Yeast Morphology Assessment through Automated Image Analysis during Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Media and Yeast
2.2. Rehydration and Propagation
2.3. Fermentation
2.4. Measurement of Sugar Concentration and Viable Cells in Suspension
2.5. Models
2.6. Image Collection
2.7. Manual Analysis of Vacuole Count
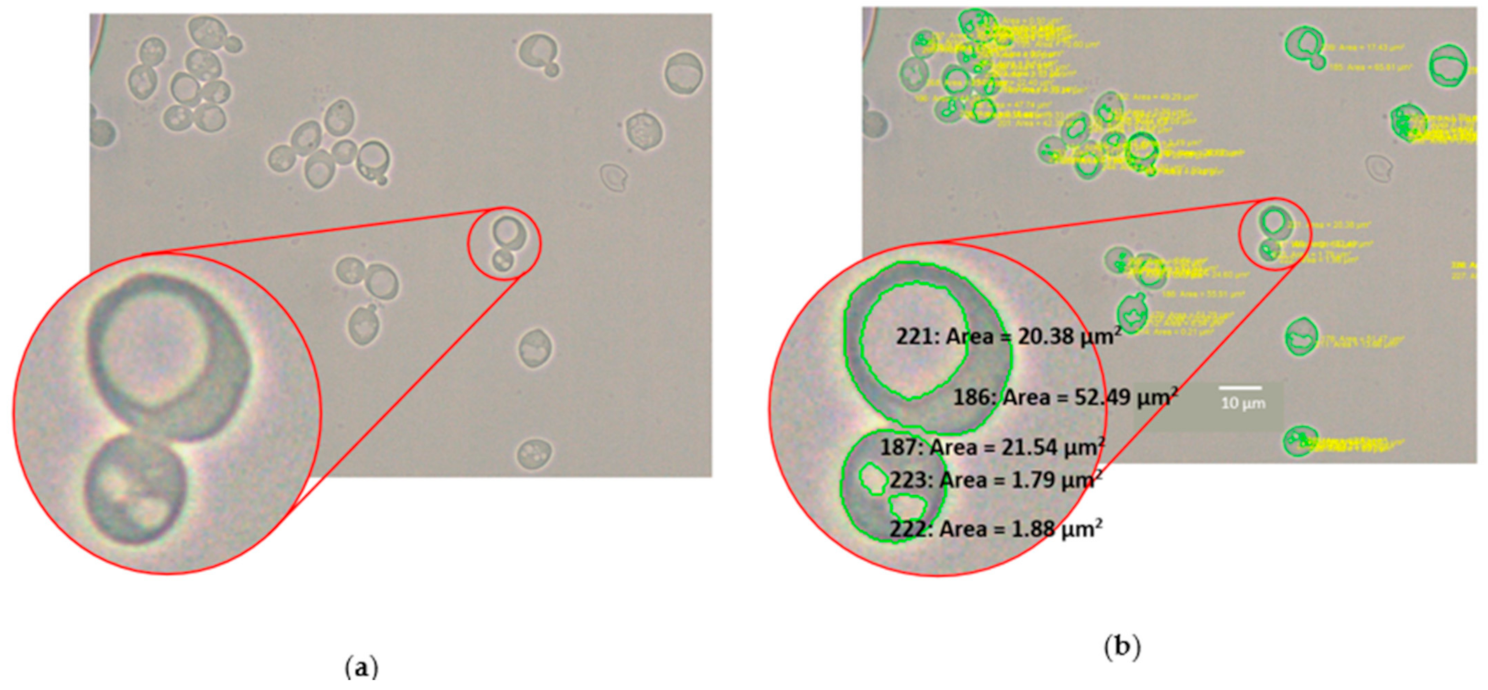
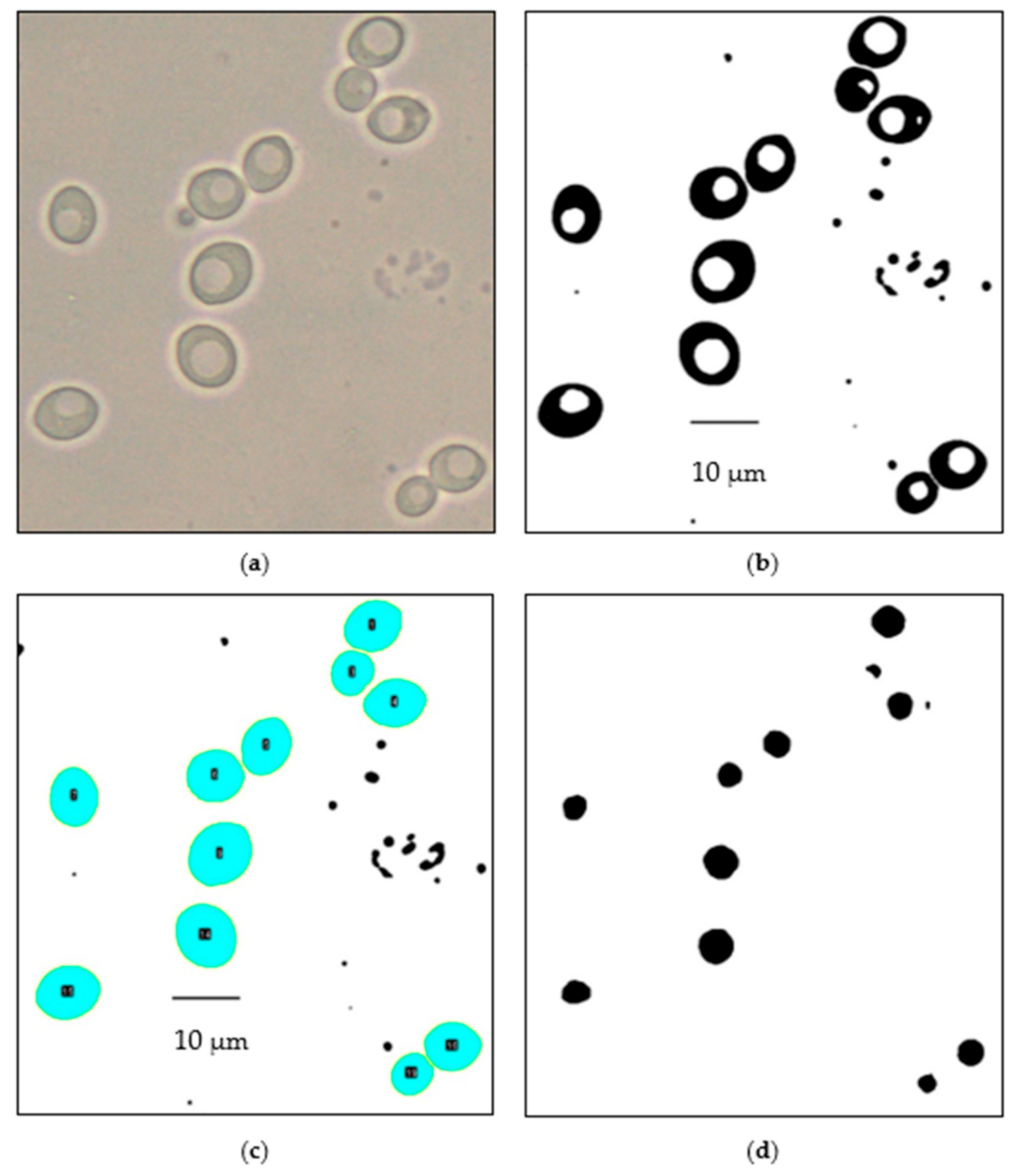
2.8. Automated Image Analysis of Yeast and Vacuole Cross-Sectional Area
2.9. Statistical Analysis
3. Results
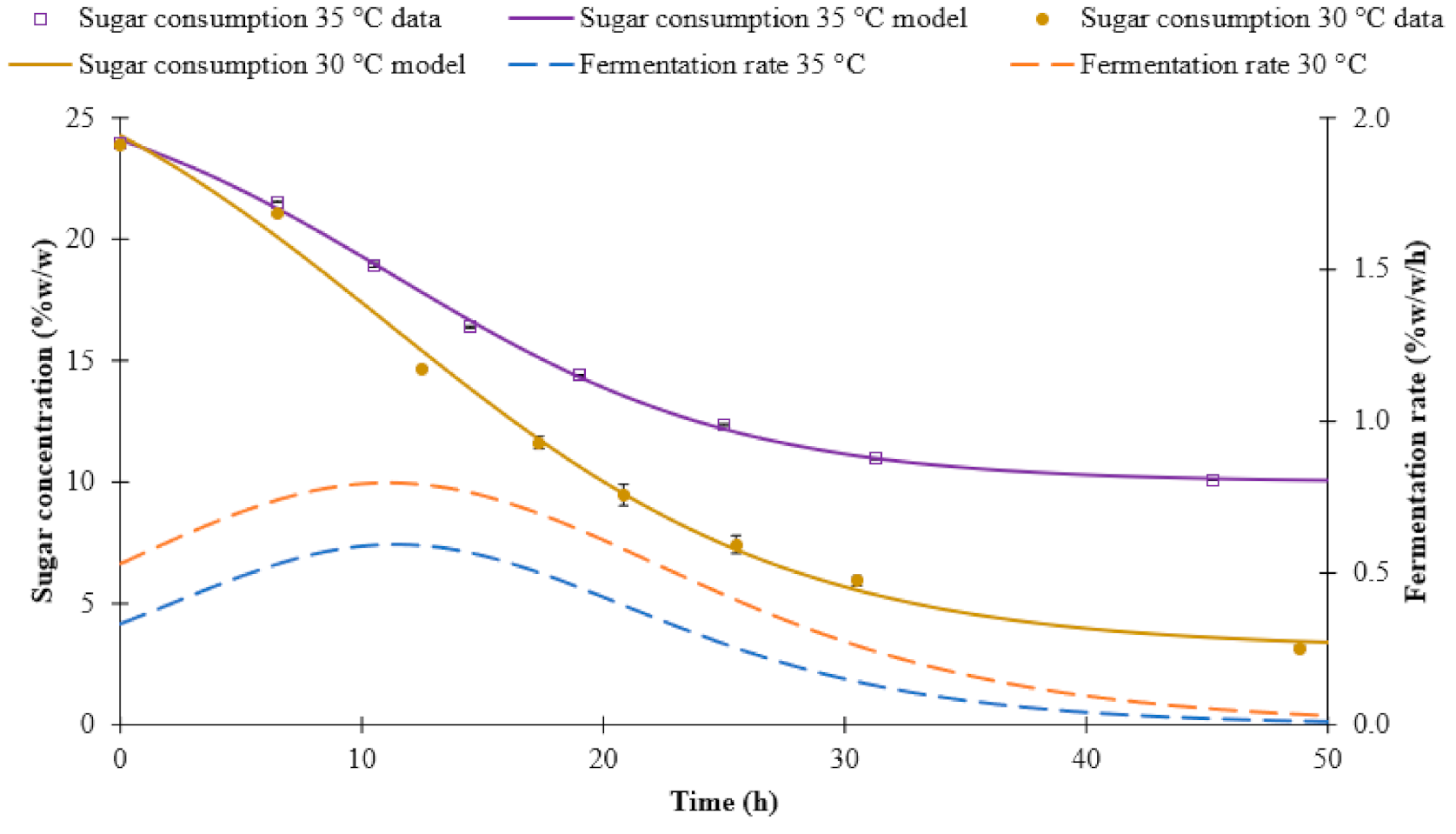
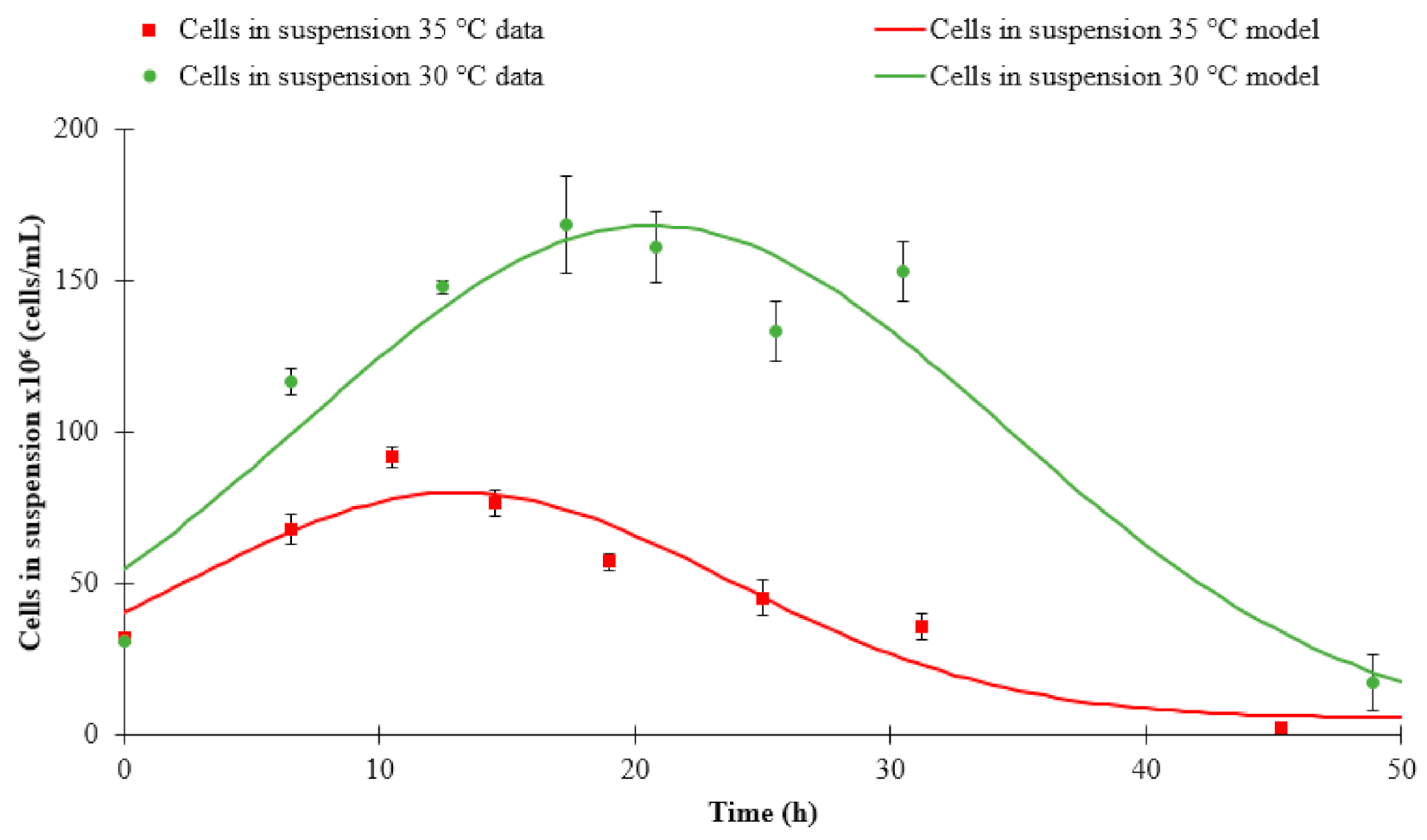
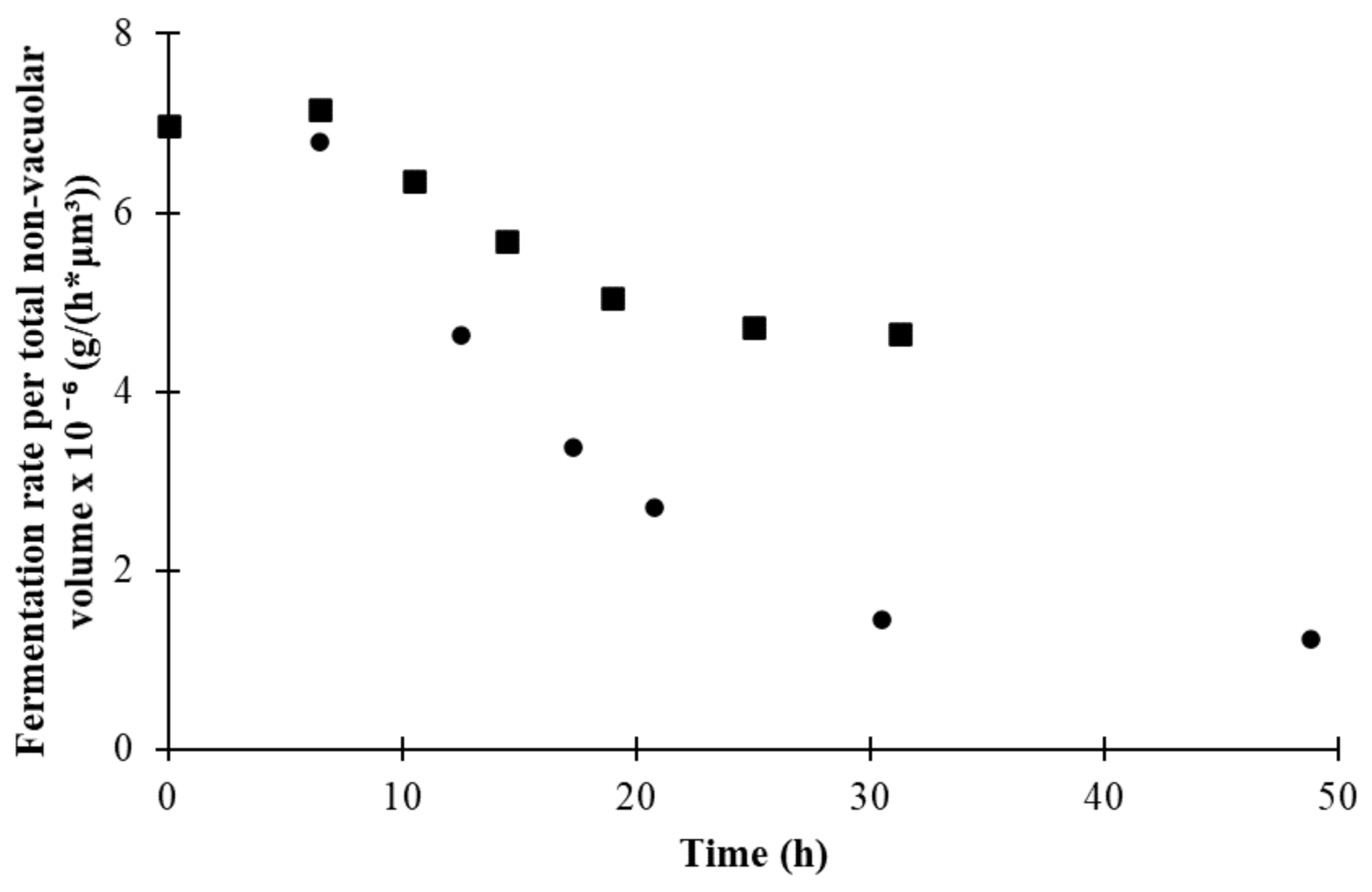
3.1. Fermentation Kinetics
3.2. Manual Image Analysis
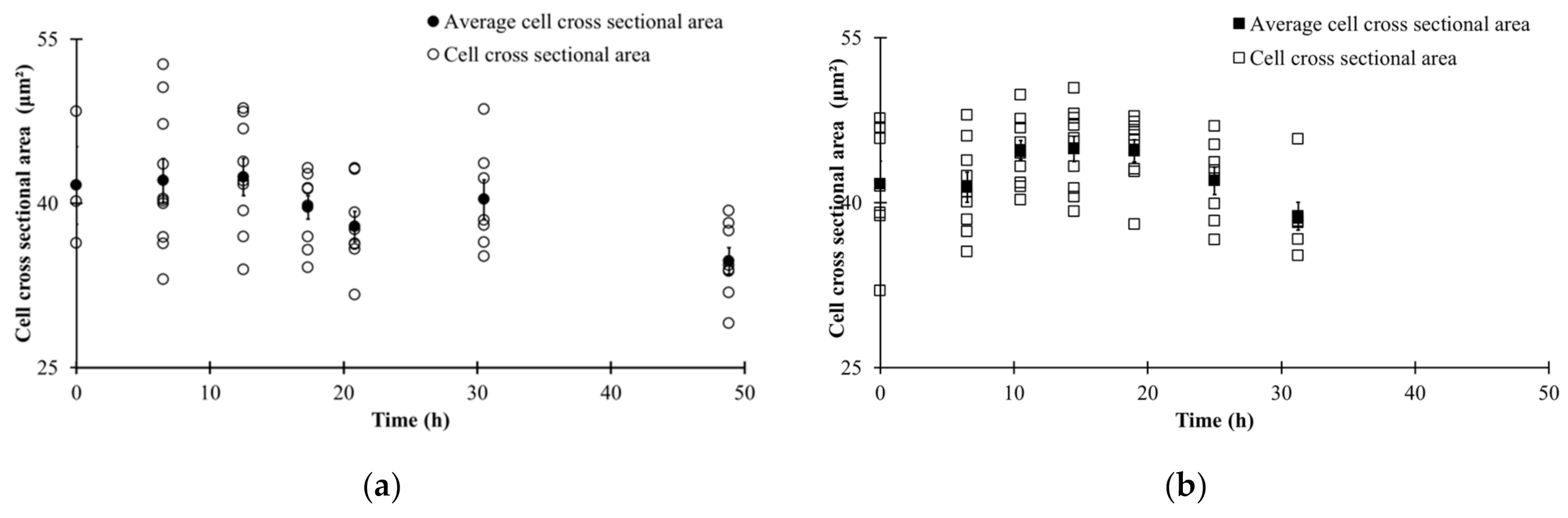
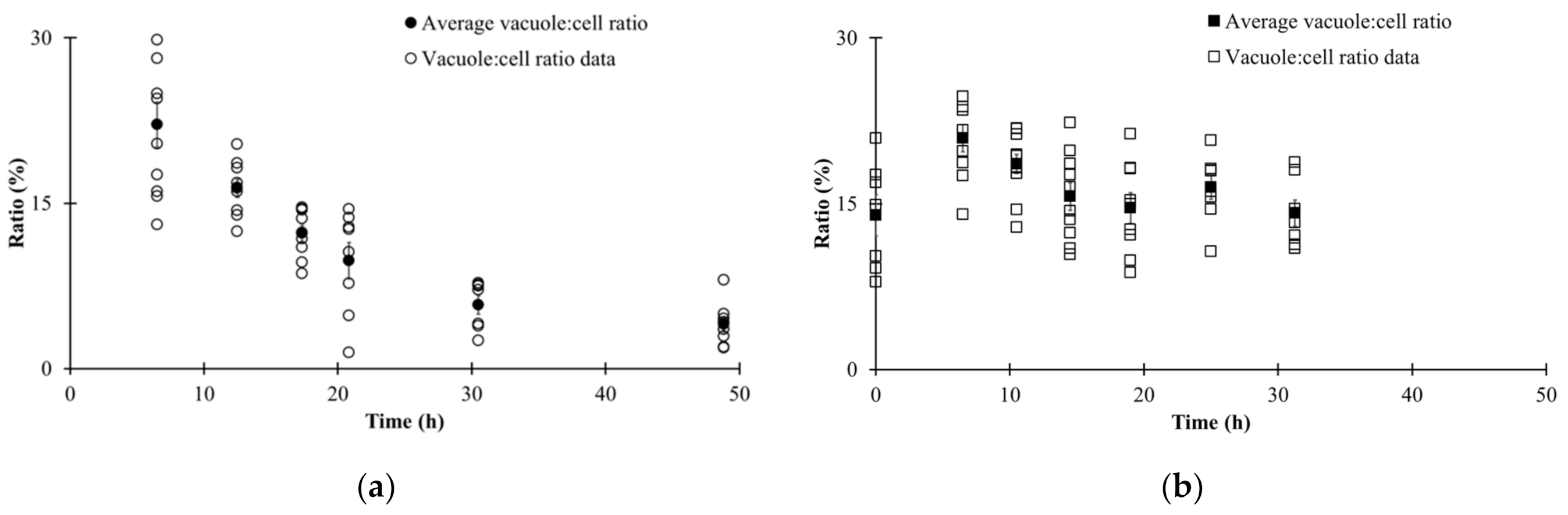
3.3. Automated Image Analysis
3.4. Changes in Morphological State over Fermentation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wunderlich, S.; Back, W. 1—Overview of Manufacturing Beer: Ingredients, Processes, and Quality Criteria. In Beer in Health and Disease Prevention; Elsevier B.V.: Burlington, MA, USA, 2009; pp. 3–16. [Google Scholar] [CrossRef]
- Tibayrenc, P.; Preziosi-Belloya, L.; Roger, J.M.; Ghommidha, C. Assessing yeast viability from cell size measurements? J. Biotechnol. 2010, 149, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, H.; Zheng, F.; Li, Y.; Liu, C.; Niu, C.; Li, Q. Physiological Changes of Beer Brewer’s Yeast during Serial Beer Fermentation. J. Am. Soc. Brew. Chem. 2019, 77, 10–20. [Google Scholar] [CrossRef]
- Zakhartsev, M.; Reuss, M. Cell size and morphological properties of yeast Saccharomyces cerevisiae in relation to growth temperature. FEMS Yeast Res. 2018, 18, foy052. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.; Belo, I.; Pinheiro, R.; Amaral, A.; Mota, M.; Coutinho, J.; Ferreira, E. Effect of hyperbaric stress on yeast morphology: Study by automated image analysis. Appl. Microbiol. Biotechnol. 2004, 66, 318–324. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pratt, P.; Bryce, J.; Stewart, G. The Effects of Osmotic Pressure and Ethanol on Yeast Viability and Morphology. J. Inst. Brew. 2003, 109, 218–228. [Google Scholar] [CrossRef]
- Pratt, P.; Bryce, J.; Stewart, G. The Yeast Vacuole—A Scanning Electron Microscopy Study during High Gravity Wort Fermentations. J. Inst. Brew. 2007, 113, 50–60. [Google Scholar] [CrossRef]
- Izawa, S.; Ikeda, K.; Miki, T.; Wakai, Y.; Inoue, Y. Vacuolar morphology of Saccharomyces cerevisiae during the process of wine making and Japanese sake brewing. Appl. Microbiol. Biotechnol. 2010, 88, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Kida, K.; Gent, D.; Slaughter, J.C. Effect of vacuoles on viability of Saccharomyces cerevisiae. J. Ferment. Bioeng. 1993, 76, 284–288. [Google Scholar] [CrossRef]
- Cahill, G.; Walsh, P.; Donnelly, D. Improved Control of Brewery Yeast Pitching Using Image Analysis. J. Am. Soc. Brew. Chem. 2018, 57, 72–78. [Google Scholar] [CrossRef]
- O’Shea, D.; Walsh, P. The effect of culture conditions on the morphology of the dimorphic yeast Kluyveromyces marxianus var. marxianus NRRLy2415: A study incorporating image analysis. Appl. Microbiol. Biotechnol. 2000, 53, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Ginovart, M.; Carbó, R.; Blanco, M.; Portell, X. Digital Image Analysis of Yeast Single Cells Growing in Two Different Oxygen Concentrations to Analyze the Population Growth and to Assist Individual-Based Modeling. Front. Microbiol. 2018, 8, 2628. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Saka, A.; Sano, F.; Ohya, Y.; Morishita, S. Development of image processing program for yeast cell morphology. J. Bioinform. Comput. Biol. 2004, 1, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Grishagin, I. Automatic cell counting with ImageJ. Anal. Biochem. 2015, 473, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Durham Brooks, T.; Wilson, E.; et al. Quantitative and Qualitative Assessment Methods for Biofilm Growth: A Mini-review. Res. Rev. J. Eng. Technol. 2017, 6. Available online: http://www.rroij.com/open-access/quantitative-and-qualitative-assessment-methods-for-biofilm-growth-a-minireview-.pdf (accessed on 5 March 2021).
- Stolze, N.; Bader, C.; Henning, C.; Mastin, J.; Holmes, A.; Sutlief, A. Automated image analysis with ImageJ of yeast colony forming units from cannabis flowers|Elsevier Enhanced Reader. J. Microbiol. Methods 2019, 164, 105681. [Google Scholar] [CrossRef] [PubMed]
- ASBC. Wort-12: Free Amino Nitrogen. Method, A. Ninhydrin method (international method). In Wort Methods; American Society of Brewing Chemists: St. Paul, MN, USA, 2011. [Google Scholar]
- ASBC. Yeast-4: Microscopic yeast cell counting. In Wort Methods; American Society of Brewing Chemists: St. Paul, MN, USA, 2004. [Google Scholar]
- Painting, K.; Kirsop, B. A quick method for estimating the percentage of viable cells in a yeast population, using methylene blue staining. World J. Microbiol. Biotechnol. 1990, 6, 346–347. [Google Scholar] [CrossRef] [PubMed]
- ASBC. Yeast-14: Miniature fermentation assay. In Wort Methods; American Society of Brewing Chemists: St. Paul, MN, USA, 2011; pp. 1–3. [Google Scholar]
- Rudolph, A.; MacIntosh, A.J.; Speers, R.A.; St. Mary, C. Modeling Yeast in Suspension during Laboratory and Commercial Fermentations to Detect Aberrant Fermentation Processes. J. Am. Soc. Brew. Chem. 2020, 78, 63–73. [Google Scholar] [CrossRef]
- Ferreira, T.; Rasband, W. ImageJ User Guide IJ 1.46r. 2012. Available online: https://imagej.nih.gov/ij/docs/guide/user-guide-A4booklet.pdf (accessed on 5 March 2021).
- Torija, M.; Rozès, N.; Poblet, M.; Guillamón, J.; Mas, A. Effects of fermentation temperature on the strain population of Saccharomyces cerevisiae. Int. J. Food Microbiol. 2003, 80, 47–53. [Google Scholar] [CrossRef]
- Reddy, L.; Reddy, O. Effect of fermentation conditions on yeast growth and volatile composition of wine produced from mango (Mangifer indica L.) fruit juice. Food Bioprod. Process. 2011, 89, 487–491. [Google Scholar] [CrossRef]
- Gervais, P.; Martínez de Marañón, I.; Evrard, C.; Ferret, E.; Moundanga, S. Cell volume changes during rapid temperature shifts. J. Biotechnol. 2003, 102, 269–279. [Google Scholar] [CrossRef]

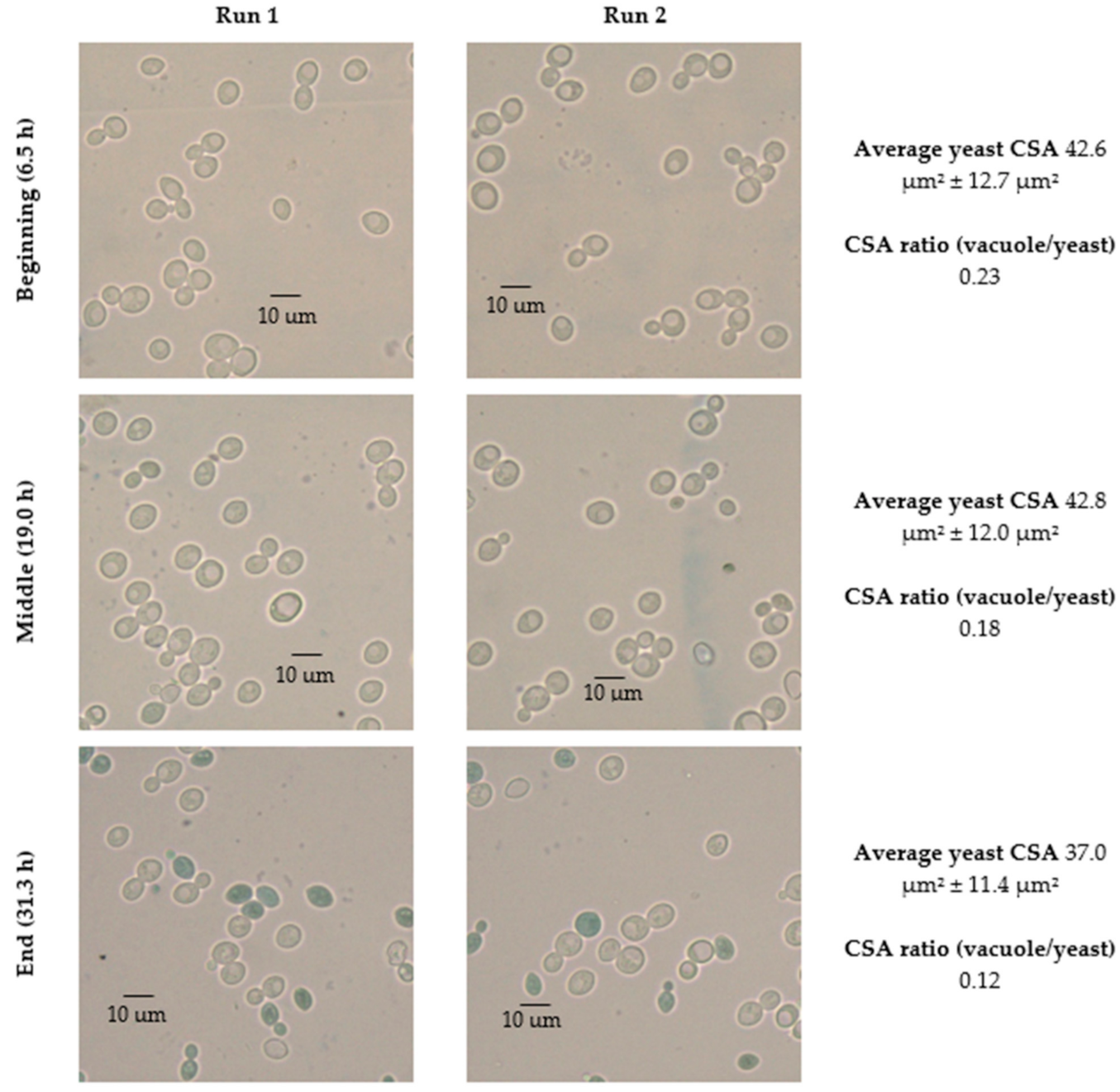

| Parameters | |||||||
|---|---|---|---|---|---|---|---|
| Attribute | Experiment | Model | A | μ | σ | S | Model Fit r2 |
| (Cells/mL) | h | h | Unitless | ||||
| Cells in suspension | 30 °C | Step model | 168.24 | 20.65 | 13.77 | 0.00 | 0.91 |
| 35 °C | Step model | 79.72 | 13.25 | 11.38 | 0.05 | 0.89 | |
| Pi | Pe | B | M | Model Fit r2 | |||
| %w/w | %w/w | %w/(w*h) | h | ||||
| Sugar consumption | 30 °C | Sigmoidal 4P | 29.92 | 3.14 | −0.12 | 11.08 | 0.99 |
| 35 °C | Sigmoidal 4P | 26.93 | 10.00 | −0.14 | 11.39 | 0.99 | |
| Temp (°C) | Time (h) | Manual | Automated | Manual | Automated | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average Yeast CSA (μm2) | Average Yeast CSA (μm2) | % Error | Average % Error | Total Yeast CSA (μm2) | Total Vacuole CSA (μm2) | % CSA Ratio (Vacuole/Yeast) | Total Yeast CSA (μm2) | Total Vacuole CSA (μm2) | % CSA Ratio (Vacuole/Yeast) | % Error | Average % Error | ||
| 30 | 6.5 | 43.3 ± 13.6 | 42.1 ± 11.2 | 2.8% | −3.9% | 6455.3 | 1362.6 | 21.1 | 5828.1 | 1225.6 | 22.1 | −4.9% | 2.0% |
| 17.3 | 37.5 ± 11.0 | 39.6 ± 10.1 | −5.8% | 9218.4 | 944.3 | 10.2 | 8711.3 | 1072.5 | 12.3 | −19.7% | |||
| 30.5 | 37.1 ± 40.4 | 40.4 ± 11.0 | −8.8% | 13,167.5 | 1096.6 | 8.3 | 11,450.7 | 661.9 | 5.8 | 30.7% | |||
| 35 | 6.5 | 42.6 ± 12.7 | 41.4 ± 12.0 | 2.7% | −2.2% | 13,999.1 | 3158.3 | 22.6 | 13,304.5 | 2766.4 | 20.9 | 7.3% | 1.2% |
| 19.0 | 42.8 ± 12.0 | 44.7 ± 11.5 | −4.4% | 15,583.6 | 2746.6 | 17.6 | 12,862.5 | 1828.6 | 15.6 | 17.2% | |||
| 31.3 | 37.0 ± 11.4 | 38.8 ± 13.6 | −4.8% | 11,169.0 | 1308.1 | 11.7 | 11,133.8 | 1569.2 | 14.1 | −20.7% | |||
| Temp (°C) | Time (h) | Average Yeast CSA (μm2) | % CSA Ratio (Vacuole/Yeast) |
|---|---|---|---|
| 30 | 6.5 | 42.1 ± 11.2 a | 22.1 ± 6.5 a |
| 12.5 | 42.4 ± 10.5 a | 16.3 ± 2.5 b | |
| 17.3 | 39.6 ± 10.1 a | 12.3 ± 2.2 bc | |
| 20.8 | 38.7 ± 9.8 a | 9l8 ± 4.7 cd | |
| 30.5 | 40.4 ± 11.0 a | 5.8 ± 2.2 d | |
| 48.8 | 34.7 ± 9.5 b | 3.8 ± 2.0 d | |
| 35 | 0.0 | 41.7 ± 12.4 abc | 13.9 ± 4.9 b |
| 6.5 | 41.4 ± 12.0 bc | 20.9 ± 3.7 a | |
| 10.5 | 44.7 ± 12.7 a | 18.6 ± 2.8 ab | |
| 14.5 | 44.9 ± 11.3 a | 15.6 ± 4.0 b | |
| 19.0 | 44.7 ± 11.5 ab | 14.6 ± 4.2 b | |
| 25.0 | 42.0 ± 12.0 abc | 16.5 ±3.0 ab | |
| 31.3 | 38.8 ± 13.6 c | 14.1 ± 3.2 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guadalupe-Daqui, M.; Chen, M.; Thompson-Witrick, K.A.; MacIntosh, A.J. Yeast Morphology Assessment through Automated Image Analysis during Fermentation. Fermentation 2021, 7, 44. https://doi.org/10.3390/fermentation7020044
Guadalupe-Daqui M, Chen M, Thompson-Witrick KA, MacIntosh AJ. Yeast Morphology Assessment through Automated Image Analysis during Fermentation. Fermentation. 2021; 7(2):44. https://doi.org/10.3390/fermentation7020044
Chicago/Turabian StyleGuadalupe-Daqui, Mario, Mandi Chen, Katherine A. Thompson-Witrick, and Andrew J. MacIntosh. 2021. "Yeast Morphology Assessment through Automated Image Analysis during Fermentation" Fermentation 7, no. 2: 44. https://doi.org/10.3390/fermentation7020044
APA StyleGuadalupe-Daqui, M., Chen, M., Thompson-Witrick, K. A., & MacIntosh, A. J. (2021). Yeast Morphology Assessment through Automated Image Analysis during Fermentation. Fermentation, 7(2), 44. https://doi.org/10.3390/fermentation7020044






