Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening
Abstract
1. Introduction
2. Materials and Methods
2.1. Cheese Sampling
2.2. Microbiological Analysis
2.3. pH Value Measurement
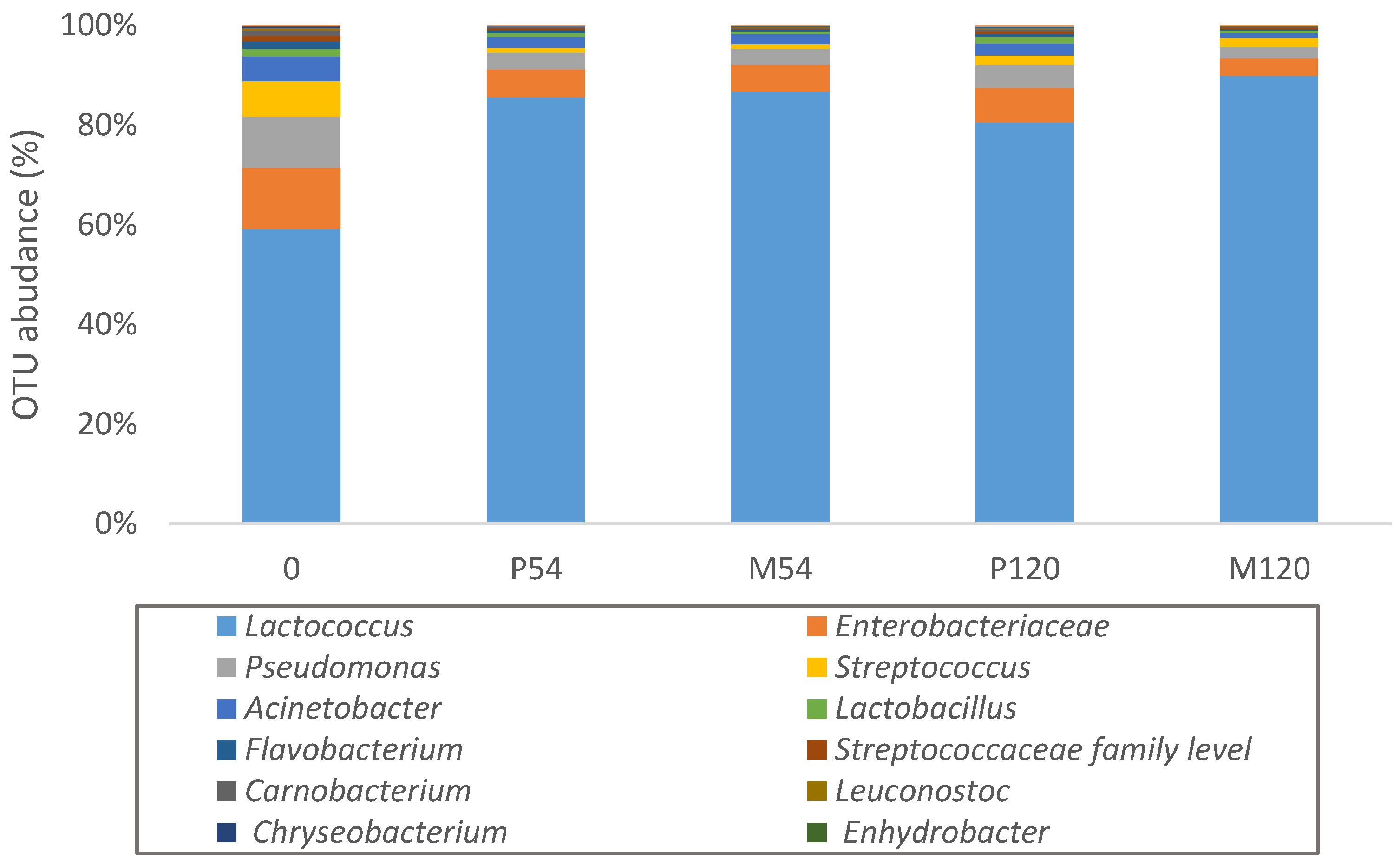
2.4. Estimation of Microbiota by Next Generation Sequencing
2.5. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- European Commission. Commission Regulation (EC) No 1829/2002 of 14 October 2002 amending the Annex to Regulation (EC) No 1107/96 with regard to the name ‘Feta’. Off. J. Eur. Commun. 2002. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2002:277:0010:0014:EN:PDF (accessed on 10 December 2019).
- Moatsou, G.; Govaris, A. White brined cheeses: A diachronic exploitation of small ruminants milk in Greece. Small Rumin. Res. 2011, 101, 113–121. [Google Scholar] [CrossRef]
- Alichanidis, E. Cheeses ripened in brine. In Cheese Problems Solved; McSweeney, P.L.H., Ed.; CRC Press: Cambridge, UK, 2007; pp. 331–342. ISBN 9781845690601. [Google Scholar]
- Rantsiou, K.; Alessandria, V.; Urso, R.; Dolci, P.; Cocolin, L. Detection, quantification and vitality of Listeria monocytogenes in food as determined by quantitative PCR. Int. J. Food Microbiol. 2008, 121, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Bozoudi, D.; Torriani, S.; Zdragas, A.; Litopoulou-Tzanetaki, E. Assessment of microbial diversity of the dominant microbiota in fresh and mature PDO Feta cheese made at three mountainous areas of Greece. LWT Food Sci. Technol. 2016, 72, 525–533. [Google Scholar] [CrossRef]
- Manolopoulou, E.; Sarantinopoulos, P.; Zoidou, E.; Aktypis, A.; Moschopoulou, E.; Kandarakis, I.G.; Anifantakis, E.M. Evolution of microbial populations during traditional Feta cheese manufacture and ripening. Int. J. Food Microbiol. 2003, 82, 153–161. [Google Scholar] [CrossRef]
- Tzanetakis, N.; Litopoulou-Tzanetaki, E. Changes in numbers and kinds of lactic acid bacteria in Feta and Teleme, two Greek cheeses from ewes’ milk. J. Dairy Sci. 1992, 75, 1389–1393. [Google Scholar] [CrossRef]
- Vassiliadis, A.; Psoni, L.; Nikolaou, S.; Arvanitis, L.; Tzanetakis, N.; Litopoulou–Tzanetaki, E. Changes in microbial populations, kinds of lactic acid bacteria and biochemical characteristics of Greek traditional feta cheese during ripening. Int. J. Dairy Technol. 2009, 62, 39–47. [Google Scholar] [CrossRef]
- Xanthopoulos, V.; Hatzikamari, M.; Adamidis, T.; Tsakalidou, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E. Heterogeneity of Lactobacillus plantarum isolates from Feta cheese throughout ripening. J. Appl. Microbiol. 2000, 88, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, O.S.; Argyri, A.A.; Varzakis, E.E.; Tassou, C.C.; Chorianopoulos, N.G. Greek functional Feta cheese: Enhancing quality and safety using a Lactobacillus plantarum strain with probiotic potential. Food Microbiol. 2018, 74, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Sarantinopoulos, P.; Kalantzopoulos, G.; Tsakalidou, E. Effect of Enterococcus faecium on microbiological, physicochemical and sensory characteristics of Greek Feta cheese. Int. J. Food Microbiol. 2002, 76, 93–105. [Google Scholar] [CrossRef]
- Litopoulou-Tzanetaki, E.; Tzanetakis, N. Microbiological characteristics of Greek traditional cheeses. Small Rumin. Res. 2011, 101, 17–32. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Nychas, G.J.E.; Sofos, J.N. Types of traditional Greek foods and their safety. Food Control. 2013, 29, 32–41. [Google Scholar] [CrossRef]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Ercolini, D.; Villani, F.; Nychas, G.J.E. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012, 157, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Bozoudi, D.; Kotzamanidis, C.; Hatzikamari, M.; Tzanetakis, N.; Menexes, G.; Litopoulou-Tzanetaki, E. A comparison for acid production, proteolysis, autolysis and inhibitory properties of lactic acid bacteria from fresh and mature Feta PDO Greek cheese, made at three different mountainous areas. Int. J. Food Microbiol. 2015, 200, 87–96. [Google Scholar] [CrossRef]

| Microorganism | Population (log CFU/g) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plastic Container | Stainless Steel Container | |||||||||||||
| 0 | 7 | 28 | 54 | 82 | 104 | 120 | 0 | 7 | 28 | 54 | 82 | 104 | 120 | |
| Total Viable Counts (TVC) | 9.05 | 7.85 | 7.93 | 8.25 | 8.65 | 7.94 | 7.79 | 9.05 | 7.74 | 9.01 | 8.25 | 9.65 | 8.94 | 7.94 |
| Lactic acid bacteria (LAB) 1 | 8.26 | 7.89 | 7.89 | 8.18 | 8.48 | 7.80 | 7.84 | 8.26 | 7.76 | 8.99 | 8.13 | 9.70 | 9.08 | 7.87 |
| Streptococci 2 | 9.22 | 8.20 | 8.98 | 5.00 | 2.80 | 4.18 | 5.12 | 9.22 | 7.89 | 9.00 | 5.57 | 4.45 | 5.45 | 4.95 |
| Enterococci | 6.25 | 6.67 | 6.77 | 7.08 | 6.71 | 6.86 | 6.67 | 6.25 | 6.86 | 7.15 | 7.01 | 6.72 | 6.66 | 6.73 |
| Yeasts and molds | 2.89 | 3.73 | 4.01 | 4.10 | 4.27 | 3.21 | 3.96 | 2.89 | 3.29 | 4.34 | 4.00 | 4.27 | 2.95 | 3.64 |
| Pseudomonas sp. | <2 | 3.49 | 3.20 | 2.35 | 2.00 | 2.00 | 3.55 | <2 | 2.00 | 3.09 | 3.25 | 2.00 | 2.00 | <2 |
| Enterobacteriaceae | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| Staphylococci | <2 | 2.15 | <2 | <2 | <2 | <2 | <2 | <2 | 2 | <2 | 4.06 | <2 | <2 | <2 |
| Enterobacteria that utilize glucose | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| pH value | 4.19 | 4.20 | 4.16 | 4.30 | 4.14 | 4.06 | 4.12 | 4.19 | 4.19 | 4.21 | 4.34 | 4.28 | 4.21 | 4.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spyrelli, E.D.; Stamatiou, A.; Tassou, C.C.; Nychas, G.-J.E.; Doulgeraki, A.I. Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening. Fermentation 2020, 6, 12. https://doi.org/10.3390/fermentation6010012
Spyrelli ED, Stamatiou A, Tassou CC, Nychas G-JE, Doulgeraki AI. Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening. Fermentation. 2020; 6(1):12. https://doi.org/10.3390/fermentation6010012
Chicago/Turabian StyleSpyrelli, Evgenia D., Anastasios Stamatiou, Chrysoula C. Tassou, George-John E. Nychas, and Agapi I. Doulgeraki. 2020. "Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening" Fermentation 6, no. 1: 12. https://doi.org/10.3390/fermentation6010012
APA StyleSpyrelli, E. D., Stamatiou, A., Tassou, C. C., Nychas, G.-J. E., & Doulgeraki, A. I. (2020). Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening. Fermentation, 6(1), 12. https://doi.org/10.3390/fermentation6010012






