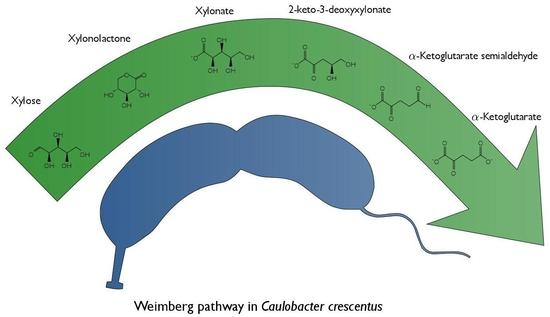
Characterization of the Weimberg Pathway in Caulobacter crescentus
Abstract
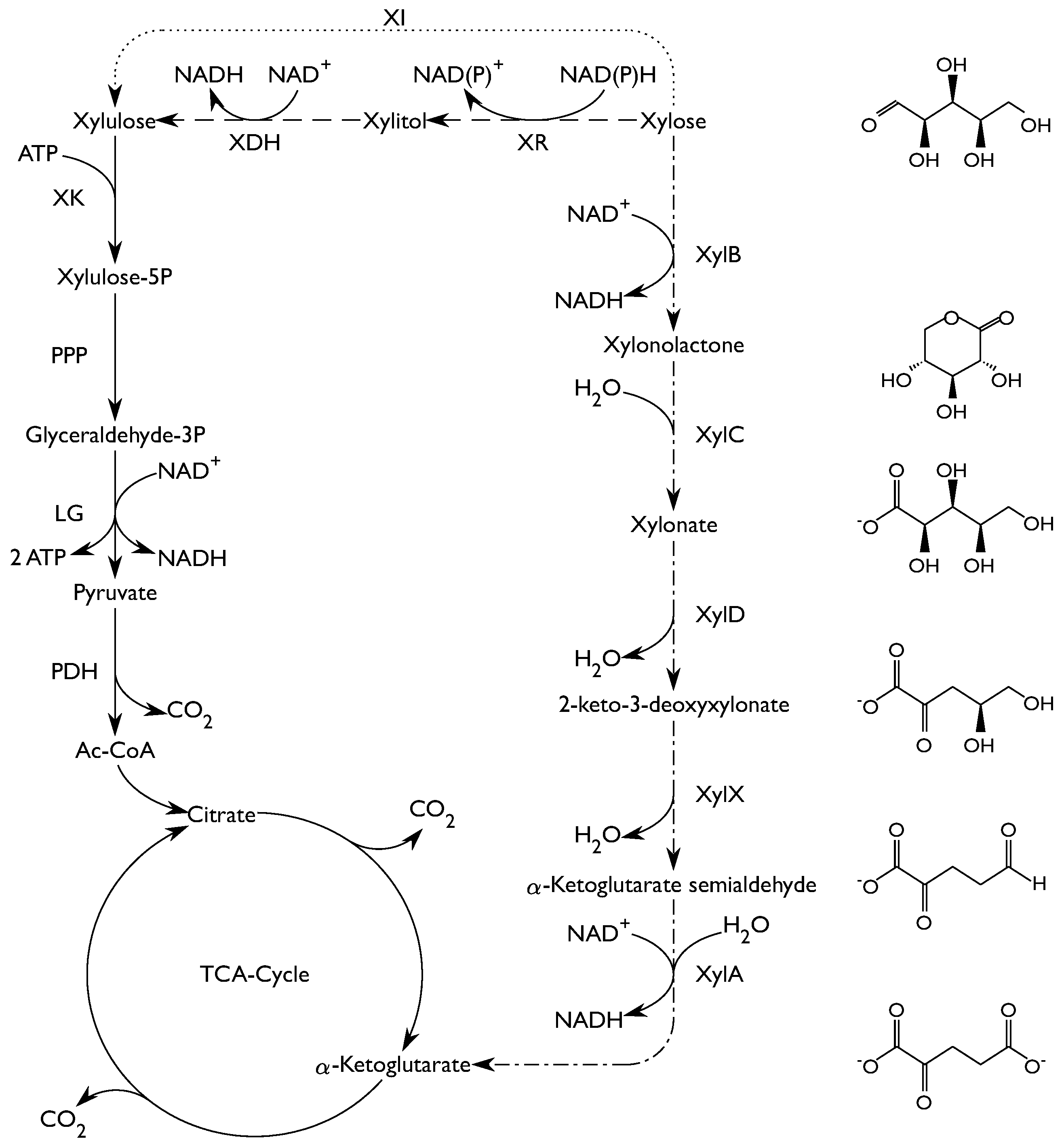
1. Introduction
2. Results
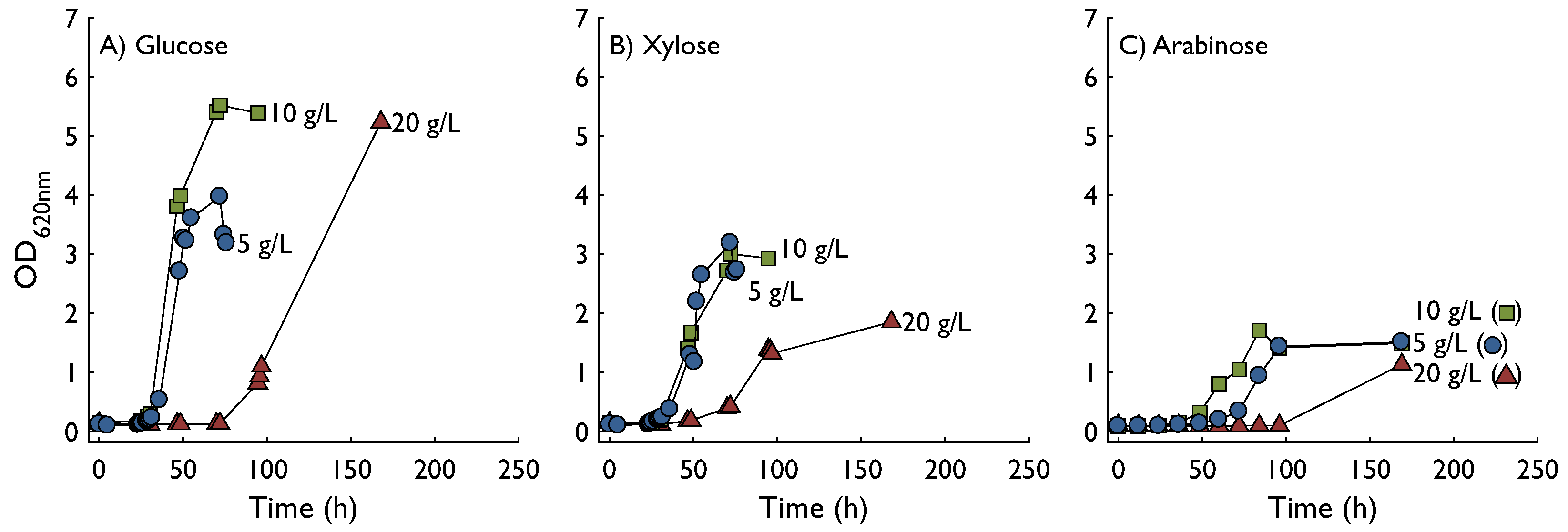
2.1. Growth in Shake Flasks
2.2. Enzyme Activity
2.3. Characterization in Bioreactors
3. Discussion
4. Materials and Methods
4.1. Microorganism
4.2. Medium Composition
4.3. Shake-Flask Cultivation
4.4. Bioreactor Cultivation
4.5. Biomass Measurements
4.6. Analyses of Sugars and Organic Acids
4.7. Enzymatic Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CDW | Cell Dry Weight |
| DOT | Dissolved Oxygen Tension |
| HPLC | High Performance Liquid Chromatography |
| LG | Lower Glycolysis |
| NADH/NAD+ | Nicotinamide Adenine Dinucleotide (Reduced/Oxidized) |
| OD | Optical Density |
| PDH | Pyruvate dehydrogenase |
| PPP | Pentose Phosphate Pathway |
| TCA-Cycle | Tricarboxylic Acid Cycle |
| UHPLC | Ultra High Performance Liquid Chromatography |
| XDH | Xylulose Dehydrogenase |
| XI | Xylose Isomerase |
| XR | Xylose Reductase |
References
- Henrici, A.T.; Johnson, D.E. Studies of Freshwater Bacteria: II. Stalked Bacteria, a New Order of Schizomycetes. J. Bacteriol. 1935, 30, 61–93. [Google Scholar] [PubMed]
- Skerker, J.M.; Laub, M.T. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2004, 2, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Curtis, P.D.; Brun, Y.V. Getting in the loop: Regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 13–41. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, C.G.; Laub, M.T. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr. Opin. Microbiol. 2012, 15, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Feldblyum, T.V.; Laub, M.T.; Paulsen, I.T.; Nelson, K.E.; Eisen, J.; Heidelberg, J.F.; Alley, M.R.K.; Ohta, N.; Maddock, J.R.; et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2001, 98, 4136–4141. [Google Scholar] [CrossRef] [PubMed]
- Hottes, A.K.; Meewan, M.; Yang, D.; Arana, N.; Romero, P.; McAdams, H.H.; Stephens, C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 2004, 186, 1448–1461. [Google Scholar] [CrossRef] [PubMed]
- Arellano, B.H.; Ortiz, J.D.; Manzano, J.; Chen, J.C. Identification of a dehydrogenase required for lactose metabolism in caulobacter crescentus. Appl. Environ. Microbiol. 2010, 76, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Zalatan, F.; Black, P. Characterization of long-chain fatty acid uptake in Caulobacter crescentus. Arch. Microbiol. 2011, 193, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Presley, G.N.; Payea, M.J.; Hurst, L.R.; Egan, A.E.; Martin, B.S.; Periyannan, G.R. Extracellular gluco-oligosaccharide degradation by Caulobacter crescentus. Microbiology 2014, 160, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Wichelecki, D.J.; Graff, D.C.; Al-Obaidi, N.; Almo, S.C.; Gerlt, J.A. Identification of the in Vivo Function of the High-Efficiency d-Mannonate Dehydratase in Caulobacter crescentus NA1000 from the Enolase Superfamily. Biochemistry 2014, 53, 4087–4089. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185. [Google Scholar] [CrossRef] [PubMed]
- Weimberg, R. Pentose oxidation by Pseudomonas fragi. J. Biol. Chem. 1961, 236, 629–635. [Google Scholar] [PubMed]
- Radek, A.; Krumbach, K.; Gätgens, J.; Wendisch, V.F.; Wiechert, W.; Bott, M.; Noack, S.; Marienhagen, J. Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of d-xylose containing substrates. J. Biotechnol. 2014, 192, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Karhumaa, K.; Garcia Sanchez, R.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 2007, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, C.; Reyes-Sosa, F.M.; Díez, B. Enzymatic hydrolysis of biomass from wood. Microb. Biotechnol. 2016, 9, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Meijnen, J.P.; De Winde, J.H.; Ruijssenaars, H.J. Establishment of oxidative d-xylose metabolism in Pseudomonas putida S12. Appl. Environ. Microbiol. 2009, 75, 2784–2791. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Valdehuesa, K.N.G.; Nisola, G.M.; Ramos, K.R.M.; Chung, W.J. High yield production of d-xylonic acid from d-xylose using engineered Escherichia coli. Bioresour. Technol. 2012, 115, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xian, M.; Zou, H.; Zhang, H. Metabolic Engineering of Escherichia coli for the Production of Xylonate. PLoS ONE 2013, 8, e67305. [Google Scholar] [CrossRef] [PubMed]
- Toivari, M.H.; Ruohonen, L.; Richard, P.; Penttilä, M.; Wiebe, M.G. Saccharomyces cerevisiae engineered to produce d-xylonate. Appl. Microbiol. Biotechnol. 2010, 88, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Lourenço, R.F.; Bernhardt, J.; Albrecht, D.; Schüler, J.; Hecker, M.; Gomes, S.L. A comprehensive genomic, transcriptomic and proteomic analysis of a hyperosmotic stress sensitive α-proteobacterium. BMC Microbiol. 2015, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Ely, B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991, 204, 372–384. [Google Scholar] [PubMed]
- Almqvist, H.; Sandahl, M.; Lidén, G. A rapid method for analysis of fermentatively produced d-xylonate using ultra-high performance liquid chromatography and evaporative light scattering detection. Biosci. Biotechnol. Biochem. 2017, 81, 1078–1080. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Johnsen, U.; Dambeck, M.; Zaiss, H.; Fuhrer, T.; Soppa, J.; Sauer, U.; Schönheit, P. d-xylose degradation pathway in the halophilic archaeon Haloferax volcanii. J. Biol. Chem. 2009, 284, 27290–27303. [Google Scholar] [CrossRef] [PubMed]
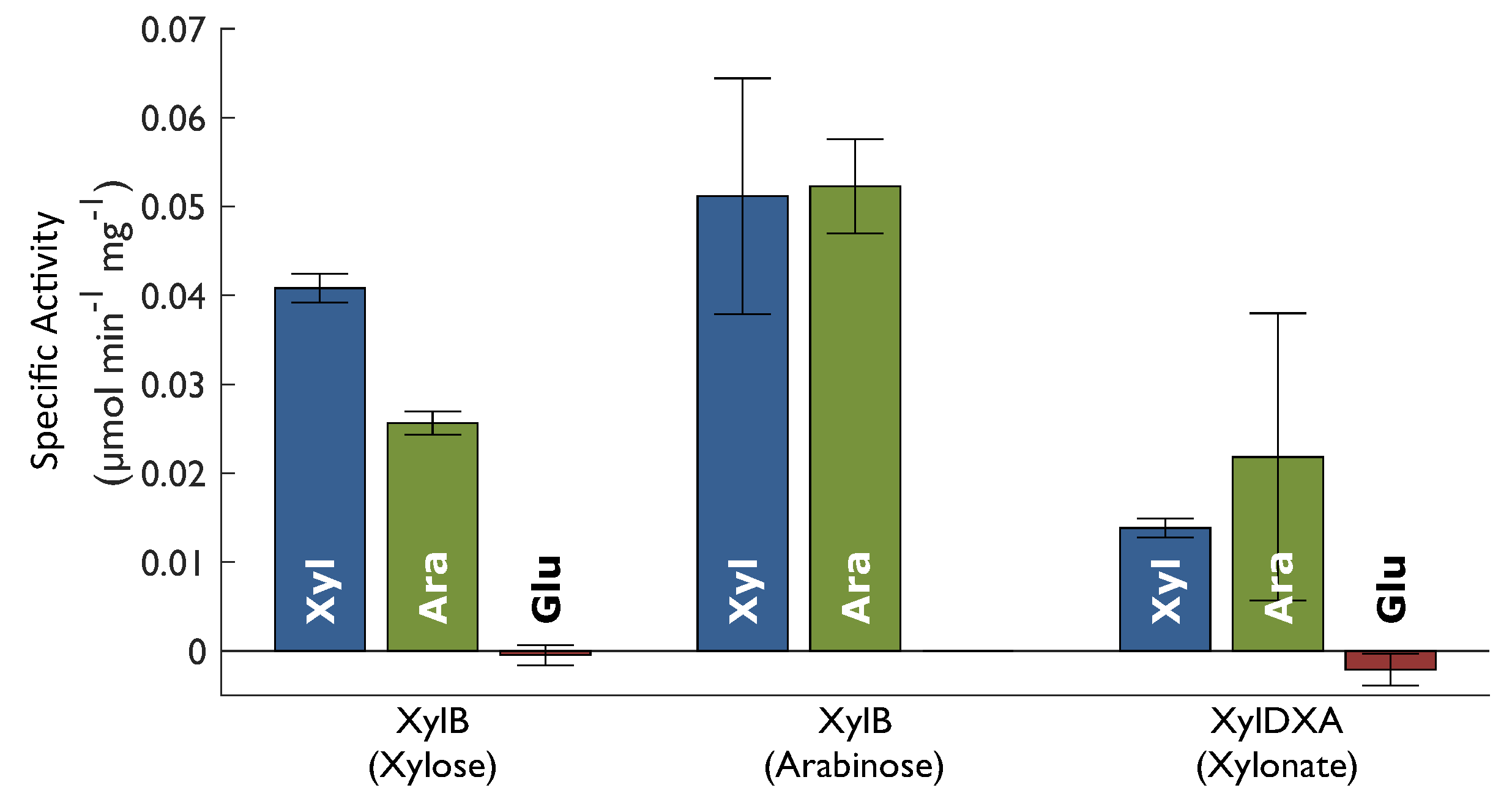

| XylB | XylB | XylDXA | |
|---|---|---|---|
| Tris-HCl, pH 8.0 (mM) | 100.0 | 100.0 | 100.0 |
| MgCl2 (mM) | 2.0 | 2.0 | 10.0 |
| NAD+ (mM) | 2.0 | 2.0 | 2.0 |
| Xylose (mM) | 83.3 | - | - |
| Arabinose (mM) | - | 83.3 | - |
| Xylonate (mM) | - | - | 80.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almqvist, H.; Jonsdottir Glaser, S.; Tufvegren, C.; Wasserstrom, L.; Lidén, G. Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation 2018, 4, 44. https://doi.org/10.3390/fermentation4020044
Almqvist H, Jonsdottir Glaser S, Tufvegren C, Wasserstrom L, Lidén G. Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation. 2018; 4(2):44. https://doi.org/10.3390/fermentation4020044
Chicago/Turabian StyleAlmqvist, Henrik, Sara Jonsdottir Glaser, Celina Tufvegren, Lisa Wasserstrom, and Gunnar Lidén. 2018. "Characterization of the Weimberg Pathway in Caulobacter crescentus" Fermentation 4, no. 2: 44. https://doi.org/10.3390/fermentation4020044
APA StyleAlmqvist, H., Jonsdottir Glaser, S., Tufvegren, C., Wasserstrom, L., & Lidén, G. (2018). Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation, 4(2), 44. https://doi.org/10.3390/fermentation4020044