Kinetic Parameters of Saccharomyces cerevisiae Alcohols Production Using Nepenthes mirabilis Pod Digestive Fluids-Mixed Agro-Waste Hydrolysates
Abstract
1. Introduction
2. Materials and Methods
2.1. Confirmatory Identification of the Commercial Yeast Used for Fermentation
2.2. Fermentation Inoculum Preparation and Yeast Cell Counts
2.3. Fermentation Medium Preparation Using Hydrolysates
2.4. Fermentation Conditions
2.5. Analytical Methods
2.5.1. Total Reducing Sugar and Phenolic Compound Quantification
2.5.2. Alcohol Determination
2.6. Kinectic Parameter Determination
2.6.1. Monod and Microbial Growth Kinetic Parameters
2.6.2. Modelling TRS Consumption for Simultaneous Biomass and Product Formation
2.6.3. Product Formation Kinetic Parameter Determination
2.7. Data Handling, Relative Differences, and other Kinetic Parameters
3. Results and Discussion
3.1. Microbial Growth Parameters Using Mixed Agro-Waste Pre-Treatment Hydrolysates
3.2. Kinetic Rate Determinations
3.3. Product Formation Kinetics, Substrate Consumption Rate, and Modeling
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Meintjes, M.M. Dissertation: Fermentation Coupled with Pervaporation: A Kinetic Study/Meintjes MM; North-West University: Potchefstroom, South Africa, 2011. [Google Scholar]
- Jiménez-Islas, D.; Páez-Lerma, J.; Soto-Cruz, N.O.; Gracida, J. Modelling of ethanol production from red beet juice by Saccharomyces cerevisiae under thermal and acid stress conditions. Food Technol. Biotechnol. 2014, 52, 93–100. [Google Scholar]
- Jung, Y.H.; Kim, I.J.; Kim, J.J.; Oh, K.K.; Han, J.I.; Choi, I.G.; Kim, K.H. Ethanol production from oil palm trunks treated with aqueous ammonia and cellulase. Bioresour. Technol. 2011, 102, 7307–7312. [Google Scholar] [CrossRef] [PubMed]
- Khamaiseh, E.I.; Abdul Hamid, A.; Abdeshahian, P.; Wan Yusoff, W.M.; Kalil, M.S. Enhanced butanol production by Clostridium acetobutylicum NCIMB 13357 grown on date fruit as carbon source in P2 medium. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Singh, S.P.; Mishra, I.M.; Adhikari, D.K. Recent advances in production of bioethanol from lignocellulosic biomass. Chem. Eng. Technol. 2009, 32, 517–526. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.A.; Malik, S.A. Determination of total phenolic and flavonoid content, antimicrobial and antioxidant activity of a root digestive fluid of Arisaema jacquemontii Blume. J. Taibah Univ. Sci. 2015, 9, 449–454. [Google Scholar] [CrossRef]
- Kim, Y.; Ximenes, E.; Mosier, N.S.; Ladisch, M.R. Soluble inhibitors/deactivators of cellulase enzymes from lignocellulosic biomass. Enzym. Microb. Technol. 2011, 48, 408–415. [Google Scholar] [CrossRef]
- Parmar, I.; Rupasinghe, H.P.V. Optimization of dilute acid-based pretreatment and application of laccase on apple pomace. Bioresour. Technol. 2012, 124, 433–439. [Google Scholar] [CrossRef]
- Jin, Q.; Zhang, H.; Yan, L.; Qu, L.; Huang, H. Kinetic characterization for hemicellulose hydrolysis of corn stover in a dilute acid cycle spray flow-through reactor at moderate conditions. Biomass Bioenergy 2011, 35, 4158–4164. [Google Scholar] [CrossRef]
- Alvira, P.; Tomás-Pejó, E.; Ballesteros, M.; Negro, M.J. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: A review. Bioresour. Technol. 2010, 101, 4851–4861. [Google Scholar] [CrossRef]
- Diaz, A.; Le Toullec, J.; Blandino, A.; De Ory, I.; Caro, I. Pretreatment of rice hulls with alkaline peroxide to enhance enzyme hydrolysis for ethanol production. Chem. Eng. Trans. 2013, 32, 949–954. [Google Scholar]
- Gao, K.; Rehmann, L. ABE fermentation from enzymatic hydrolysate of NaOH-pretreated corncobs. Biomass Bioenergy 2014, 66, 110–115. [Google Scholar] [CrossRef]
- Procentese, A.; Raganati, F.; Olivieri, G.; Elena Russo, M.; Marzocchella, A. Pre-treatment and enzymatic hydrolysis of lettuce residues as feedstock for bio-butanol production. Biomass Bioenergy 2017, 96, 172–179. [Google Scholar] [CrossRef]
- Qing, Q.; Wyman, C.E. Hydrolysis of different chain length xylooliogmers by cellulase and hemicellulase. Bioresour. Technol. 2011, 102, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Narayanaswamy, N.; Dheeran, P.; Verma, S.; Kumar, S. Biological pretreatment of lignocellulosic biomass for enzymatic saccharification. In Pretreatment Techniques for Biofuels and Biorefineries; Fang, Z., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 3–34. [Google Scholar]
- Chan, X.Y.; Hong, K.W.; Yin, W.F.; Chan, K.G. Microbiome and biocatalytic bacteria in monkey cup (Nepenthes pitcher) digestive fluid. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.K.; Hiligsmann, S.; Outili, N.; Cherfia, R.; Chaouche, N.K. Kinetic models and parameters estimation study of biomass and ethanol production from inulin by Pichia caribbica (KC977491). Afr. J. Biotechnol. 2017, 16, 124–131. [Google Scholar]
- Dhavale, A.; Vhanmarathi, A.; Deshmukh, S.; Dabeer, S. Unstructured kinetic modeling of glutathione production by Saccharomyces cerevisiae NCIM 3345. In Biotechnology and Biochemical Engineering; Gummadi, S., Vadlani, P., Eds.; Springer: Singapore, 2016; pp. 11–20. [Google Scholar]
- Garnier, A.; Gaillet, B. Analytical solution of Luedeking–Piret equation for a batch fermentation obeying Monod growth kinetics. Biotechnol. Bioeng. 2015, 112, 2468–2474. [Google Scholar] [CrossRef]
- Ng, C.Y.C.; Takahashi, K.; Liu, Z. Isolation, characetrization and optimization of an aerobic butanol producing bacteria from Singapore. Biotechnol. Appl. Biochem. 2015, 63, 86–91. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Sánchez-Femat, E.; Cruz-Leija, R.; Torres-Hernández, M.; Herrera-Mayorga, E. Mobile application for automatic counting of bacterial colonies. In International Conference on Software Process Improvement; Springer: Cham, Germany, 2016; pp. 221–230. [Google Scholar]
- Cheng, C.L.; Che, P.Y.; Chen, B.Y.; Lee, W.J.; Lin, C.Y.; Chang, J.S. Biobutanol production from agricultural waste by an acclimated mixed bacterial microflora. Appl. Energy 2012, 100, 3–9. [Google Scholar] [CrossRef]
- Shi, J.; Ebrik, M.A.; Yang, B.; Garlock, R.J.; Balan, V.; Dale, B.E.; Ramesh Pallapolu, V.; Lee, Y.Y.; Kim, Y.; Mosier, N.S.; et al. Application of cellulase and hemicellulase to pure xylan, pure cellulose, and switchgrass solids from leading pretreatments. Bioresour. Technol. 2011, 102, 11080–11088. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.K.; Kim, Y.; Ximenes, E.; Ladisch, M.R. Effect of liquid hot water pretreatment severity on properties of hardwood lignin and enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2015, 112, 252–262. [Google Scholar] [CrossRef]
- Taherzadeh, M.; Karimi, K. Pretreatment of Lignocellulosic wastes to improve ethanol and biogas production: A Review. Int. J. Mol. Sci. 2008, 9, 1621–1651. [Google Scholar] [CrossRef] [PubMed]
- Mewa-Ngongang, M.; Du Plessis, H.W.; Hutchinson, U.F.; Mekuto, L.; Ntwampe, S.K. Kinetic modelling and optimisation of antimicrobial compound production by Candida pyralidae KU736785 for control of Candida guilliermondii. Food Sci. Technol. Int. 2017, 23, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Makkar, H.P.; Blümmel, M.; Borowy, N.K.; Becker, K. Gravimetric determination of tannins and their correlations with chemical and protein precipitation methods. J. Sci. Food Agric. 1993, 61, 161–165. [Google Scholar] [CrossRef]
- Lavate, S.M.; Kale, A.; Patil, J.; Deshpande, N.R. Spectroscopic determination of total phenolic and flavonoid contents of Aglaia lawii leaves. Int. J. Pharm. Pharm. Sci. 2013, 5, 851–853. [Google Scholar]
- Rossouw, D.; Bauer, F.F. Exploring the phenotypic space of non-Saccharomyces wine yeast biodiversity. Food Microbiol. 2016, 55, 32–46. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Kim, Y.; Kreke, T.; Hendrickson, R.; Parenti, J.; Ladisch, M.R. Fractionation of cellulase and fermentation inhibitors from steam pretreated mixed hardwood. Bioresour. Technol. 2013, 135, 30–38. [Google Scholar] [CrossRef]
- Lian, J.; Si, T.; Nair, N.U.; Zhao, H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab. Eng. 2014, 24, 139–149. [Google Scholar] [CrossRef] [PubMed]
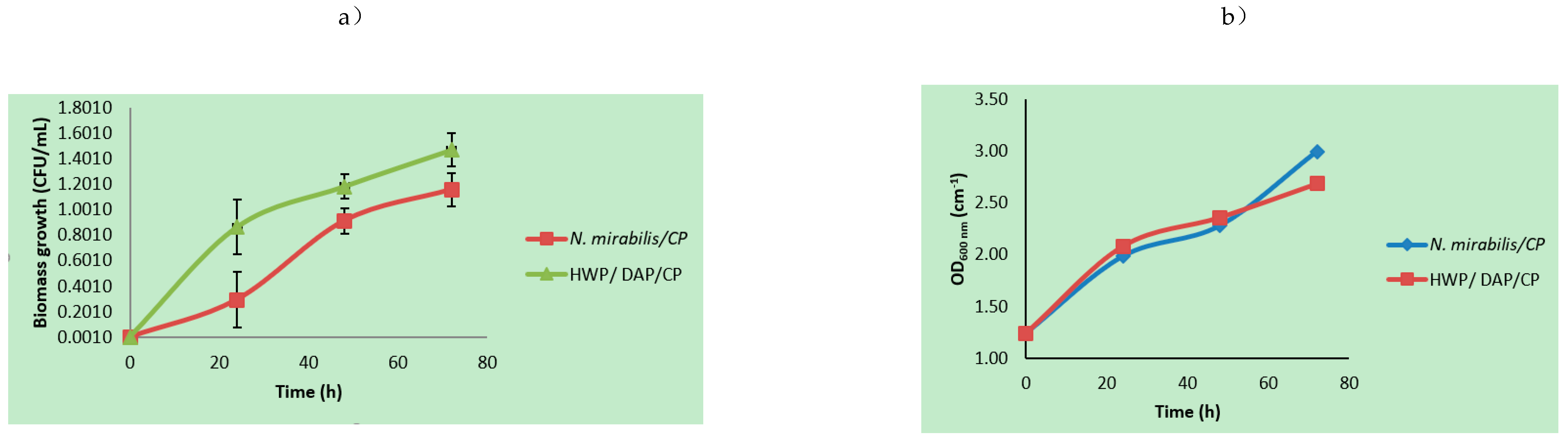
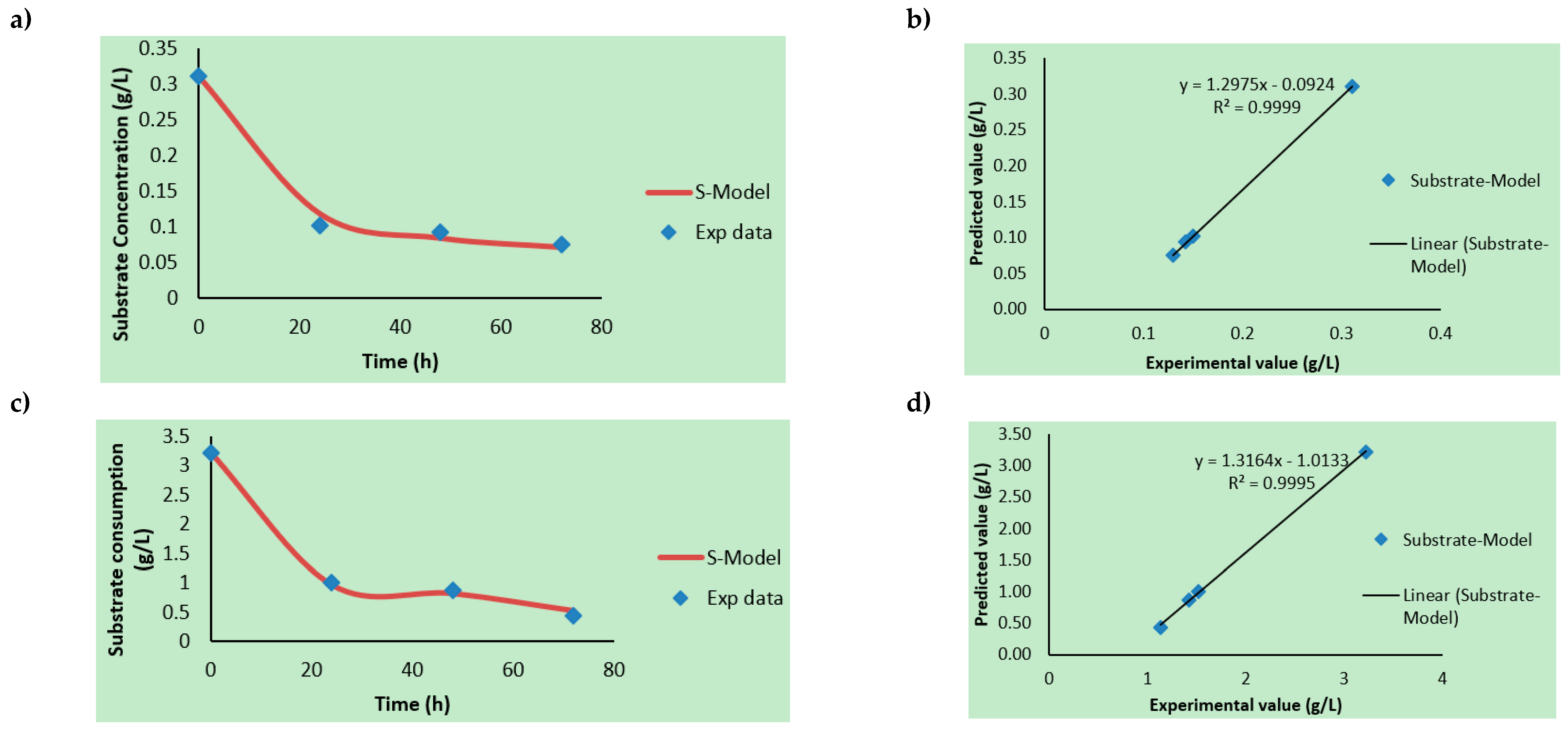
| Hydrolysates | Xmax (×1010 CFU/mL) | So/S (g/L) | P (%Area) (Ethanol; Butanol; 2-Phenylethanol) | Yx/s (CFU/g × 10−13) | Yp/x (Area % mL/CFU × 10−10) | µmax/µ (h−1) | Ks (g/L) | TRPCs (mg/L) |
|---|---|---|---|---|---|---|---|---|
| N. mirabilis/CP | 1.16 | 0.311/0.075 | 1.83 (1.23; 0.23; 0.38) | 4.92 | 1.58 | 1.76/0.095 | 1.32 | 4.26 |
| HWP/DAP/CP | 1.47 | 3.22/0.439 | 1.93 (1.02; 0.53; 0.38) | 0.53 | 1.31 | 1.58/0.088 | 7.46 | 5.65 |
| Absolute difference | 0.31 | n/d | - | 4.39 | 0.3 | 0.2/0.01 | 6.1 | 1.4 |
| Relative difference (%) | 21.1 | n/d | - | 829.9 | 20.2 | 12/9.1 | 82.3 | 24.6 |
| Parameter | Description (Units) | N. mirabilis/CP | HWP/DAP/CP | Relative Difference (%) |
|---|---|---|---|---|
| Biomass formation rate (×108 CFU/mL/h) | 1.61 | 2.04 | 21.1 | |
| Product formation rate (area %/h) | 0.025 | 0.027 | 5.26 | |
| Substrate utilization rate (g/(L·h)) | 0.0033 | 0.0387 | 91.5 |
| Parameter | Description (Units) | N. Mirabilis/CP | HWP/DAP/CP |
|---|---|---|---|
| R2 | Correlation coefficient | 0.941 | 0.4981 |
| m | Slope (Area % mL/CFU) | 1.0035 | 0.4848 |
| *p | g/CFU × 1012 | 2.0 | 18.9 |
| *q | 1/h × 10−5 | 7.84 | 0.185 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dlangamandla, N.; Ntwampe, S.K.O.; Angadam, J.O.; Chidi, B.S.; Mewa-Ngongang, M. Kinetic Parameters of Saccharomyces cerevisiae Alcohols Production Using Nepenthes mirabilis Pod Digestive Fluids-Mixed Agro-Waste Hydrolysates. Fermentation 2019, 5, 10. https://doi.org/10.3390/fermentation5010010
Dlangamandla N, Ntwampe SKO, Angadam JO, Chidi BS, Mewa-Ngongang M. Kinetic Parameters of Saccharomyces cerevisiae Alcohols Production Using Nepenthes mirabilis Pod Digestive Fluids-Mixed Agro-Waste Hydrolysates. Fermentation. 2019; 5(1):10. https://doi.org/10.3390/fermentation5010010
Chicago/Turabian StyleDlangamandla, Nkosikho, Seteno K. O. Ntwampe, Justine O. Angadam, Boredi S. Chidi, and Maxwell Mewa-Ngongang. 2019. "Kinetic Parameters of Saccharomyces cerevisiae Alcohols Production Using Nepenthes mirabilis Pod Digestive Fluids-Mixed Agro-Waste Hydrolysates" Fermentation 5, no. 1: 10. https://doi.org/10.3390/fermentation5010010
APA StyleDlangamandla, N., Ntwampe, S. K. O., Angadam, J. O., Chidi, B. S., & Mewa-Ngongang, M. (2019). Kinetic Parameters of Saccharomyces cerevisiae Alcohols Production Using Nepenthes mirabilis Pod Digestive Fluids-Mixed Agro-Waste Hydrolysates. Fermentation, 5(1), 10. https://doi.org/10.3390/fermentation5010010







