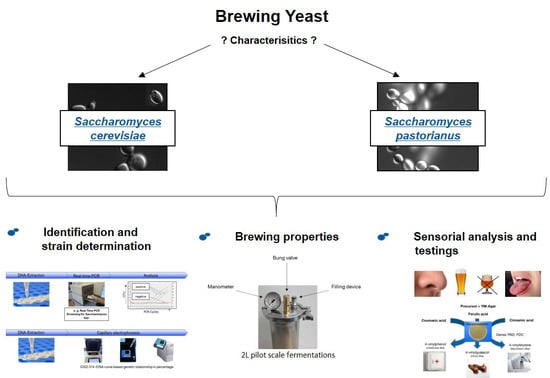
The Importance of a Comparative Characterization of Saccharomyces Cerevisiae and Saccharomyces Pastorianus Strains for Brewing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Isolates and Strains
2.2. Genetic Identification and Strain Determination
2.2.1. Real-Time Polymerase Chain Reaction
2.2.2. DNA Fingerprinting (PCR-Capillary Electrophoresis of the IGS2-314 Fragment)
2.3. Phenolic Off-Flavor Test
2.4. Brewing Trials
2.4.1. Wort
2.4.2. Propagation
2.4.3. Fermentation
2.5. Analytical Methods
2.5.1. Determining the Cell Count (Cells in Suspension and Total Cell Count)
2.6. Sensory Evaluation
3. Results
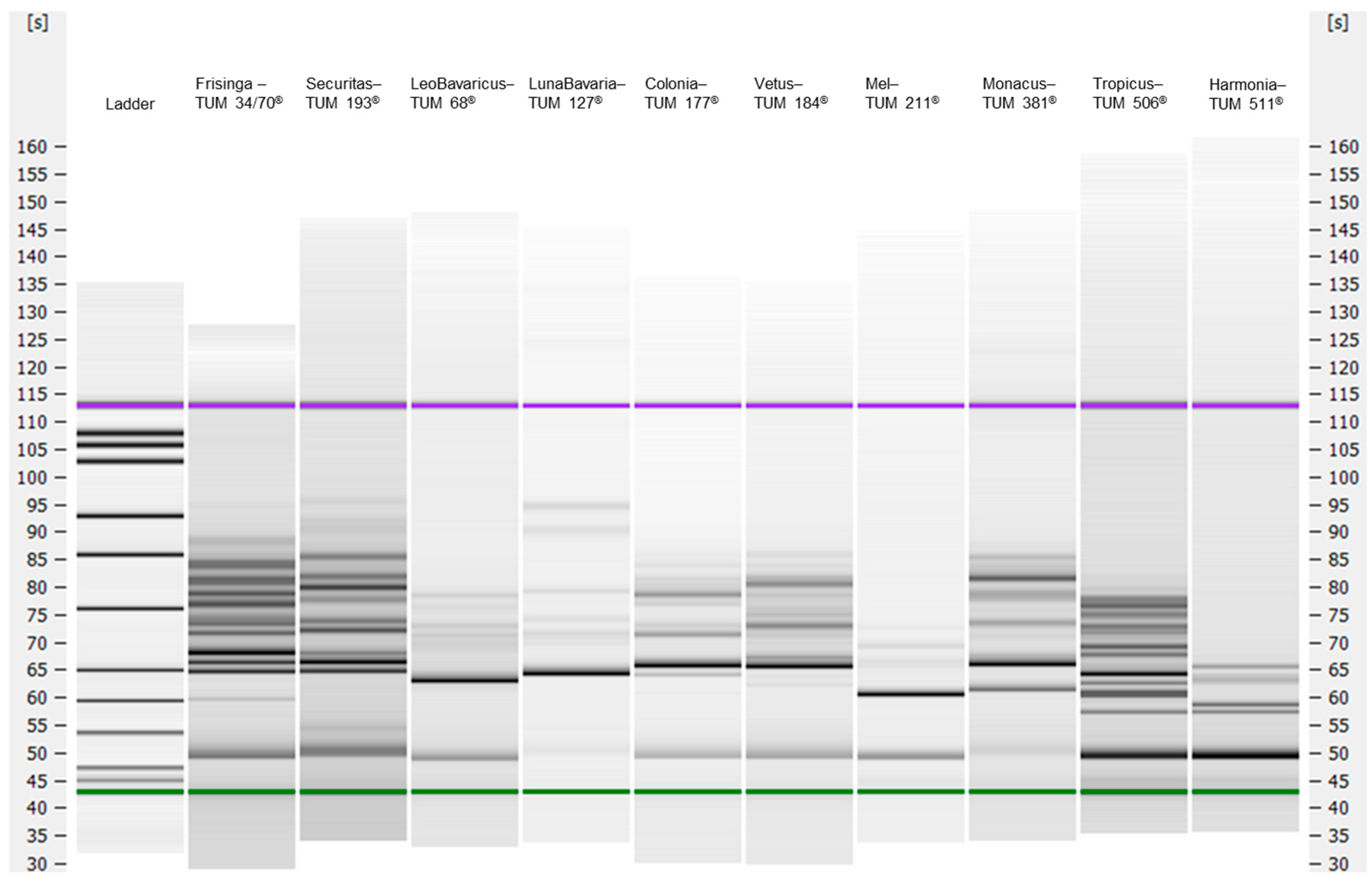
3.1. Genetic Analysis
3.1.1. Real-Time PCR Assays and IGS2-314 PCR-Capillary Electrophoresis
3.2. Brewing Trials
3.2.1. Sugar Utilization
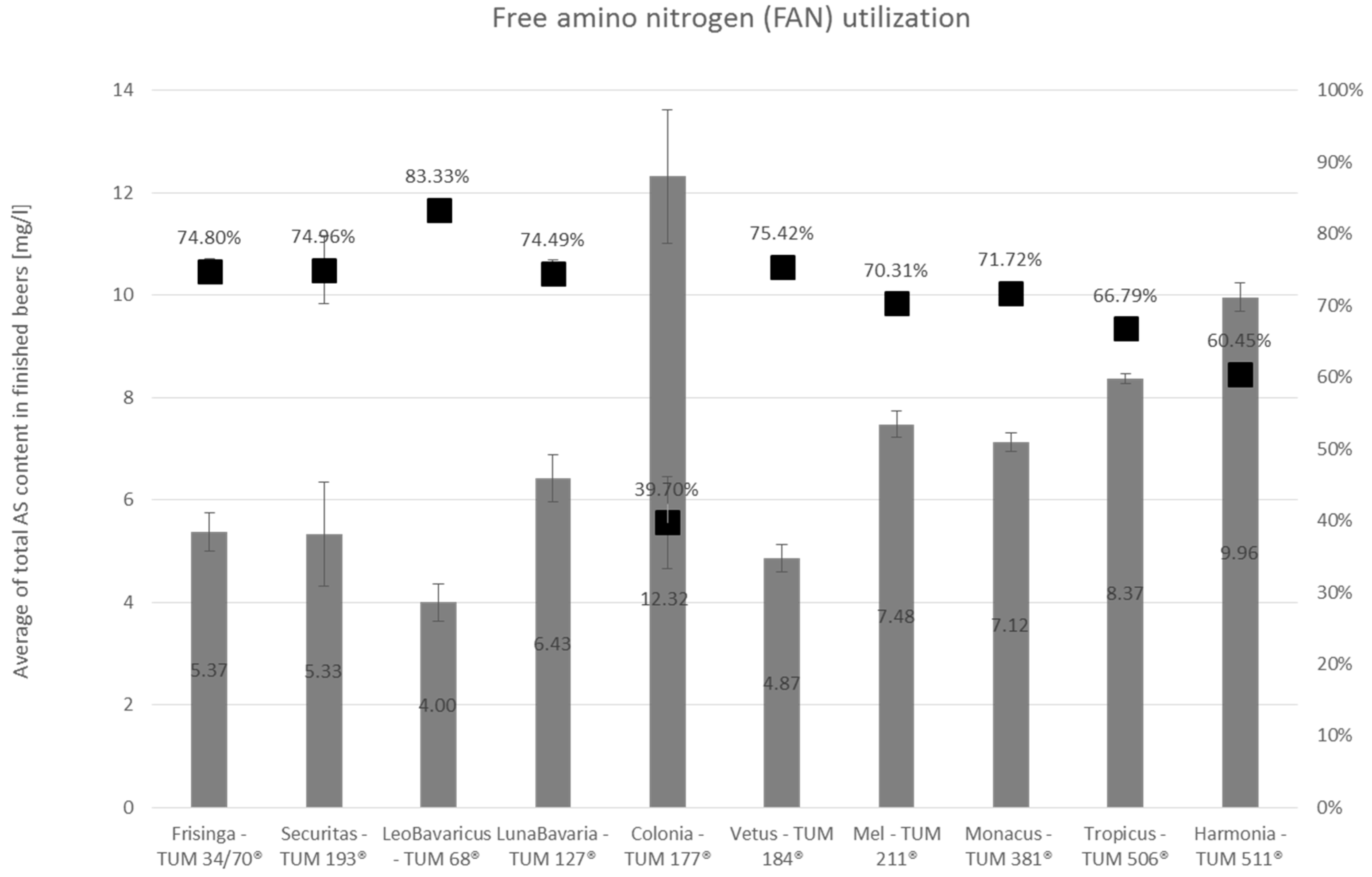
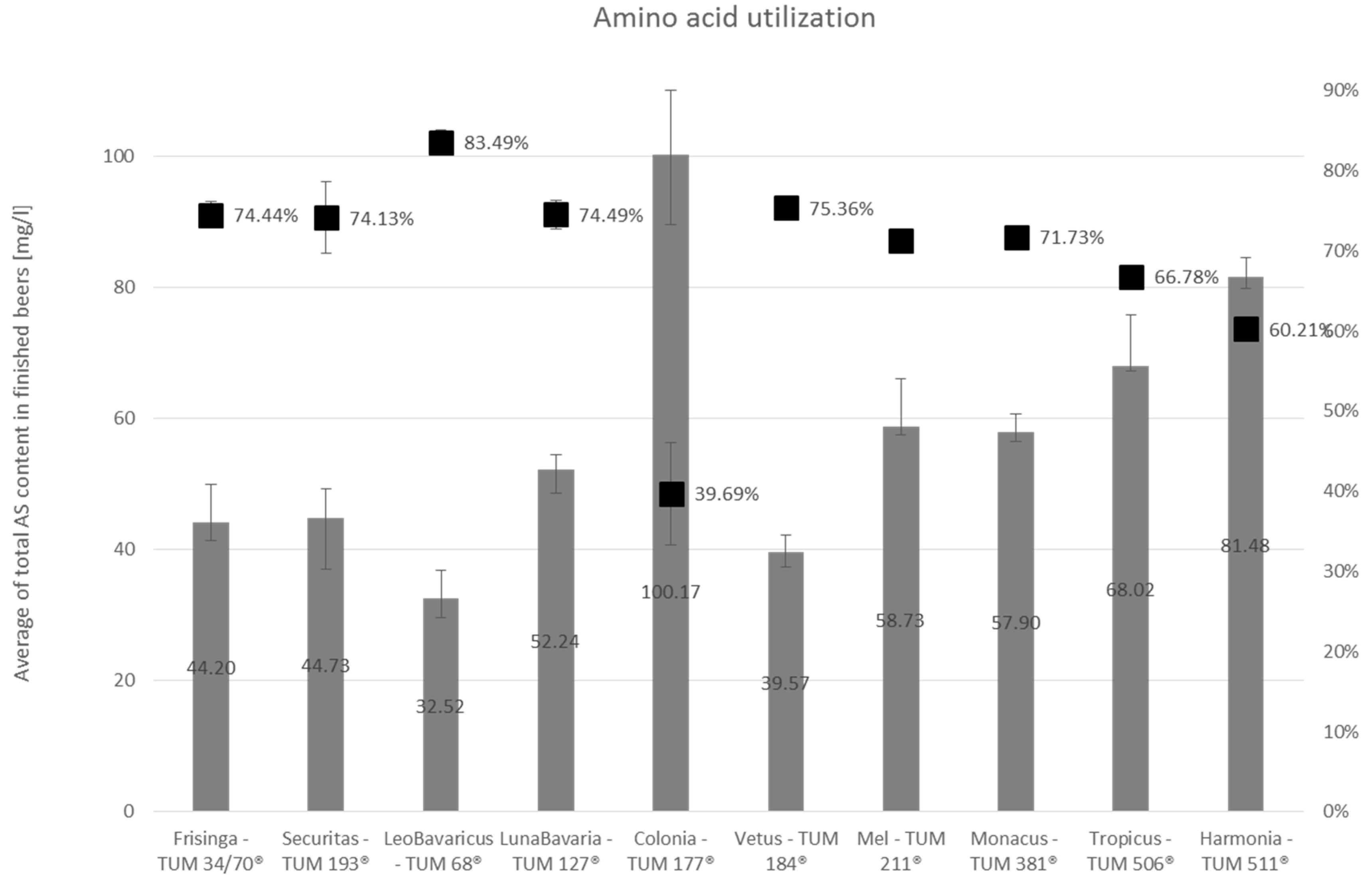
3.2.2. Amino Acid Utilization
3.2.3. Fermentation Kinetics
3.2.4. Flocculation (Cell Count)
3.2.5. Change in pH Value
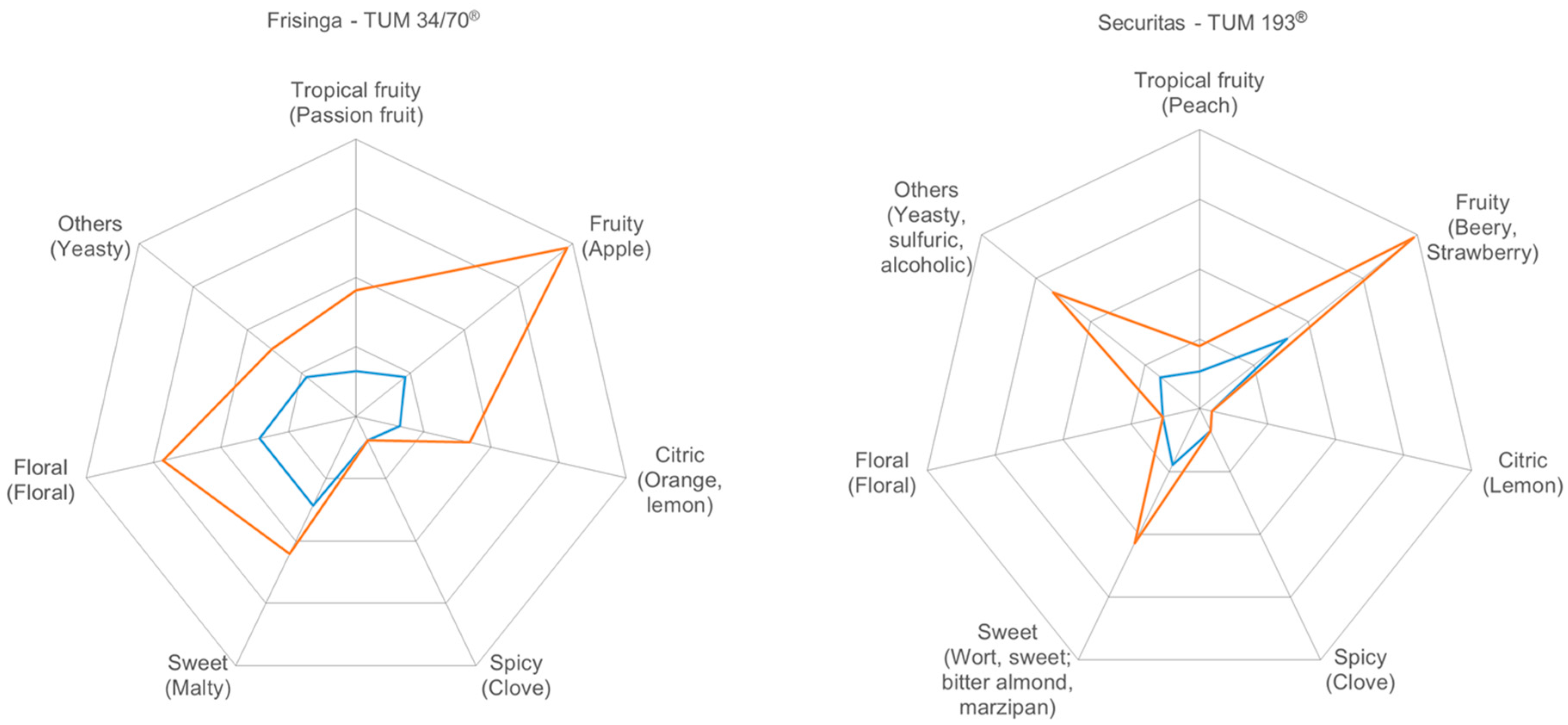
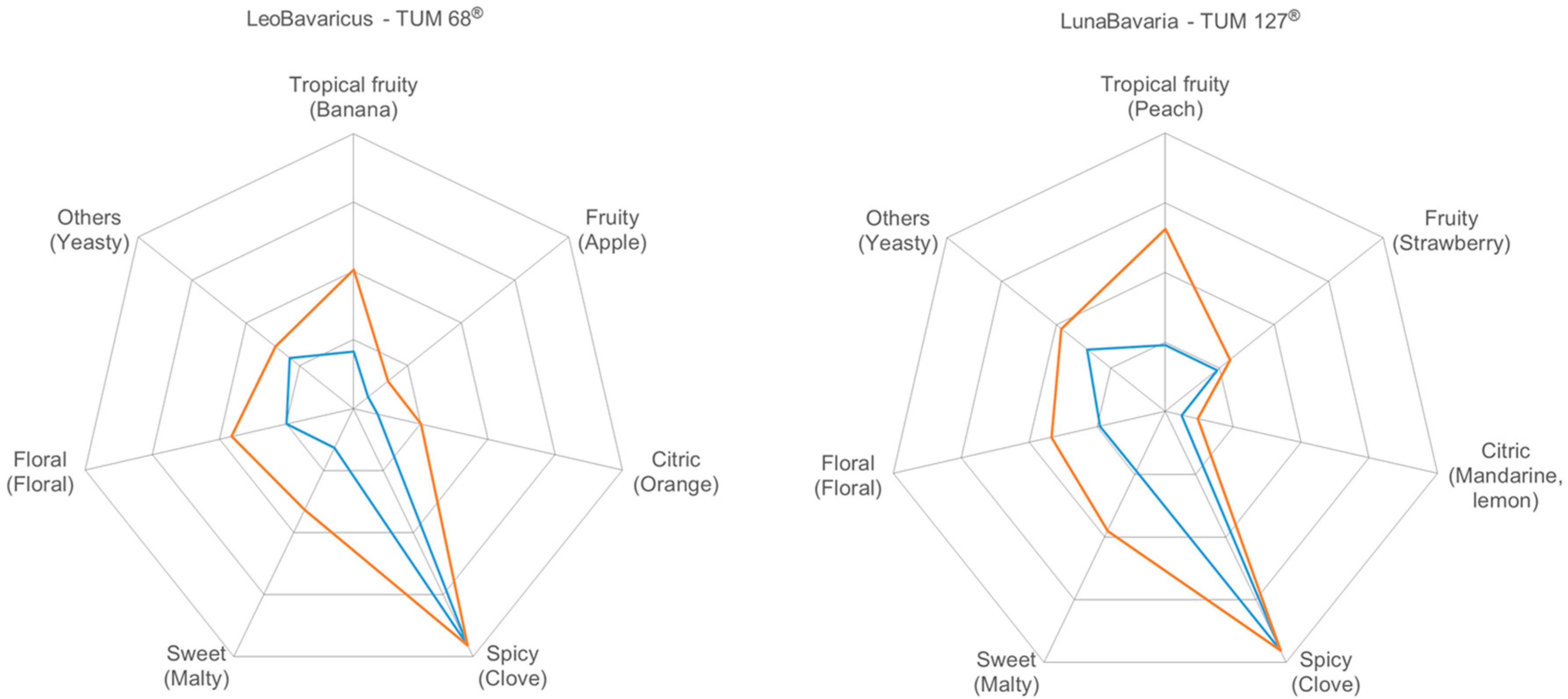
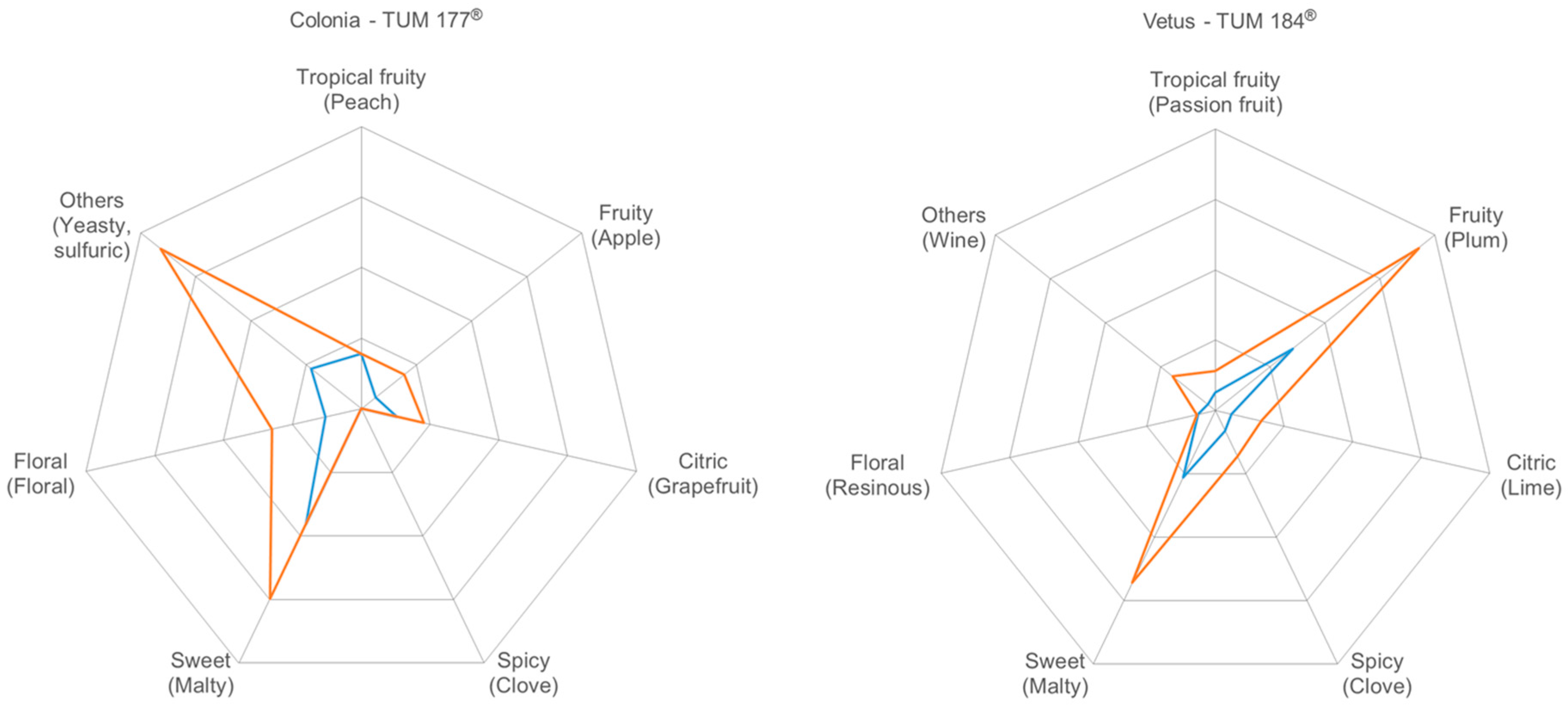
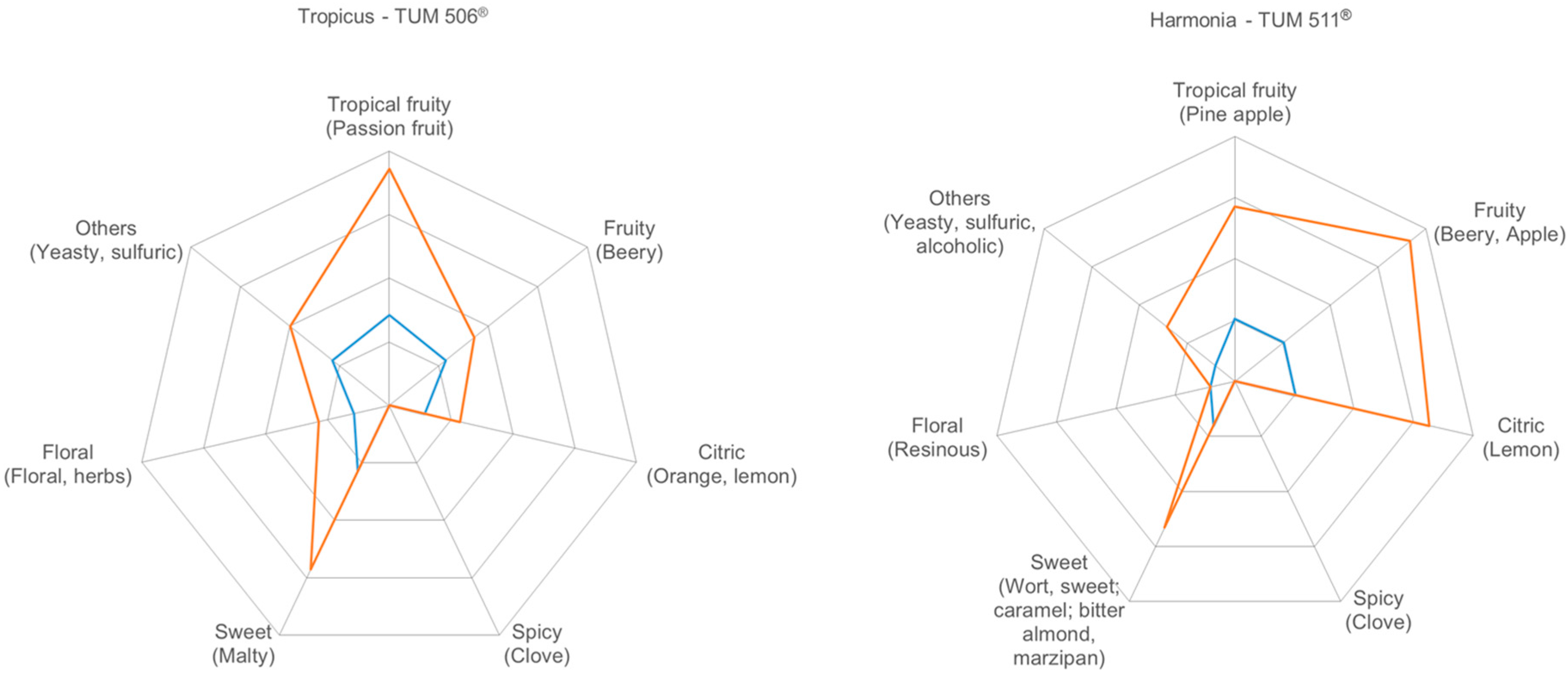
3.3. Flavor Characterization
3.3.1. Phenolic Off-Flavor
3.3.2 Fermentation by-Products
3.3.3. Sulfur Dioxide
3.3.4. Sensory Evaluation
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cotterchio, D.; Zarnkow, M.; Jacob, F. Europäische Interpretationen zur Reinheit des Bieres. Brauwelt 2016, 2016, 50–53. [Google Scholar]
- Meier-Dörnberg, T. Genetic and phenotypic characterization of different top-fermenting Saccharomyces cerevisiae ale yeast isolates. In Proceedings of the 36th European Brewery Convention, Ljubljana, Slovenia, 14–18 May 2017. [Google Scholar]
- Sachon, V.W. The science of brewing in the heart of slovenia: 36th EBC Convention held in Ljubljana. Brew. Bever. Ind. Int. 2017, 102, 28–31. [Google Scholar]
- Hutzler, M.; Meier-Dörnberg, T.; Stretz, D.; Englmann, J.; Zarnkow, M.; Jacob, F. TUM-Hefen—die Geburtsstunde von LeoBavaricus-TUM 68®. Brauwelt 2017, 157, 266–269. [Google Scholar]
- White, C.; Zainasheff, J. Yeast: The Practical Guide to Beer Fermentation; Brewers Publications: Boulder, CO, USA, 2010. [Google Scholar]
- Gábor, P.; Rosa, C. Biodiversity and Ecophysiology of Yeasts; Springer-Verlag: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Rainieri, S.; Zambonelli, C.; Kaneko, Y. Saccharomyces sensu stricto: Systematics, genetic diversity and evolution. J. Biosci. Bioeng. 2003, 96, 1–9. [Google Scholar] [CrossRef]
- Vaughan-Martini, A.; Martini, A. Saccharomyces Meyen ex Reess. In The yeasts, a taxonomic study, 5th ed.; Kurtzman, C.P., Fell, J.W., Eds.; Elsevier: Amsterdam, Netherlands, 2011; pp. 733–746. [Google Scholar]
- Naseeb, S.; James, S.A.; Alsammar, H.; Michaels, C.J.; Gini, B.; Nueno-Palop, C.; Bond, C.J.; McGhie, H.; Roberts, I.N.; Delneri, D. Saccharomyces jurei sp. nov., isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. Syst. Evol. Microbiol. 2017, 67, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Rainieri, S.; Kodama, Y.; Kaneko, Y.; Mikata, K.; Nakao, Y.; Ashikari, T. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 2006, 72, 3968–3974. [Google Scholar] [CrossRef] [PubMed]
- Bamforth, C.W. Food, fermentation, and micro-organisms; Blackwell Science: Oxford, Ames, IA, USA, 2005. [Google Scholar]
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.A.; Bai, F.Y. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 2008, 58, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Dunn, B.; Sherlock, G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008, 18, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Través, L.; Lopes, C.A.; Querol, A.; Barrio, E. On the complexity of the Saccharomyces bayanus taxon: Hybridization and potential hybrid speciation. PLoS ONE 2014, 9, e93729. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Agarbati, A.; Comitini, F.; Ciani, M. Torulaspora delbrueckii in the brewing process: A new approach to enhance bioflavour and to reduce ethanol content. Food Microbiol. 2016, 56, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Comitini, F.; Ciani, M. Torulaspora delbrueckii contribution in mixed brewing fermentations with different Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 2017, 259, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.; Meier-Dörnberg, T.; Jacob, F.; Schneiderbanger, H.; Haslbeck, K.; Zarnkow, M.; Hutzler, M. Optimization of Beer Fermentation with a Novel Brewing Strain Torulaspora delbrueckii using Response Surface Methodology. Tech. Q. 2017, 54, 23–33. [Google Scholar] [CrossRef]
- Spitaels, F.; Wieme, A.D.; Janssens, M.; Aerts, M.; Daniel, H.-M.; van Landschoot, A.; Vuyst, L.; de Vandamme, P. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS ONE 2014, 9, e95384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Bamforth, C.W.; Mills, D.A. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PLoS ONE 2012, 7, e35507. [Google Scholar] [CrossRef] [PubMed]
- Nakao, Y.; Kanamori, T.; Itoh, T.; Kodama, Y.; Rainieri, S.; Nakamura, N.; Shimonaga, T.; Hattori, M.; Ashikari, T. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 2009, 16, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Müller-Auffermann, K.; Caldera, A.; Jacob, F.; Hutzler, M. Characterization of Different Bottom Fermenting Saccharomyces pastorianus Brewing Yeast Strains. BrewingScience 2015, 68, 46–57. [Google Scholar]
- Meier-Dörnberg, T.; Michel, M.; Wagner, R.S.; Jacob, F.; Hutzler, M. Genetic and Phenotypic Characterization of Different Top-fermenting Saccharomyces cerevisiae Ale Yeast Isolates. BrewingScience 2017, 70, 9–25. [Google Scholar] [CrossRef]
- Zocher, H. Bierspezialitäten beleben den Biermarkt. Brauindustrie 2017, 102, 9. [Google Scholar]
- Gibson, B.; Geertman, J.-M.A.; Hittinger, C.T.; Krogerus, K.; Libkind, D.; Louis, E.J.; Magalhães, F.; Sampaio, J.P. New yeasts—new brews: Modern approaches to brewing yeast design and development. FEMS yeast res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Schneiderbanger, H.; Koob, J.; Poltinger, S.; Jacob, F.; Hutzler, M. Gene expression in wheat beer yeast strains and the synthesis of acetate esters. J. Inst. Brew. 2016, 122, 403–411. [Google Scholar] [CrossRef]
- Schneiderbanger, H.; Hutzler, M.; Jacob, F. Aromaprofile ausgewählter Weizenbier-Hefestämme. Brauwelt 2013, 153, 267–270. [Google Scholar]
- Hutzler, M.; Geiger, E.; Jacob, F. Use of PCRDHPLC (Polymerase Chain Reaction Denaturing High Performance Liquid Chromatography) for the Rapid Differentiation of Industrial Saccharomyces pastorianus and Saccharomyces cerevisiae Strains. J. Inst. Brew. 2010, 116, 464–474. [Google Scholar] [CrossRef]
- Hutzler, M. Entwicklung und Optimierung von Methoden zur Identifizierung und Differenzierung von getränkerelevanten Hefen. Ph.D. Thesis, Technische Universität München, Weihenstephan, July 2009. Available online: https://mediatum.ub.tum.de/doc/796815/796815.pdf (accessed on 17 August 2017).
- Hutzler, M. Getränkerelevante Hefen—Identifizierung und Differenzierung: Wie Können Hefen Praxisrelevant Unterschieden Werden, und Wie Können Identifizierungsergebnisse Technologisch bewertet Werden? Südwestdeutscher Verlag für Hochschulschriften: Saarbrücken, Germany, 2009. (In German) [Google Scholar]
- Bamforth, C.W.; Bokulich, N.A. Brewing Microbiology; Caister Academic Press: Poole, UK, 2017; Available online: https://books.google.com.hk/books/about/Brewing_Microbiology.html?id=LS1FMQAACAAJ&redir_esc=y (accessed on 17 August 2017).
- Brandl, A. Entwicklung und Optimierung von PCR-Methoden zur Detektion und Identifizierung von brauereirelevanten Mikroorganismen zur Routine-Anwendung in Brauereien. Ph.D. Thesis, Technische Universität München, Weihenstephan, March 2006. Available online: http://d-nb.info/105814118X/34 (accessed on 17 August 2017).
- Josepa, S.; Guillamon, J.-M.; Cano, J. PCR differentiation of Saccharomyces cerevisiae from Saccharomyces bayanus/Saccharomyces pastorianus using specific primers. FEMS Microbiol. Lett. 2000, 193, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Büchl, N.-R.; Hutzler, M.; Mietke-Hofmann, H.; Wenning, M.; Scherer, S. Differentiation of probiotic and environmental Saccharomyces cerevisiae strains in animal feed. J. Appl. Microbiol. 2010, 109, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Pierce, J.S. Absorption of amino acids from wort by yeasts. J. Inst. Brew. 1964, 2, 307–315. [Google Scholar] [CrossRef]
- Procopio, S.; Brunner, M.; Becker, T. Differential transcribed yeast genes involved in flavour formation and its associated amino acid metabolism during brewery fermentation. Eur. Food Res. Technol. 2014, 239, 421–439. [Google Scholar] [CrossRef]
- Zepf, M. Die Bedeutung des Absetzverhaltens der Hefe: Flokkulationsverhalten. Der Doemensianer 2010, 1, 51. [Google Scholar]
- Vidgren, V. Yeast Flocculation and Sedimentation in Brewing. J. Inst. Brew. 2011, 117, 475–487. [Google Scholar] [CrossRef]
- Soares, E.V. Flocculation in Saccharomyces cerevisiae: A review. J. Appl. Microbiol. 2011, 110, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Annemüller, G.; Manger, H.-J.; Lietz, P. Die Hefe in der Brauerei. Hefemanagement, Kulturhefe-Hefereinzucht, Hefepropagation im Bierherstellungsprozess, 3rd ed.; VLB: Berlin, Germany, 2013; Available online: https://www.amazon.de/Hefe-Brauerei-Hefemanagement-Hefepropagation-Bierherstellungsprozess/dp/3921690501 (accessed on 17 August 2017).
- Bühligen, F.; Sträuber, H.; Ondruschka, J.; Harms, H.; Müller, S. Monitoring von Bruch- und Staubhefeanteilen in Brauprozessen: Prozessoptimierung. BRAUWELT 2010, 150, 18–21. [Google Scholar]
- Annemüller, G.; Manger, H.-J. Gärung und Reifung des Bieres. Grundlagen-Technologie-Anlagentechnik, 2nd ed.; VLB: Berlin, Germany, 2013; Available online: https://www.vlb-berlin.org/publikationen/fachbuecher/gaerung-und-reifung-des-bieres-0 (accessed on 17 August 2017).
- European Bioinformatic Institut Cambridge. Available online: http://www.ebi.ac.uk/interpro/IEntry?ac=IPR004507 (accessed on 15 November 2016).
- Richard, P.; Viljanen, K.; Penttilä, M. Overexpression of PAD1 and FDC1 results in significant cinnamic acid decarboxylase activity in Saccharomyces cerevisiae. AMB Express 2015, 5, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Mukai, N.; Masaki, K.; Fujii, T.; Lefuji, H. Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J. Biosci. Bioeng. 2014, 118, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Meilgaard, M.C. Flavour chemistry of beer. Part 2. Flavour and threshold of 239 aroma volatiles. Tech. Qual. Master Brew. Assoc. Am. 1975, 12, 151–168. [Google Scholar]
- Back, W. Ausgewählte Kapitel der Brauereitechnologie; Hans Carl: Nürnberg, 2005; Available online: https://www.amazon.de/Ausgew%C3%A4hlte-Kapitel-Brauereitechnologie-BRAUWELT-Wissen/dp/341800802X (accessed on 17 August 2017).
- Swiegers, J.H.; Pretorius, I.-S. Modulation of volatile sulfur compounds by wine yeast. Appl. Microbiol. Biotechnol. 2007, 74, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Narziss, L.; Back, W. Abriss der Bierbrauerei, 7., aktualisierte und erweiterte Aufl; Wiley-VCH: Weinheim, Germany, 2005; Available online: http://eu.wiley.com/WileyCDA/WileyTitle/productCd-3527310355,subjectCd-HO40.html (accessed on 17 August 2017).




| TUM Yeast Strains | |||
|---|---|---|---|
| TUM Yeast Strain | Yeast Species | Industrial Application | Origin |
| Frisinga-TUM 34/70® | Saccharomyces pastorianus | lager beer production | Freising-Weihenstephan, Germany |
| Securitas-TUM 193® | Saccharomyces pastorianus | lager beer production | Freising-Weihenstephan, Germany |
| LeoBavaricus-TUM 68® | Saccharomyces cerevisiae | wheat beer production | Freising-Weihenstephan, Germany |
| LunaBavara-TUM 127® | Saccharomyces cerevisiae | wheat beer production | Freising-Weihenstephan, Germany |
| Colonia-TUM 177® | Saccharomyces cerevisiae | kölsch beer production | Krefeld, Germany |
| Vetus-TUM 184® | Saccharomyces cerevisiae | alt beer production | Dusseldorf, Germany |
| Mel-TUM 211® | Saccharomyces cerevisiae | ale and stout beer production | region unknown, Great Britain |
| Monacus-TUM 381® | Saccharomyces cerevisiae | trappist beer production | region unknown, Germany |
| Tropicus-TUM 506® | Saccharomyces cerevisiae | ale beer production | region unknown, Great Britain |
| Harmonia-TUM 511® | Saccharomyces cerevisiae | ale beer production | region unknown, United States of America |
| Real-Time PCR Systems, Primer and Probe Sequences (5´–3´) | System Name | Reference | S. cer. | S. cer. var. dia. | S. past. |
|---|---|---|---|---|---|
| Sbp-f CTTGCTATTCCAAACAGTGAGACT | Sbp | [32,33] | − | − | + |
| Sbp-r1 TTGTTACCTCTGGGCGTCGA | |||||
| Sbp-r2 GTTTGTTACCTCTGGGCTCG | |||||
| Sbp ACTTTTGCAACTTTTTCTTTGGGTTTCGAGCA | |||||
| Sc-f CAAACGGTGAGAGATTTCTGTGC | Sce | [32,33] | + | + | + |
| Sc-r GATAAAATTGTTTGTGTTTGTTACCTCTG | |||||
| Scer FAM-ACACTGTGGAATTTTCATATCTTTGCAACTT-BHQ1 | |||||
| Sc-GRC-f CACATCACTACGAGATGCATATGCA | Sc-GRC3 | [30] | + | + | + |
| Sc-GRC-r GCCAGTATTTTGAATGTTCTCAGTTG | |||||
| Sc-GRC FAM-TCCAGCCCATAGTCTGAACCACACCTTATCT-BHQ1 | |||||
| TF-f TTCGTTGTAACAGCTGCTGATGT | TF-COXII | [30] | + | + | − |
| TF-r ACCAGGAGTAGCATCAACTTTAATACC | |||||
| TF-MGB FAM-ATGATTTTGCTATCCCAAGTT-BHQ1 (MGB probe) | |||||
| BF300E CTCCTTGGCTTGTCGAA | BF-300 | [32] | − | − | + |
| BF300M GGTTGTTGCTGAAGTTGAGA | |||||
| BF300 FAM-TGCTCCACATTTGATCAGCGCCA-BHQ1 | |||||
| BF-LRE-f ACTCGACATTCAACTACAAGAGTAAAATTT | BF-LRE1 | [30] | − | − | + |
| BF-LRE-r TCTCCGGCATATCCTTCATCA | |||||
| BF-LRE FAM-ATCTCTACCGTTTTCGGTCACCGGC-BHQ1 | |||||
| Sd-f TTCCAACTGCACTAGTTCCTAGAGG | Sdia | [32] | − | + | − |
| Sd-r GAGCTGAATGGAGTTGAAGATGG | |||||
| Sdia FAM-CCTCCTCTAGCAACATCACTTCCTCCG-BHQ1 |
| Real-Time PCR Internal Amplification Control (IAC135) | ||
|---|---|---|
| System Name | Primer | Primer Sequence (5′–3′) |
| IAC135 | IAC135-f | TGGATAGATTCGATGACCCTAGAAC |
| IAC135-r | TGAGTCCATTTTCGCAGATAACTT | |
| Probe | Probe Sequence (5′–3′) | |
| IAC135-S | HEX-TGGGAGGATGCATTAGGAGCATTGTAAGAGAG-BHQ1 | |
| Target DNA | DNA Sequence (5′–3′) | |
| IAC135 | TGCTAGAGAATGGATAGATTCGATGACCCTAGAACTAGTGGGAGGATGCATTAGGAGCATTGTAAGAGAGTCGGAAGTTATCTGCGAAAATGGACTCATTCGAGTGGCCTATTGACGGTCGCCCAAGGTGTCGCA | |
| IAC135-rev | TGCGACACCTTGGGCGACCGTCAATAGGCCACTCGAATGAGTCCATTTTCGCAGATAACTTCCGACTCTCTTACAATGCTCCTAATGCATCCTCCCACTAGTTCTAGGGTCATCGAATCTATCCATTCTCTAGCA | |
| Wort composition | |
|---|---|
| Parameter | Amount |
| Original gravity (°P) | 12.40 |
| pH | 5.19 |
| Spec. weight SL 20/20 °C | 1.05 |
| Zinc (mg/L) | 0.15 |
| Free amino nitrogen (FAN) (mg/100 mL) | 25.00 |
| Total amino acid content (AS) (mg/100 mL) | 203.22 |
| Total sugar (g/L) | 83.78 |
| EBC-Bittering units | 20.20 |
| Glucose (g/L) | 11.46 |
| Fructose (g/L) | 2.57 |
| Saccharose (g/L) | 1.12 |
| Maltose (g/L) | 53.65 |
| Maltotriose (g/L) | 14.98 |
| RT-PCR-System | ||||||||
|---|---|---|---|---|---|---|---|---|
| Species Confirmation | TUM Yeast Strain | Sc-GRC3 | Sce | TF-COXII | Sbp | BF-LRE1 | BF-300 | Sdia |
| S. pastorianus (S. cerevisiae hybrid strain) | Frisinga-TUM 34/70® | + | + | − | + | + | + | − |
| Securitas-TUM 193® | + | + | − | + | + | + | − | |
| S. cerevisiae | LeoBavaricus-TUM 68® | + | + | + | − | − | − | − |
| LunaBavaricus-TUM 127® | + | + | + | − | − | − | − | |
| Colonia-TUM 177® | + | + | + | − | − | − | − | |
| Vetus-TUM 184® | + | + | + | − | − | − | − | |
| Mel-TUM 211® | + | + | + | − | − | − | − | |
| Monacus-TUM 381® | + | + | + | − | − | − | − | |
| Tropicus-TUM 506® | + | + | + | − | − | − | − | |
| Harmonia-TUM 511® | + | + | + | − | − | − | − | |
| TUM Yeast Strain | Glucose | Fructose | Sucrose | Maltose | Maltotriose |
|---|---|---|---|---|---|
| Frisinga-TUM 34/70® | 98.61 ± 0.00 | 96.15 ± 0.00 | 100.00 ± 0.00 | 99.02 ± 0.12 | 94.06 ± 0.91 |
| Securitas-TUM 193® | 98.70 ± 0.14 | 96.15 ± 0.00 | 100.00 ± 0.00 | 96.89 ± 0.12 | 86.60 ± 0.52 |
| LeoBavaricus-TUM 68® | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.92 ± 0.02 | 99.65 ± 0.28 |
| LunaBavaria-TUM 127® | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.07 ± 0.10 | 01.05 ± 2.89 |
| Colonia-TUM 177® | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.85 ± 0.02 | 94.80 ± 0.78 |
| Vetus-TUM 184® | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 98.00 ± 0.53 | 60.92 ± 5.87 |
| Mel-TUM 211® | 98.55 ± 0.47 | 98.57 ± 0.21 | 98.51 ± 0.48 | 87.23 ± 0.82 | 26.66 ± 0.26 |
| Monacus-TUM 381® | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.65 ± 0.14 | 97.75 ± 0.09 |
| Tropicus-TUM 506® | 98.23 ± 0.12 | 98.05 ± 0.36 | 99.11 ± 0.00 | 98.48 ± 0.93 | 59.28 ± 0.81 |
| Harmonia-TUM 511® | 99.42 ± 0.12 | 98.05 ± 0.00 | 95.54 ± 0.00 | 99.28 ± 0.09 | 83.91 ± 0.71 |
| Apparent attenuation (AA %) of the final beer | ||
|---|---|---|
| TUM Yeast Strain | AA (%) | Fermentation time (hours) |
| Frisinga-TUM 34/70® | 81.63 ± 0.51 | 216 |
| Securitas-TUM 193® | 79.30 ± 0.51 | 216 |
| LeoBavaricus-TUM 68® | 86.17 ± 0.05 | 96 |
| LunaBavaria-TUM 127® | 76.20 ± 1.76 | 144 |
| Colonia-TUM 177® | 84.93 ± 0.37 | 96 |
| Vetus-TUM 184® | 80.97 ± 3.02 | 264 |
| Mel-TUM 211® | 66.13 ± 0.51 | 240 |
| Monacus-TUM 381® | 86.17 ± 0.11 | 168 |
| Tropicus-TUM 506® | 77.37 ± 1.34 | 216 |
| Harmonia-TUM 511® | 82.70 ± 0.42 | 144 |
| Yeast Cell Sedimentation at the end of Primary Fermentation | ||||
|---|---|---|---|---|
| TUM Yeast Strain | Max. Cell Conc. | Cell Conc. FG | Difference | Flocculation Behavior |
| Frisinga-TUM 34/70® | 35.90 | 1.80 | −34.10 | flocculent |
| Securitas-TUM 193® | 48.80 | 1.30 | −47.50 | flocculent |
| LeoBavaricus-TUM 68® | 62.81 | 32.29 | −30.52 | powdery |
| LunaBavara-TUM 127® | 39.38 | 7.50 | −31.88 | flocculent |
| Colonia-TUM 177® | 23.00 | 17.20 | −5.8 | powdery |
| Vetus-TUM 184® | 17.83 | 0.50 | −17.33 | flocculent |
| Mel-TUM 211® | 34.30 | 17.80 | −16.50 | powdery |
| Monacus-TUM 381® | 57.65 | 18.12 | −39.53 | powdery |
| Tropicus-TUM 506® | 36.45 | 19.36 | −17.09 | powdery |
| Harmonia-TUM 511® | 61.30 | 43.07 | −18.23 | powdery |
| pH Value Decrease during Primary Fermentation | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TUM Yeast Strain | 0 h | 24 h | 48 h | 72 h | 96 h | After primary fermentation | After maturation | Final beer (after lagering) | ∆pH |
| Frisinga-TUM 34/70® | 5.2 | 4.6 | 4.5 | 4.5 | 4.5 | 4.5 | 4.5 | 4.4 ± 0.04 | 0.8 |
| Securitas-TUM 193® | 5.2 | 4.6 | 4.5 | 4.5 | 4.5 | 4.5 | 4.5 | 4.5 ± 0.01 | 0.7 |
| LeoBavaricus-TUM 68® | 5.2 | 4.8 | 4.3 | 4.3 | 4.2 | 4.2 | 4.2 | 4.4 ± 0.01 | 0.8 |
| LunaBavaria-TUM 127® | 5.2 | 4.6 | 4.5 | 4.5 | 4.4 | 4.4 | 4.4 | 4.6 ± 0.01 | 0.6 |
| Colonia-TUM 177® | 5.2 | 5 | 4.8 | 4.8 | 4.7 | 4.7 | 4.7 | 4.6 ± 0.02 | 0.6 |
| Vetus-TUM 184® | 5.2 | 4.7 | 4.6 | 4.6 | 4.5 | 4.5 | 4.6 | 4.5 ± 0.04 | 0.7 |
| Mel-TUM 211® | 5.2 | 4.5 | 4.4 | 4.4 | 4.4 | 4.4 | 4.5 | 4.7 ± 0.01 | 0.5 |
| Monacus-TUM 381® | 5.2 | 4.6 | 4.5 | 4.5 | 4.5 | 4.5 | 4.5 | 4.5 ± 0.01 | 0.7 |
| Tropicus-TUM 506® | 5.2 | 4.6 | 4.5 | 4.5 | 4.5 | 4.5 | 4.6 | 4.7 ± 0.01 | 0.5 |
| Harmonia-TUM 511® | 5.2 | 4.6 | 4.4 | 4.4 | 4.4 | 4.4 | 4.4 | 4.4 ± 0.01 | 0.8 |
| POF-Test/Sniffing Perception of: | |||
|---|---|---|---|
| TUM Yeast Strain | 4-Vinylguajacol/ Ferulic Acid | 4-Vinylphenol/ Coumaric Acid | 4-Vinylstyrene/ Cinnamic Acid |
| Frisinga - TUM 3470® | − | − | − |
| Securitas-TUM 193® | − | − | − |
| LeoBavaricus-TUM 68® | + | + | + |
| LunaBavaria-TUM 127® | + | + | + |
| Colonia-TUM 177® | − | − | − |
| Vetus-TUM 184® | − | − | − |
| Mel-TUM 211® | − | − | − |
| Monacus-TUM 381® | + | + | + |
| Tropicus-TUM 506® | − | − | − |
| Harmonia-TUM 511® | + | + | + |
| Fermentation by-Products (mg/L) | |||||
|---|---|---|---|---|---|
| FBP | Frisinga-TUM 3470® | Securitas-TUM 193® | LeoBavaricus-TUM 68® | LunaBavaria-TUM 127® | Colonia-TUM 177® |
| Isoamyl acetate | 0.63 ± 0.19 | 1.38 ± 0.05 | 4.07 ± 0.46 | 3.97 ± 2.30 | 2.40 ± 0.16 |
| Ethyl acetate | 19.77 ± 2.50 | 26.07 ± 1.89 | 32.50 ± 2.97 | 36.97 ± 2.93 | 32.87 ± 1.68 |
| ∑ Ester (E) | 20.40 ± 2.69 | 27.90 ± 1.94 | 36.57 ± .3.43 | 40.93 ± 3.16 | 35.27 ± 1.83 |
| n-Propanol | 11.23 ± 0.77 | 13.43 ± 0.72 | 22.77 ± 2.37 | 15.93 ± 0.70 | 21.30 ± 1.25 |
| i-Butanol | 10.63 ± 0.71 | 14.27 ± 0.47 | 62.30 ± 3.51 | 43.70 ± 3.42 | 10.53 ± 0.11 |
| Amyl alcohols | 60.53 ± 3.31 | 82.60 ± 2.68 | 127.00 ± 7.33 | 91.60 ± 3.34 | 80.77 ± 1.06 |
| ∑ Higher alcohols (HE) | 82.40 ± 4.76 | 110.30 ± 3.86 | 212.07 ± 13.15 | 151.23 ± 6.25 | 112.60 ± 2.32 |
| 4-Vinylguajacol | 0.10 ± 0.00 | 0.10 ± 0.00 | 3.10 ± 0.40 | 1.23 ± 0.30 | 0.10 ± 0.00 |
| Diacetyl | 0.18 ± 0.07 | 0.10 ± 0 01 | 0.11 ± 0.01 | 0.17 ± 0.03 | 0.14 ± 0 03 |
| 2,3-Pentandione | 0.00 ± 0.01 | 0.03 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.01 |
| ∑ Vicinal diketones | 0.18 ± 0.07 | 0.12 ± 0.02 | 0.12 ± 0.01 | 0.19 ± 0.03 | 0.15 ± 0.04 |
| Acetaldehyde | 1.33 ± 0.47 | 11.17 ± 1.46 | 7.27 ± 1.80 | 2.80 ± 0.94 | 8.23 ± 2.02 |
| Ratio (∑E:∑HE) | 4.04 | 3.95 | 5.80 | 3.69 | 3.19 |
| Fermentation by-Products (mg/L) | |||||
|---|---|---|---|---|---|
| FBP | Vetus- TUM 184® | Mel- TUM 211® | Monacus- TUM 381® | Tropicus- TUM 506® | Harmonia- TUM 511® |
| Isoamyl acetate | 1.80 ± 0.09 | 2.03 ± 0.05 | 4.33 ± 0.05 | 1.53 ± 0.11 | 2.93 ± 0.05 |
| Ethyl acetate | 37.40 ± 0.58 | 37.93 ± 0.11 | 50.60 ± 0.85 | 22.57 ± 1.31 | 54.40 ± 0.61 |
| ∑ Ester (E) | 39.20 ± 0.61 | 39.97 ± 0.14 | 54.93 ± 0.84 | 24.10 ± 1.36 | 57.33 ± 0.65 |
| n-Propanol | 15.40 ± 0.46 | 18.30 ± 0.16 | 18.30 ± 0.16 | 20.67 ± 1.01 | 20.77 ± 0.53 |
| i-Butanol | 11.37 ± 0.14 | 16.20 ± 0.37 | 24.17 ± 0.66 | 20.90 ± 1.40 | 13.13 ± 0.28 |
| Amyl alcohols | 67.37 ± 0.91 | 59.27 ± 1.32 | 101.00 ± 4.23 | 88.50 ± 5.90 | 74.97 ± 1.24 |
| ∑ Higher alcohols (HE) | 94.13 ± 1.43 | 93.77 ± 1.83 | 142.23 ± 5.26 | 130.07 ± 8.12 | 108.87 ± 1.23 |
| 4-Vinylguajacol | 0.10 ± 0.00 | 0.10 ± 0.00 | 3.27 ± 0.05 | 0.10 ± 0.00 | 3.33 ± 0.05 |
| Diacetyl | 0.06 ± 0.01 | 0.10 ± 0.01 | 0.07 ± 0.01 | 0.12 ± 0.01 | 0.06 ± 0.00 |
| 2,3-Pentandione | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.00 |
| ∑ Vicinal diketones | 0.06 ± 0.01 | 0.12 ± 0.01 | 0.08 ± 0.01 | 0.14 ± 0.01 | 0.07 ± 0.00 |
| Acetaldehyde | 6.80 ± 0.80 | 4.60 ± 0.48 | 6.23 ± 0.77 | 5.93 ± 0.93 | 4.03 ± 0.46 |
| Ratio (∑E:∑HE) | 2.40 | 2.35 | 2.59 | 5.40 | 1.90 |
| SO2 Concentration of the Finished Beers | |
|---|---|
| TUM Yeast Strain | SO2 (mg/L) |
| Frisinga-TUM 34/70® | 6.03 ± 0.30 |
| Securitas-TUM 193® | 9.47 ± 0.68 |
| LeoBavaricus-TUM 68® | 2.87 ± 0.30 |
| LunaBavaria-TUM 127® | 1.63 ± 0.51 |
| Colonia-TUM 177® | 3.80 ± 0.79 |
| Vetus-TUM 184® | 3.10 ± 0.16 |
| Mel-TUM 211® | 2.60 ± 0.98 |
| Monacus-TUM 381® | 0.50 ± 0.00 |
| Tropicus-TUM 506® | 2.23 ± 1.02 |
| Harmonia-TUM 511® | 0.50 ± 0.00 |
| TUM Yeast Strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TUM Yeast Strain | Yeast Species | Recommended Main Beer Style | Phenolic Off-Flavor | Flocculation Behavior | Maltotriose-Utilization (%) | ∆pH | SO2(mg/L) | AA(%) | Fermentation Time (days) |
| Frisinga-TUM 34/70® | S. pastorianus | lager beer | − | flocculent | 94.06 ± 0.91 | 0.8 | 6.03 ± 0.30 | 81.63 ± 0.51 | 9 |
| Securitas-TUM 193® | S. pastorianus | lager bee | − | flocculent | 86.60 ± 0.52 | 0.7 | 9.47 ± 0.68 | 79.30 ± 0.51 | 9 |
| LeoBavaricus-TUM 68® | S. cerevisiae | wheat beer | + | powdery | 99.65 ± 0.28 | 0.8 | 2.87 ± 0.30 | 86.17 ± 0.05 | 4 |
| LunaBavaria-TUM 127® | S. cerevisiae | wheat beer | + | flocculent | 01.05 ± 2.89 | 0.6 | 1.63 ± 0.51 | 76.20 ± 1.76 | 6 |
| Colonia-TUM 177® | S. cerevisiae | kölsch and alt beer | − | powdery | 94.80 ± 0.78 | 0.6 | 3.80 ± 0.79 | 84.93 ± 0.37 | 4 |
| Vetus-TUM 184® | S. cerevisiae | alt beer | − | flocculent | 60.92 ± 5.87 | 0.7 | 3.10 ± 0.16 | 80.97 ± 3.02 | 11 |
| Mel-TUM 211® | S. cerevisiae | ale beer | − | powdery | 26.66 ± 0.26 | 0.5 | 2.60 ± 0.98 | 66.13 ± 0.51 | 10 |
| Monacus-TUM 381® | S. cerevisiae | wheat beer | + | powdery | 97.75 ± 0.09 | 0.7 | 0.50 ± 0.00 | 86.17 ± 0.11 | 7 |
| Tropicus-TUM 506® | S. cerevisiae | ale beer | − | powdery | 59.28 ± 0.81 | 0.5 | 2.23 ± 1.02 | 77.37 ± 1.34 | 9 |
| Harmonia-TUM 511® | S. cerevisiae | ale and wheat beer | + | powdery | 83.91 ± 0.71 | 0.8 | 0.50 ± 0.00 | 82.70 ± 0.42 | 6 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meier-Dörnberg, T.; Hutzler, M.; Michel, M.; Methner, F.-J.; Jacob, F. The Importance of a Comparative Characterization of Saccharomyces Cerevisiae and Saccharomyces Pastorianus Strains for Brewing. Fermentation 2017, 3, 41. https://doi.org/10.3390/fermentation3030041
Meier-Dörnberg T, Hutzler M, Michel M, Methner F-J, Jacob F. The Importance of a Comparative Characterization of Saccharomyces Cerevisiae and Saccharomyces Pastorianus Strains for Brewing. Fermentation. 2017; 3(3):41. https://doi.org/10.3390/fermentation3030041
Chicago/Turabian StyleMeier-Dörnberg, Tim, Mathias Hutzler, Maximilian Michel, Frank-Jürgen Methner, and Fritz Jacob. 2017. "The Importance of a Comparative Characterization of Saccharomyces Cerevisiae and Saccharomyces Pastorianus Strains for Brewing" Fermentation 3, no. 3: 41. https://doi.org/10.3390/fermentation3030041
APA StyleMeier-Dörnberg, T., Hutzler, M., Michel, M., Methner, F.-J., & Jacob, F. (2017). The Importance of a Comparative Characterization of Saccharomyces Cerevisiae and Saccharomyces Pastorianus Strains for Brewing. Fermentation, 3(3), 41. https://doi.org/10.3390/fermentation3030041