Biological Demalication and Deacetification of Musts and Wines: Can Wine Yeasts Make the Wine Taste Better?
Abstract
1. General Introduction
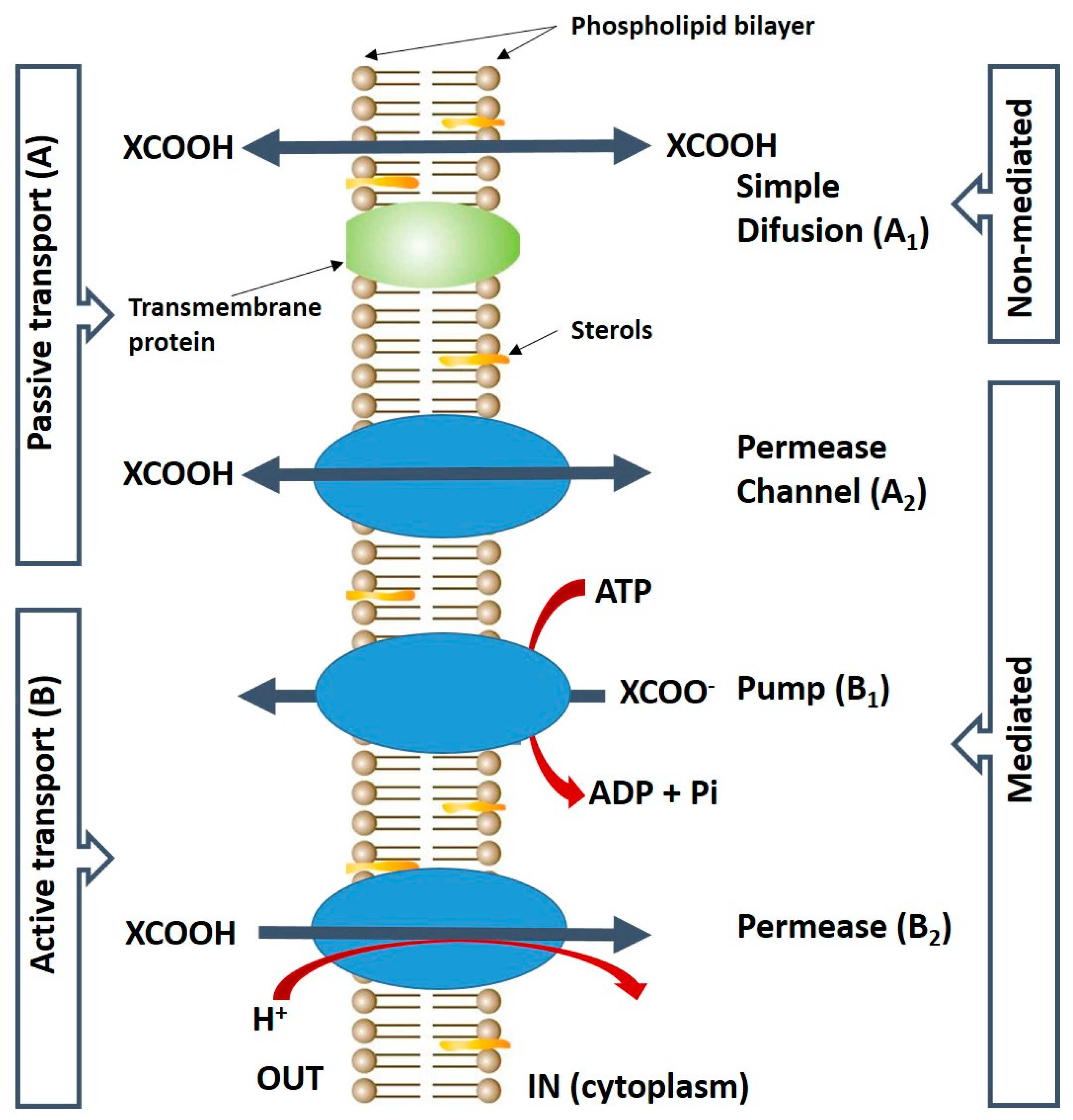
2. Uptake of Malic Acid into the Yeast Cell
3. The Demalication Activity of Saccharomyces Strains
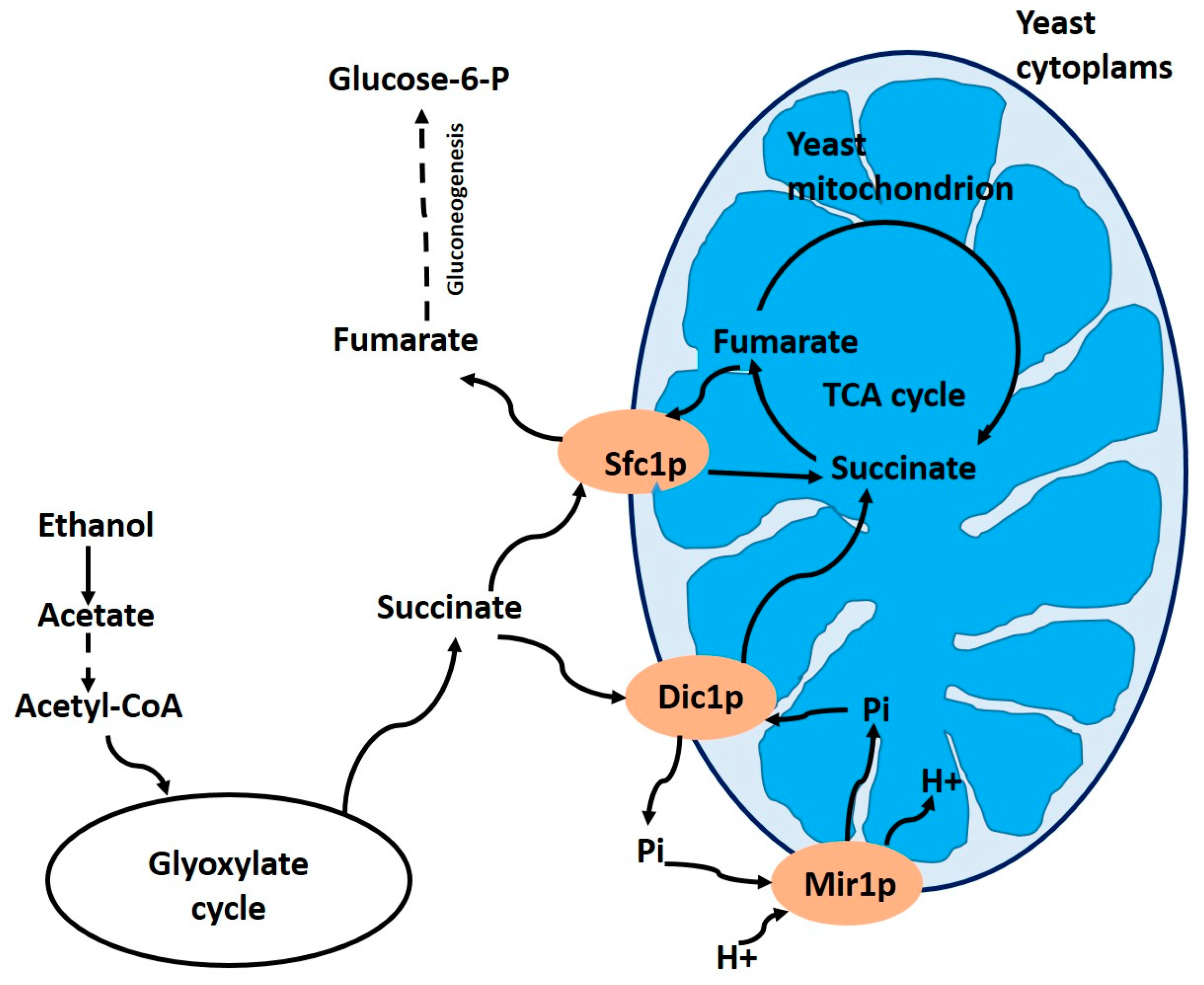
3.1. Dicarboxylate Carrier Dic1p (YLR348c) in Yeasts
3.2. The Aspartate/Glutamate Carrier Agc1p (YPR021c) in Yeasts
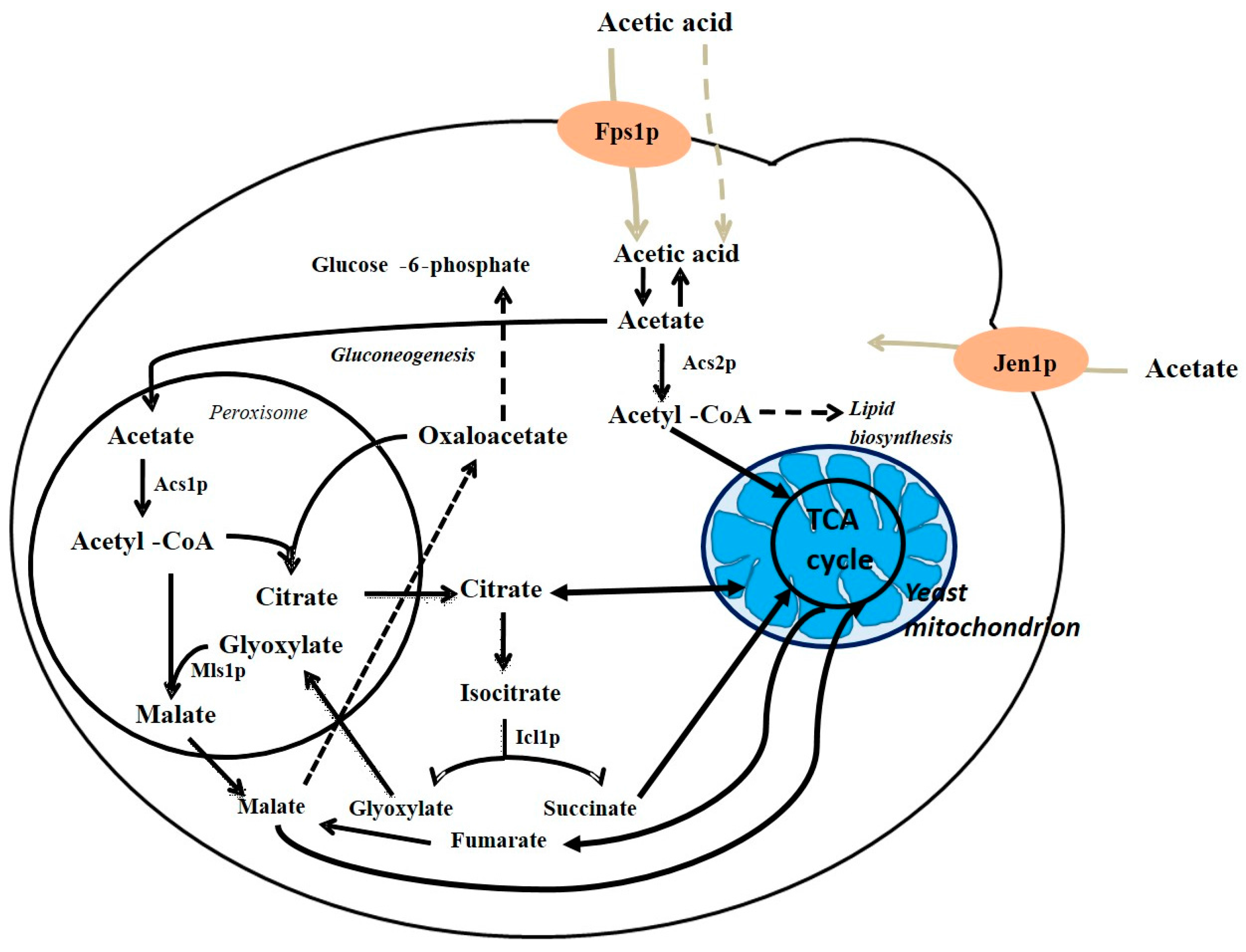
4. Acetic Acid Metabolism in S. cerevisiae
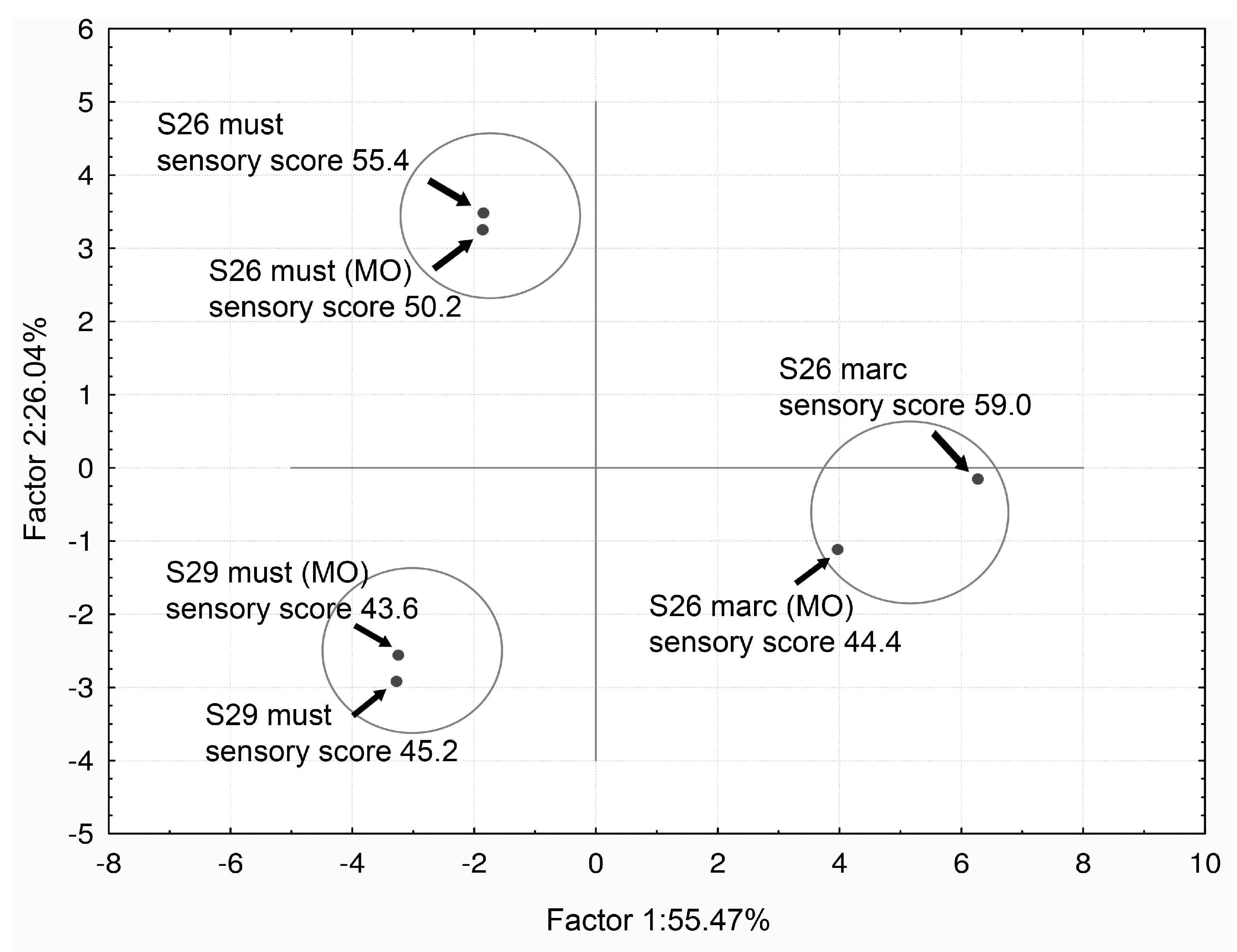
VA Bio-Reduction by S. cerevisiae Yeasts Strains and its Limitations
5. Final Remarks
Acknowledgments
Conflicts of Interest
References
- Vilela, A.; Inês, A.; Cosme, F. Is wine savory? Umami taste in wine. SDRP J. Food Sci. Technol. 2016, 1, 1–6. [Google Scholar]
- Vilela, A.; Jordão, A.M.; Cosme, F. Wine phenolics: Looking for a smooth mouthfeel. SDRP J. Food Sci. Technol. 2016, 1, 1–8. [Google Scholar] [CrossRef]
- Jordão, A.M.; Vilela, A.; Cosme, F. From Sugar of Grape to Alcohol of Wine: Sensorial Impact of Alcohol in Wine. Beverages 2015, 1, 292–310. [Google Scholar] [CrossRef]
- Torija, M.J.; Beltran, G.; Novo, M.; Poblet, M.; Rozès, N.; Mas, A.; Guillamón, J.M. Effect of organic acids and nitrogen source on alcoholic fermentation: Study of their buffering capacity. J. Agric. Food Chem. 2003, 51, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.F.; Wu, B.H.; Fan, P.G.; Li, S.H.; Li, L.S. Sugar and acid concentrations in 98 grape cultivars analyzed by principal component analysis. J. Sci. Food Agric. 2006, 86, 1526–1536. [Google Scholar] [CrossRef]
- Lamikanra, O.; Inyang, I.; Leong, S. Distribution and effect of grape maturity on organic acid content of red Muscadine grapes. J. Agric. Food Chem. 1995, 43, 3026–3028. [Google Scholar] [CrossRef]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Silva, R.F.; Chaves, S.R.; Sousa, M.J.; Côrte-Real, M. The impact of acetate metabolism on yeast fermentative performance and wine quality: Reduction of volatile acidity of grape-musts and wines—Minireview. Appl. Microbiol. Biotechnol. 2011, 89, 271–280. [Google Scholar] [CrossRef]
- Inês, A.; Tenreiro, T.; Tenreiro, R.; Mendes-Faia, A. Revisão: As bactérias do ácido láctico do vinho-Parte I. Ciênc. Téc. Vitiviníc. 2008, 23, 81–96. [Google Scholar]
- Van Rooyen, T.J.; Tracel, R.P. Biological Deacidification of Musts Induced by Yeasts or Malolactic Bacteria and the Effect on Wine Quality. S. Afr. J. Enol. Vitic. 1987, 8, 60–69. [Google Scholar] [CrossRef]
- Camarasa, C.; Bidard, F.; Bony, M.; Barre, P.; Dequin, S. Characterization of Schizosaccharomyces pombe Malate Permease by Expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2001, 67, 4144–4151. [Google Scholar] [CrossRef] [PubMed]
- Benito, A.; Calderón, F.; Palomero, F.; Benito, S. Combined use of selected Schizosaccharomyces pombe and Lachancea thermotolerans yeast strains as an alternative to the traditional malolactic fermentation in red wine production. Molecules 2015, 20, 9510–9523. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, L.; Capozzi, V.; Berbegal, C.; Corbo, M.R.; Bevilacqua, A.; Spano, G.; Sinigaglia, M. Microbial resources and enological significance: Opportunities and benefits. Front. Microbiol. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zoecklein, B.W.; Fugelsang, K.C.; Gump, B.H.; Nury, F.S. Volatile acidity. In Wine Analysis and Production, 1st ed.; Chapman & Hall: New York, NY, USA, 1995; pp. 192–198. ISBN 978-1-4757-6967-8. [Google Scholar]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. The microbiology of wine and vinifications. In Handbook of Enology, 1st ed.; John Wiley & Sons Ltd.: Chichester, UK, 2000; Volume 1, p. 454. ISBN 13:978-0-470-01034-1 (HB). [Google Scholar]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Côrte-Real, M. Reduction of volatile acidity of wines by selected yeast strains. Appl. Microbiol. Biotechnol. 2008, 80, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Côrte-Real, M. Effects of acetic acid, ethanol and SO2 on the removal of volatile acidity from acidic wines by two Saccharomyces cerevisiae commercial strains. Appl. Microbiol. Biotechnol. 2010, 87, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
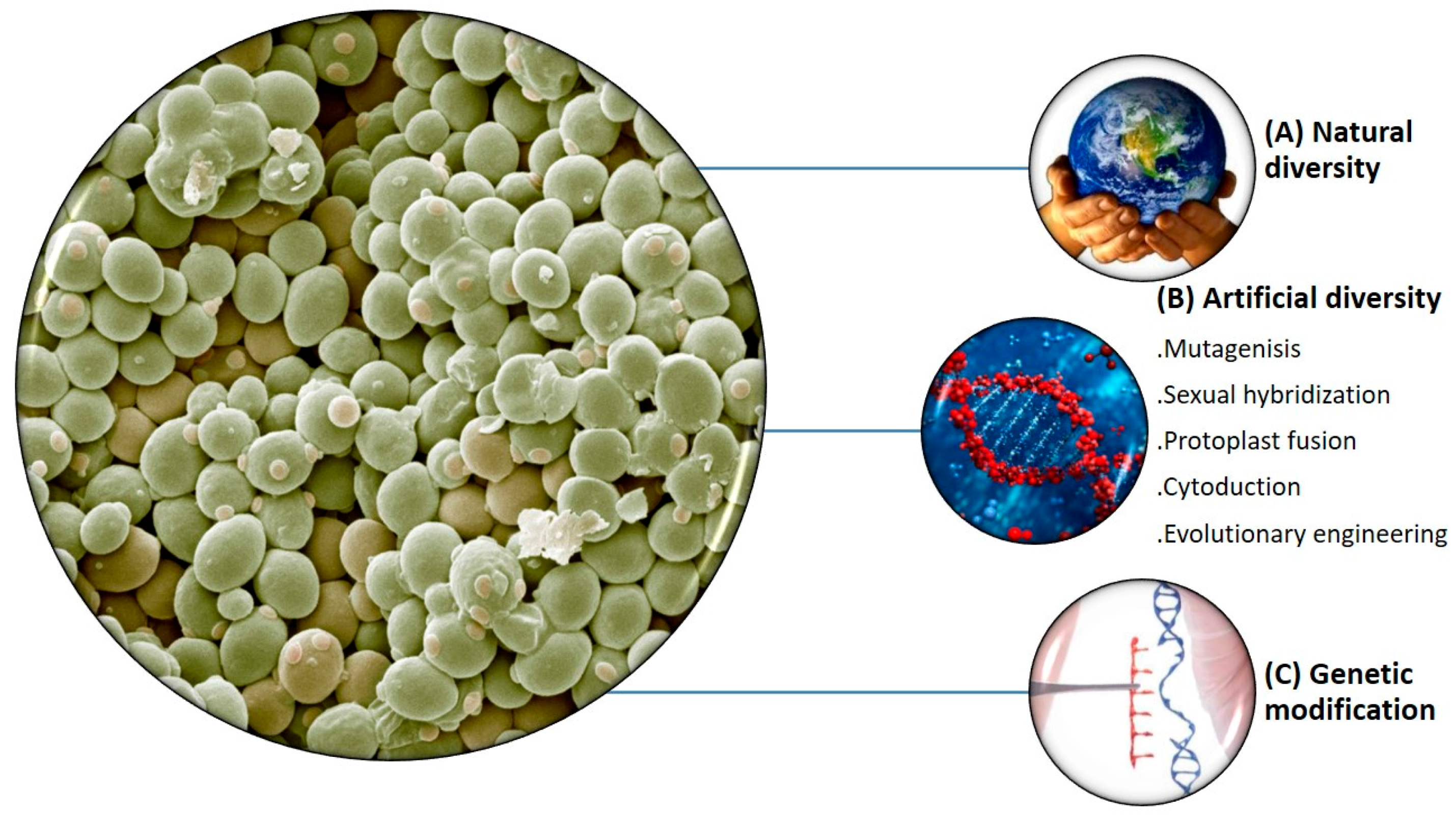
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving industrial yeast strains: Exploiting natural and artificial diversity. Fems Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef] [PubMed]
- Schuller, D. Better yeast for better wine—Genetic improvement of Saccharomyces cerevisiae winemaking strains. In Progress in Mycology; Rai, M., Kövics, G., Eds.; Scientific Publishers: Jodhpur, India, 2010; pp. 1–51. ISBN 978-90-481-3713-8. [Google Scholar]
- Husnik, J.I.; Volschenk, H.; Bauer, J.; Colavizza, D.; Luo, Z.; van Vuuren, H.J. Metabolic engineering of malolactic wine yeast. Metab. Eng. 2006, 8, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Saayman, M.; van Zyl, W.H.; Viljoen-Bloom, M. Cloning, characterization, and heterologous expression of the Candida utilis malic enzyme gene. Curr. Genet. 2006, 49, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Saayman, M.; Viljoen-Bloom, M. The biochemistry of malic acid metabolism by wine yeasts—A review. S. Afr. J. Enol. Vitic. 2006, 27, 113–122. [Google Scholar] [CrossRef]
- Kunicka-Styczynska, A.; Rajkowska, K. Phenotypic and genotypic diversity of wine yeasts used for acidic musts. World J. Microbiol. Biotechnol. 2012, 28, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Kunicka-Styczynska, A.; Rajkowska, K. Physiological and genetic stability of hybrids of industrial wine yeasts Saccharomyces sensu stricto complex. J. Appl. Microbiol. 2011, 110, 1538–1549. [Google Scholar] [CrossRef] [PubMed]
- Redzepovic, S.; Orlic, S.; Majdak, A.; Kozina, B.; Volschenk, H.; Viljoen-Bloom, M. Differential malic acid degradation by selected strains of Saccharomyces during alcoholic fermentation. Int. J. Food Microbiol. 2003, 83, 49–61. [Google Scholar] [CrossRef]
- Bony, M.; Bidart, F.; Camarasa, C.; Ansanay, V.; Dulau, L.; Barre, P.; Dequin, S. Metabolic analysis of S. cerevisiae strains engineered for malolactic fermentation. FEBS Lett. 1997, 410, 452–456. [Google Scholar] [CrossRef]
- Husnik, J.I.; Delaquis, P.J.; Cliff, M.A.; van Vuuren, H.J.J. Functional analyses of the malolactic wine yeast ml01. Am. J. Enol. Vitic. 2007, 58, 42–52. [Google Scholar]
- Main, G.L.; Threlfall, R.T.; Morris, J.R. Reduction of malic acid in wine using natural and genetically enhanced microorganisms. Am. J. Enol. Vitic. 2007, 58, 341–345. [Google Scholar]
- Silva, S.; Ramon-Portugal, F.; Andrade, P.; Abreu, S.; Texeira, M.D.; Strehaiano, P. Malic acid consumption by dry immobilized cells of Schizosaccharomyces pombe. Am. J. Enol. Vitic. 2003, 54, 50–55. [Google Scholar]
- Sousa, M.J.; Teixeira, J.A.; Mota, M. Must deacidification with an induced flocculent yeast strain of Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 1993, 39, 189–193. [Google Scholar] [CrossRef]
- Remize, F.; Andrieu, E.; Dequin, S. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl. Environ. Microbiol. 2000, 66, 3151–3159. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.J.; Luo, Z.; Madilao, L.L.; van Vuuren, H.J.J. The Fermentation Stress Response Protein Aaf1p/Yml081Wp Regulates Acetate Production in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e51551. [Google Scholar] [CrossRef] [PubMed]
- Cordente, A.G.; Cordero-Bueso, G.; Pretorius, I.S.; Curtin, C.D. Novel wine yeast with mutations in YAP1 that produce less acetic acid during fermentation. FEMS Yeast Res. 2013, 13, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Walkey, C.J.; Madilao, L.L.; Measday, V.; Van Vuuren, H.J.J. Functional improvement of Saccharomyces cerevisiae to reduce volatile acidity in wine. FEMS Yeast Res. 2013, 13, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Casal, M.; Paiva, S.; Queirós, O.; Soares-Silva, I. Transport of carboxylic acids in yeasts. FEMS Microbiol. Rev. 2008, 32, 974–994. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.M. L-Malic acid permeation in resting cells of anaerobically grown Saccharomyces cerevisiae. Biochim. Biophys. Acta 1987, 901, 30–34. [Google Scholar] [CrossRef]
- Benito, Á.; Jeffares, D.; Palomero, F.; Calderón, F.; Bai, F.-Y.; Bähler, J.; Benito, S. Selected Schizosaccharomyces pombe strains have characteristics that are beneficial for winemaking. PLoS ONE 2016, 11, e0151102. [Google Scholar] [CrossRef] [PubMed]
- Rodriquez, S.B.; Thornton, R.J. A malic acid dependent mutant of Schizosaccharomyces malidevorans. Arch. Microbiol. 1989, 152, 564–566. [Google Scholar] [CrossRef]
- Gao, C.; Fleet, G.H. Degradation of malic and tartaric acids by high density cell suspensions of wine yeasts. Food Microbiol. 1995, 12, 65–71. [Google Scholar] [CrossRef]
- Côrte-Real, M.; Leão, C.; Van Uden, N. Transport of L(−)malic acid and other dicarboxylic acids in the yeast Candida sphaerica. Appl. Microbiol. Biotechnol. 1989, 31, 551–555. [Google Scholar] [CrossRef]
- Côrte-Real, M.; Leão, C. Transport of malic acid and other dicarboxylic acids in the yeast Hansenula anomala. Appl. Environ. Microbiol. 1990, 56, 1109–1113. [Google Scholar] [PubMed]
- Queirós, O.; Casal, M.; Althoff, S.; Moradas-Ferreira, P.; Leão, C. Isolation and characterization of Kluyveromyces marxianus mutants deficient in malate transport. Yeast 1998, 14, 401–407. [Google Scholar] [CrossRef]
- Zmijewski, M.J.; Macquillan, A.M. Dual effects of glucose on dicarboxylic acid transport in Kluyveromyces lactis. Can. J. Microbiol. 1975, 21, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Baranowski, K.; Radler, F. The glucose dependent transport of l-malate in Zygosaccharomyces bailii. Antonie Van Leeuwenhoek 1984, 50, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S.; Bauer, F.F. Meeting the consumer challenge through genetically customized wine-yeast strains. Trends Biotechnol. 2002, 20, 426–432. [Google Scholar] [CrossRef]
- Rodriguez, S.B.; Thornton, R.J. Factors influencing the utilization of l-malate by yeasts. FEMS Microbiol. Lett. 1990, 60, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Volschenk, H.; van Vuuren, H.J.J.; Viljoen-Bloom, M. Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces. Curr. Genet. 2003, 43, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Osothsilp, C.; Subden, R.E. Isolation and characterization of Schizosaccharomyces pombe mutants with defective NAD-dependent malic enzyme. Can. J. Microbiol. 1986, 32, 481–486. [Google Scholar] [CrossRef]
- Volschenk, H.; Viljoen, M.; Grobler, J.; Petzold, B.; Bauer, F.; Subden, R.E.; Young, R.A.; Lonvaud, A.; Denayrolles, M.; van Vuuren, H.J.J. Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat. Biotechnol. 1997, 15, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Lancar-Benba, J.; Foucher, B.; Saint-Macary, M. Characterization, purification and properties of the yeast mitochondrial dicarboxylate carrier (Saccharomyces cerevisiae). Biochimie 1996, 78, 195–200. [Google Scholar] [CrossRef]
- Palmieri, L.; Palmieri, F.; Runswick, M.J.; Walker, J.E. Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. FEBS Lett. 1996, 399, 299–302. [Google Scholar] [CrossRef]
- Palmieri, F.; Agrimi, G.; Blanco, E.; Castegna, A.; Di Noia, M.-A.; Iacobazzi, V.; Lasorsa, F.-M.; Marobbio, C.M.T.; Palmieri, L.; Scarcia, P.; et al. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim. Biophys. Acta 2006, 1757, 1249–1262. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Vozza, A.; Honlinger, A.; Dietmeier, K.; Palmisano, A.; Zara, V.; Palmieri, F. The mitochondrial dicarboxylate carrier is essential for the growth of Saccharomyces cerevisiae on ethanol or acetate as the sole carbon source. Mol. Microbiol. 1999, 31, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Cavero, S.; Vozza, A.; del Arco, A.; Palmieri, L.; Villa, A.; Blanco, E.; Runswick, M.J.; Walker, J.E.; Cerdan, S.; Palmieri, F.; et al. Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol. Microbiol. 2003, 50, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Ribéreau-Gayon, P.; Glories, Y.; Maujean, A.; Dubourdieu, D. Alcohols and other volatile compounds. The chemistry of wine stabilization and treatments. In Handbook of Enology, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2006; Volume 2, pp. 51–64. [Google Scholar] [CrossRef]
- Office Internationale de la Vigne et du Vin. International Code of Oenological Practices; OIV: Paris, France, 2010. [Google Scholar]
- Vilela-Moura, A.; Schuller, D.; Falco, V.; Mendes-Faia, A.; Côrte-Real, M. Effect of refermentation conditions and micro-oxygenation on the reduction of volatile acidity by commercial S. cerevisiae strains and their impact on the aromatic profile of wines. Int. J. Food Microbiol. 2010, 141, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Vasserot, Y.; Mornet, F.; Jeandet, P. Acetic acid removal by Saccharomyces cerevisiae during fermentation in oenological conditions. Metabolic consequences. Food Chem. 2010, 119, 1220–1223. [Google Scholar] [CrossRef]
- Schüller, H.J. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 2003, 43, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, M.M.; Gombert, A.K.; Christensen, B.; Olsson, L.; Nielsen, J. Identification of in vivo enzyme activities in the cometabolism of glucose and acetate by Saccharomyces cerevisiae by using 13C-labeled substrates. Eukaryot. Cell 2003, 2, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Casal, M.; Cardoso, H.; Leão, C. Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology 1996, 142, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Casal, M.; Cardoso, H.; Leão, C. Effects of ethanol and other alkanols on transport of acetic acid in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1998, 64, 665–668. [Google Scholar] [PubMed]
- Casal, M.; Paiva, S.; Andrade, R.P.; Gancedo, C.; Leão, C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 1999, 181, 2620–2623. [Google Scholar] [PubMed]
- Paiva, S.; Devaux, F.; Barbosa, S.; Jacq, C.; Casal, M. Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast 2004, 21, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Vilela-Moura, A. Isolation and Characterization of Yeasts: Application in Controlled Processes of Volatile Acidity Bio-Reduction in Wines. Ph.D. Thesis, University os Trás-os-Montes e Alto Douro (UTAD), Vila Real, Portugal, 2010. [Google Scholar]
- Vilela, A.; Amaral, C.; Shuller, D.; Mendes-Faia, A.; Corte-Real, M. Combined use of Wallerstein and Zygosaccharomyces bailii modified differential media to isolate yeasts for the controlled reduction of volatile acidity of grape musts and wines. J. Biotech. Res. 2015, 6, 43–53. [Google Scholar]
- Pizarro, F.; Vargas, F.A.; Agosin, E. A systems biology perspective of wine fermentations. Yeast 2007, 24, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Moreno, R.; Morales, P.; Gonzalez, R.; Mas, A.; Beltran, G. Biomass production and alcoholic fermentation performance of Saccharomyces cerevisiae as a function of nitrogen source. FEMS Yeast Res. 2012, 12, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Eglinton, J.M.; Henschke, P.A. Restarting incomplete fermentations: The effect of high concentrations of acetic acid. Aust. J. Grape Wine Res. 1999, 52, 71–78. [Google Scholar] [CrossRef]
- Lilly, M.; Lambrechts, M.G.; Pretorius, I.S. Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl. Environ. Microbiol. 2000, 66, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Carlton, W.K.; Gump, B.; Fugelsang, K.; Hasson, A.S. Monitoring acetaldehyde concentrations during MO of red wine by headspace solid-phase micro extraction with on-fiber derivatization. J. Agric. Food Chem. 2007, 55, 5620–5625. [Google Scholar] [CrossRef] [PubMed]
- Genisheva, Z.; Teixeira, J.A.; Oliveira, J.M. Immobilized cell systems for batch and continuous winemaking. Trends Food Sci. Technol. 2014, 40, 33–47. [Google Scholar] [CrossRef]
- Vilela, A.; Schuller, D.; Mendes-Faia, A.; Côrte-Real, M. Reduction of volatile acidity of acidic wines by immobilized Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2013, 97, 4991–5000. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilela, A. Biological Demalication and Deacetification of Musts and Wines: Can Wine Yeasts Make the Wine Taste Better? Fermentation 2017, 3, 51. https://doi.org/10.3390/fermentation3040051
Vilela A. Biological Demalication and Deacetification of Musts and Wines: Can Wine Yeasts Make the Wine Taste Better? Fermentation. 2017; 3(4):51. https://doi.org/10.3390/fermentation3040051
Chicago/Turabian StyleVilela, Alice. 2017. "Biological Demalication and Deacetification of Musts and Wines: Can Wine Yeasts Make the Wine Taste Better?" Fermentation 3, no. 4: 51. https://doi.org/10.3390/fermentation3040051
APA StyleVilela, A. (2017). Biological Demalication and Deacetification of Musts and Wines: Can Wine Yeasts Make the Wine Taste Better? Fermentation, 3(4), 51. https://doi.org/10.3390/fermentation3040051