; Ladakis, D.; Lidén, G.; Koutinas, A.A.; Vlysidis, A. Modelling succinic acid fermentation using a xylose based substrate. Biochem. Eng. J. 2016, 114, 26–41. [Google Scholar] [CrossRef]
- Vlysidis, A.; Du, C.; Webb, C.; Theodoropoulos, C. Experimental and Modelling Studies of the Bioconversion of Glycerol to Succinic Acid by Actinobacillus Succinogenes. In Proceedings of the AIChE Annual Meeting, Philadelphia, PA, USA, 16–21 November 2008. [Google Scholar]
- McKinlay, J.B.; Zeikus, J.G.; Vieille, C. Insights into Actinobacillus succinogenes fermentative metabolism in a chemically defined growth medium. Appl. Environ. Microbiol. 2005, 71, 6651–6656. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Shachar-Hill, Y.; Zeikus, J.G.; Vieille, C. Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metab. Eng. 2007, 9, 177–192. [Google Scholar] [CrossRef] [PubMed]
- McKinlay, J.B.; Vieille, C. 13C-metabolic flux analysis of Actinobacillus succinogenes fermentative metabolism at different NaHCO3 and H2 concentrations. Metab. Eng. 2008, 10, 55–68. [Google Scholar] [CrossRef]
- Edwards, J.S.; Palsson, B.O. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 1999, 274, 17410–17416. [Google Scholar] [CrossRef] [PubMed]
- Tibocha-Bonilla, J.D.; Zuñiga, C.; Godoy-Silva, R.D.; Zengler, K. Advances in metabolic modeling of oleaginous microalgae. Biotechnol. Biofuels 2018, 11, 241. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).