Green Tea Modulates Temporal Dynamics and Environmental Adaptation of Microbial Communities in Daqu Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Daqu Blocks and Sample Collection
2.2. Endogenous Factor Determination
2.3. Total DNA Extraction, PCR Amplification, and Sequencing
2.4. Sequence Data Processing
2.5. Statistical Analysis
3. Results
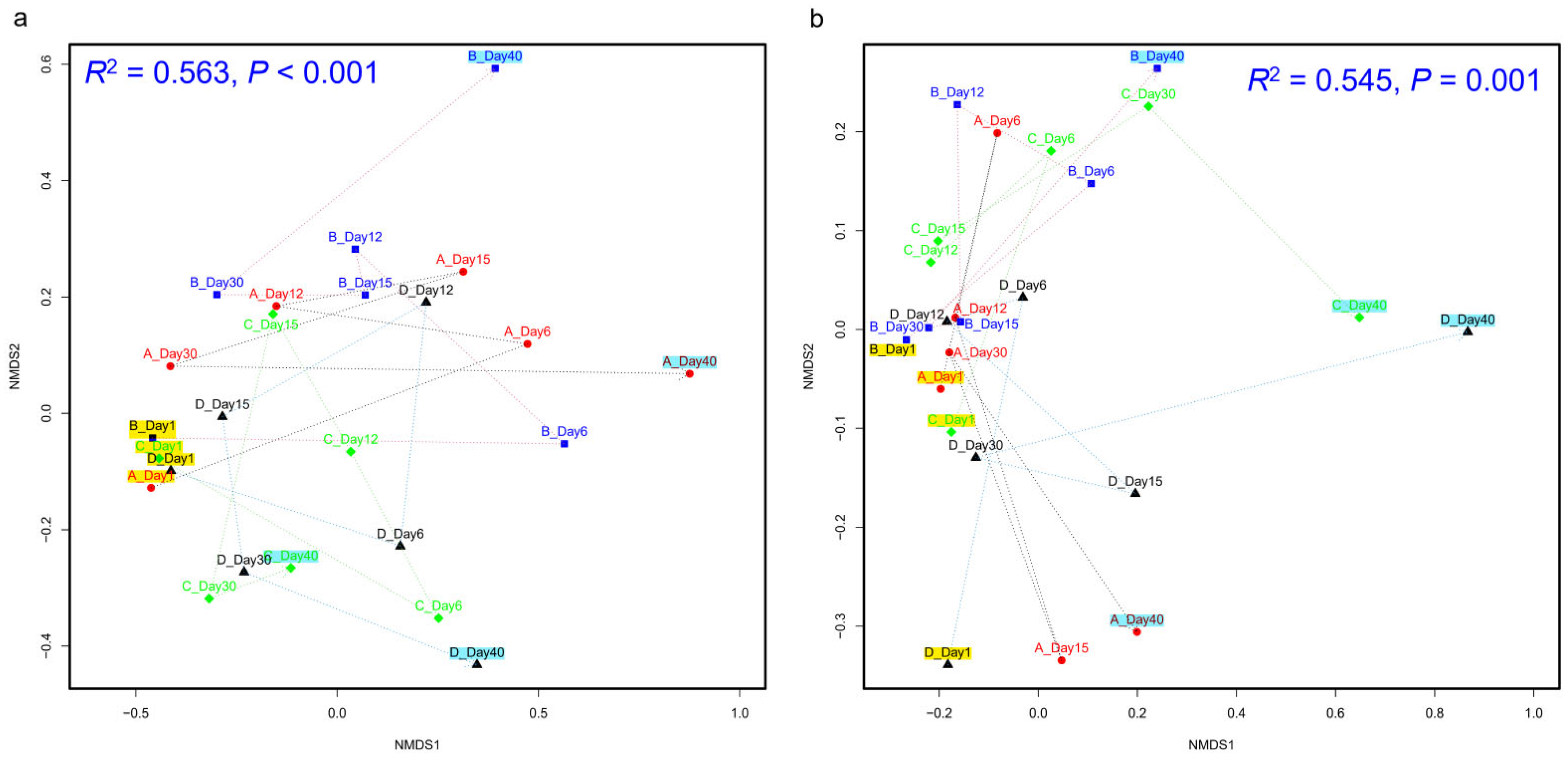
3.1. Microbial Community Dynamics During Daqu Fermentation
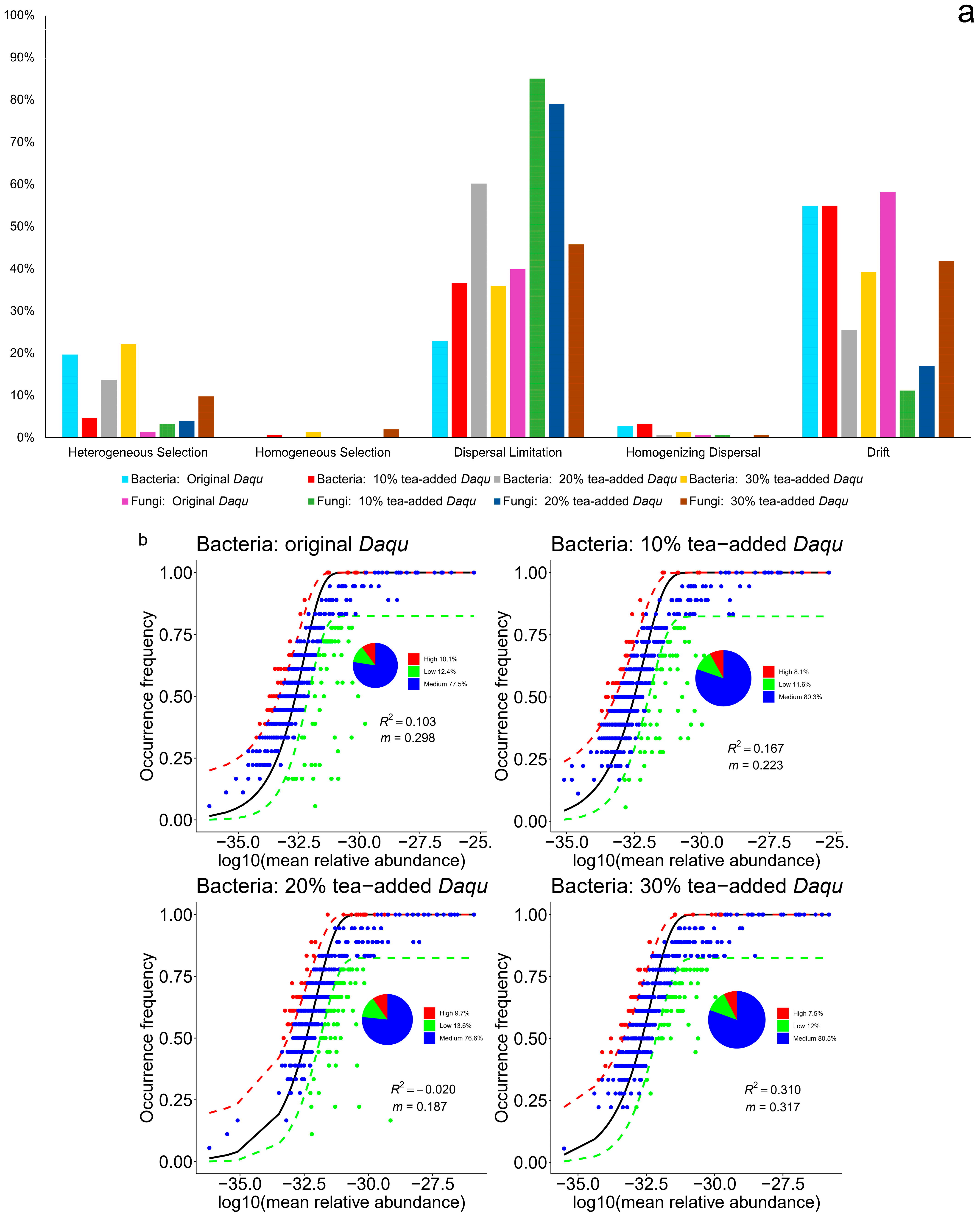
3.2. Community Assembly Process Mechanisms
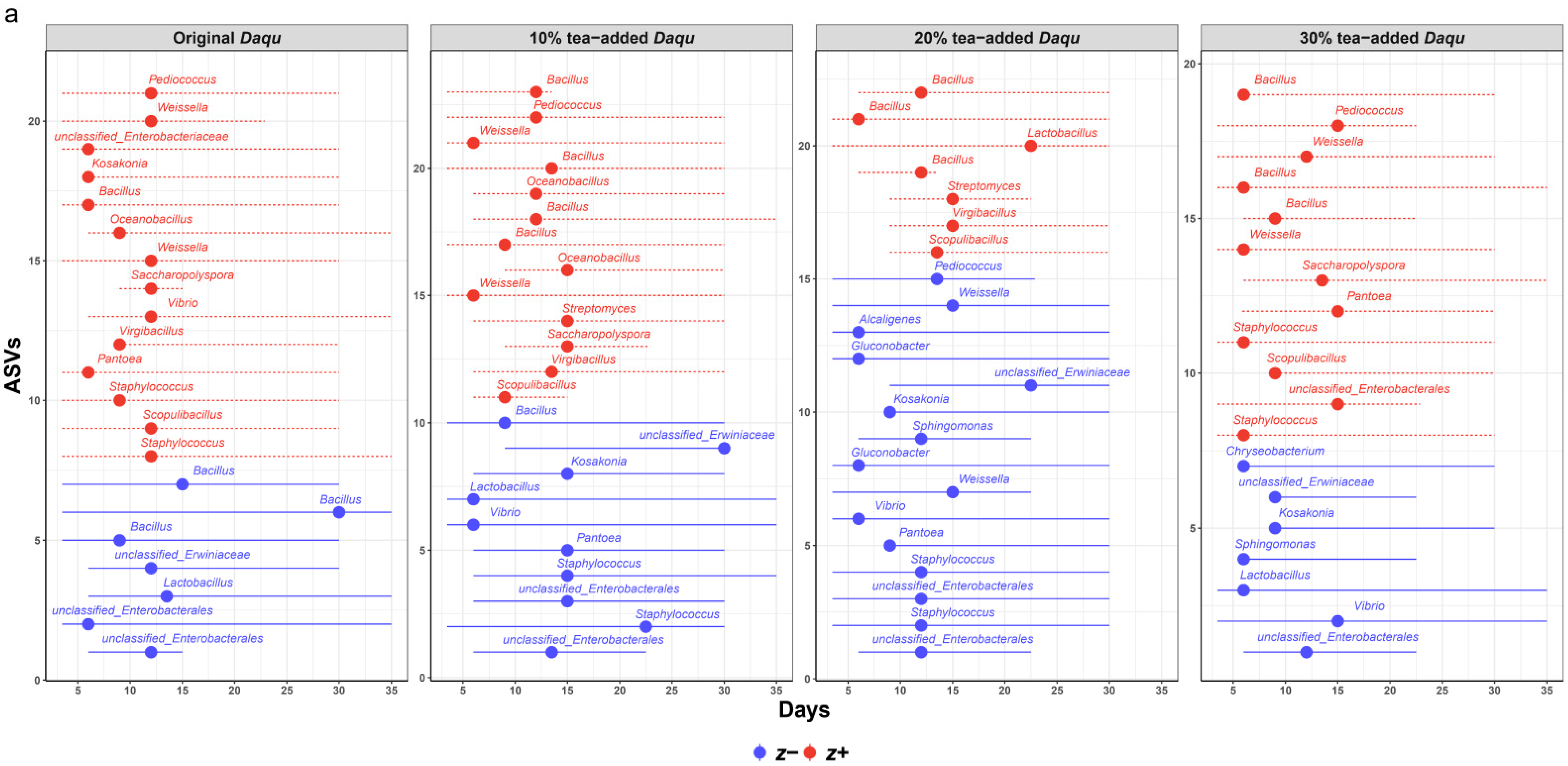
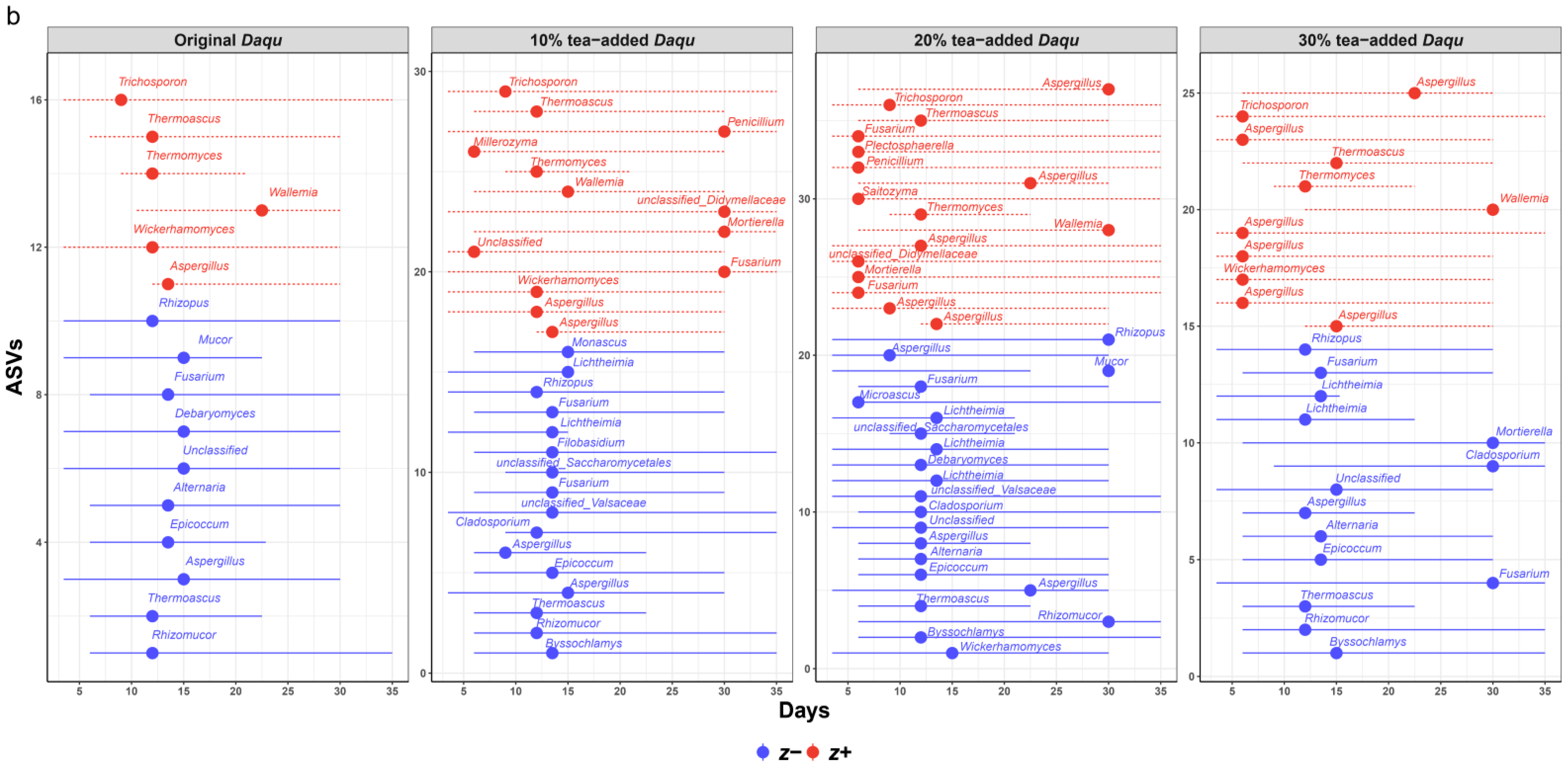
3.3. Microbial Phylogenetic Dynamics and Adaptation Conservatism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.X.; Wang, L.; Tan, Y.W.; Wang, H.Y.; Yang, F.; Chen, L.Q.; Hao, F.; Lv, X.B.; Du, H.; Xu, Y. Effect of Pichia on shaping the fermentation microbial community of sauce-flavor Baijiu. Int. J. Food Microbiol. 2021, 336, 11. [Google Scholar] [CrossRef]
- Wang, X.S.; Du, H.; Zhang, Y.; Xu, Y. Environmental Microbiota Drives Microbial Succession and Metabolic Profiles during Chinese Liquor Fermentation. Appl. Environ. Microbiol. 2018, 84, 13. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.Y.; Shang, Z.C.; Chen, J.; Shen, Y.J.; Li, Z.J.; Huang, D.; Luo, H.B. Differences in structure, volatile metabolites, and functions of microbial communities in Nongxiangxing daqu from different production areas. LWT-Food Sci. Technol. 2022, 166, 10. [Google Scholar] [CrossRef]
- Wang, X.S.; Du, H.; Xu, Y. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int. J. Food Microbiol. 2017, 244, 27–35. [Google Scholar] [CrossRef]
- Sakandar, H.A.; Hussain, R.; Khan, Q.F.; Zhang, H.P. Functional microbiota in Chinese traditional Baijiu and Mijiu Qu (starters): A review. Food Res. Int. 2020, 138, 10. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Chai, L.J.; Fang, G.Y.; Mei, J.L.; Lu, Z.M.; Zhang, X.J.; Xiao, C.; Wang, S.T.; Shen, C.H.; Shi, J.S.; et al. Spatial heterogeneity of the microbiome and metabolome profiles of high-temperature Daqu in the same workshop. Food Res. Int. 2022, 156, 13. [Google Scholar] [CrossRef]
- Wei, J.; Jie, W.; Baosheng, L.; Jianhui, D.; Xiaojun, W.; Zhihua, S.; Xinglin, H.; Pujie, H.; Chunguang, L.; Jianqin, H.; et al. Analysis of characteristic flavor compounds in single-grain chinese baijiu brewed from different raw materials. Food Sci. 2020, 41, 234–238, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Huang, H.; Chen, Y.Y.; Hong, J.X.; Chen, H.; Zhao, D.R.; Wu, J.H.; Li, J.C.; Sun, J.Y.; Sun, X.T.; Huang, M.Q.; et al. Unraveling the chemosensory characteristics on different types of spirits based on sensory contours and quantitative targeted flavoromics analysis. Food Chem. X 2024, 23, 12. [Google Scholar] [CrossRef]
- Qiao, Z.; Shenghua, L.; Xiang, L.; Xin, H.; Yang, L.; Cheng, F.; Jie, D.; Tongling, D. Bioactive ingredients and microbial diversity of fuzhuan tea produced from different raw materials. Food Sci. 2020, 41, 69–77, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Shang, A.; Li, J.H.; Zhou, D.D.; Gan, R.Y.; Li, H.B. Molecular mechanisms underlying health benefits of tea compounds. Free Radic. Biol. Med. 2021, 172, 181–200. [Google Scholar] [CrossRef]
- Ng, K.W.; Cao, Z.J.; Chen, H.B.; Zhao, Z.Z.; Zhu, L.; Yi, T. Oolong tea: A critical review of processing methods, chemical composition, health effects, and risk. Crit. Rev. Food Sci. Nutr. 2018, 58, 2957–2980. [Google Scholar] [CrossRef]
- Dai, Y.; Li, Z.; Tian, Z. Analysis of bacterial diversity in daqu and fermented grains for maotai-flavor liquor. Food Sci. 2019, 40, 152–159, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Du, H.; Wang, X.S.; Zhang, Y.H.; Xu, Y. Exploring the impacts of raw materials and environments on the microbiota in Chinese Daqu starter. Int. J. Food Microbiol. 2019, 297, 32–40. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, X.; Luo, Z.; Gan, G.; Wu, F.; Zhong, Y.; Wang, X. Optimization of the fermentation process of Moutai-flavored Daqu with the addition of green tea: Impacts on bacterial succession and endogenous factors. J. Food Sci. Technol. 2025; in press. [Google Scholar] [CrossRef]
- Tang, J.; Rao, J.; Zou, Y.; Liao, L.; Huang, D.; Luo, H. The community assembly patterns determined differences between the surface and the core microbial communities of Nongxiangxing Daqu. LWT-Food Sci. Technol. 2023, 183, 9. [Google Scholar] [CrossRef]
- Mondal, S.; Karande, M.; Srivastava, S.; Sharma, A.; Sharma, S.; Ghosh, A. Unravelling the microbiome perspective to variations in tea metabolome. Ind. Crop. Prod. 2025, 223, 14. [Google Scholar] [CrossRef]
- Bai, X. A Technical Study on the Tea-Flower Daqu Preparation. Ph.D. Thesis, Anhui Agricultural University, Hefei, China, 2016. (In Chinese with English Abstract). [Google Scholar]
- Ge, Y.H.; Wang, L.; Huang, Y.J.; Jia, L.Y.; Wang, J.H. Characteristic flavor compounds in Guizhou green tea and the environmental factors influencing their formation: Investigation using stable isotopes, electronic nose, and headspace-gas chromatography ion migration spectrometry. LWT-Food Sci. Technol. 2024, 196, 13. [Google Scholar] [CrossRef]
- Zuo, Q.; Huang, Y.; Guo, M. Evaluation of bacterial diversity during fermentation process: A comparison between handmade and machine-made high-temperature daqu of maotai-flavor liquor. Ann. Microbiol. 2020, 13, 14–23. [Google Scholar] [CrossRef]
- QB/T 4257-2011; General Methods of Analysis for Daqu. Ministry of Industry and Information Technology of the People’s Republic of China: Beijing, China, 2011; pp. 1–16. (In Chinese)
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 7. [Google Scholar] [CrossRef]
- Lei, L.; Wu, T.; Wang, C. Effect of Gastrodia elata and Fagopyrum tataricum compound solution on synthesis of extracellular polysaccharide from Grifola frondosa and its fermentation kinetics. J. Food Sci. Technol. 2020, 38, 53–59, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- White, E.P.; Adler, P.B.; Lauenroth, W.K.; Gill, R.A.; Greenberg, D.; Kaufman, D.M.; Rassweiler, A.; Rusak, J.A.; Smith, M.D.; Steinbeck, J.R.; et al. A comparison of the species-time relationship across ecosystems and taxonomic groups. Oikos 2006, 112, 185–195. [Google Scholar] [CrossRef]
- Dengler, J. Which function describes the species-area relationship best? A review and empirical evaluation. J. Biogeogr. 2009, 36, 728–744. [Google Scholar] [CrossRef]
- Liang, Z.; Zhuo, M.; Fuyong, W.; Lu, J.; Fangxiang, L.; Yanxia, Z.; Yang, Z.; Xinliang, M.; Hui, H.; Xinye, W. Roles of rare and abundant taxa in bacterial succession during pile fermentation in moutai-flavored baijiu making. Food Sci. 2022, 43, 168–175, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Baker, M.E.; King, R.S. A new method for detecting and interpreting biodiversity and ecological community thresholds. Methods Ecol. Evol. 2010, 1, 25–37. [Google Scholar] [CrossRef]
- Ning, D.L.; Yuan, M.T.; Wu, L.W.; Zhang, Y.; Guo, X.; Zhou, X.S.; Yang, Y.F.; Arkin, A.P.; Firestone, M.K.; Zhou, J.Z. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, 12. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.J.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 15. [Google Scholar] [CrossRef]
- Sloan, W.T.; Lunn, M.; Woodcock, S.; Head, I.M.; Nee, S.; Curtis, T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006, 8, 732–740. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Change Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef]
- Zhao, L.; Luo, Z.; Hu, Z.; Zhang, Y.; Zhao, T.; Zhong, Y.; Wang, X. Linking phylogenetic niche conservatism in bacterial communities in sorghum root compartments revealed by the Hongyingzi cultivar. Plant Biol. 2025, 27, 134–145. [Google Scholar] [CrossRef]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef]
- Pagel, M. Inferring the historical patterns of biological evolution. Nature 1999, 401, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Fritz, S.A.; Purvis, A. Selectivity in Mammalian Extinction Risk and Threat Types: A New Measure of Phylogenetic Signal Strength in Binary Traits. Conserv. Biol. 2010, 24, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.E.; Adamowicz, S.J. Community Phylogenetics: Assessing Tree Reconstruction Methods and the Utility of DNA Barcodes. PLoS ONE 2015, 10, 18. [Google Scholar] [CrossRef]
- Goberna, M.; Verdú, M. Predicting microbial traits with phylogenies. ISME J. 2016, 10, 959–967. [Google Scholar] [CrossRef]
- Preston, F. Time and space and the variation of species. Ecology 1960, 41, 611–627. [Google Scholar] [CrossRef]
- McNaughton, S. Diversity and stability of ecological communities: A comment on the role of empiricism in ecology. Am. Nat. 1977, 111, 515–525. [Google Scholar] [CrossRef]
- Ives, A.R.; Dennis, B.; Cottingham, K.L.; Carpenter, S.R. Estimating community stability and ecological interactions from time-series data. Ecol. Monogr. 2003, 73, 301–330. [Google Scholar] [CrossRef]
- Ives, A.R.; Carpenter, S.R. Stability and diversity of ecosystems. Science 2007, 317, 58–62. [Google Scholar] [CrossRef]
- Shade, A.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013, 7, 1493–1506. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Paszkiewicz, K.; Field, D.; Knight, R.; Gilbert, J.A. The Western English Channel contains a persistent microbial seed bank. ISME J. 2012, 6, 1089–1093. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.M.; Wang, E.T.; Wang, J.M.; Liu, Z.S.; Li, Y.N.; Wei, G.H. Microbial succession in response to pollutants in batch-enrichment culture. Sci. Rep. 2016, 6, 11. [Google Scholar] [CrossRef]
- Jiao, S.; Zhang, Z.Q.; Yang, F.; Lin, Y.B.; Chen, W.M.; Wei, G.H. Temporal dynamics of microbial communities in microcosms in response to pollutants. Mol. Ecol. 2017, 26, 923–936. [Google Scholar] [CrossRef]
- Nekola, J.C.; White, P.S. The distance decay of similarity in biogeography and ecology. J. Biogeogr. 1999, 26, 867–878. [Google Scholar] [CrossRef]
- Oliver, A.; Lilley, A.; van der Gast, C. Species-Time Relationships for Bacteria; Horizon Scientific Press: Norwich, UK, 2012. [Google Scholar]
- Hatosy, S.M.; Martiny, J.B.H.; Sachdeva, R.; Steele, J.; Fuhrman, J.A.; Martiny, A.C. Beta diversity of marine bacteria depends on temporal scale. Ecology 2013, 94, 1898–1904. [Google Scholar] [CrossRef]
- Liang, Y.T.; Jiang, Y.J.; Wang, F.; Wen, C.Q.; Deng, Y.; Xue, K.; Qin, Y.J.; Yang, Y.F.; Wu, L.Y.; Zhou, J.Z.; et al. Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. ISME J. 2015, 9, 2561–2572. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Montoya, L.; Xu, L.; Madera, M.; Hollingsworth, J.; Purdom, E.; Singan, V.; Vogel, J.; Hutmacher, R.B.; Dahlberg, J.A.; et al. Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nat. Commun. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, D.; Mu, Y.; Zhu, Z.Y.; Wu, Y.Z.; Qi, Q.; Mu, Y.C.; Su, W. Exploring the heterogeneity of community and function and correspondence of “species-enzymes” among three types of Daqu with different fermentation peak-temperature via high-throughput sequencing and metagenomics. Food Res. Int. 2024, 176, 11. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Lu, Y. Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 2020, 22, 1052–1065. [Google Scholar] [CrossRef]
- Jiao, S.; Yang, Y.F.; Xu, Y.Q.; Zhang, J.; Lu, Y.H. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef]
- Liang, Z.; Xiangyong, W.; Zhenbiao, L.; Lu, J.; Fuyong, W.; Yanxia, Z.; Xinye, W. Effect of adding green tea on physicochemical characteristics and bacterial diversity of daqu during fermentation. Food Sci. 2024, 45, 59–68, (In Chinese with English Abstract). [Google Scholar] [CrossRef]

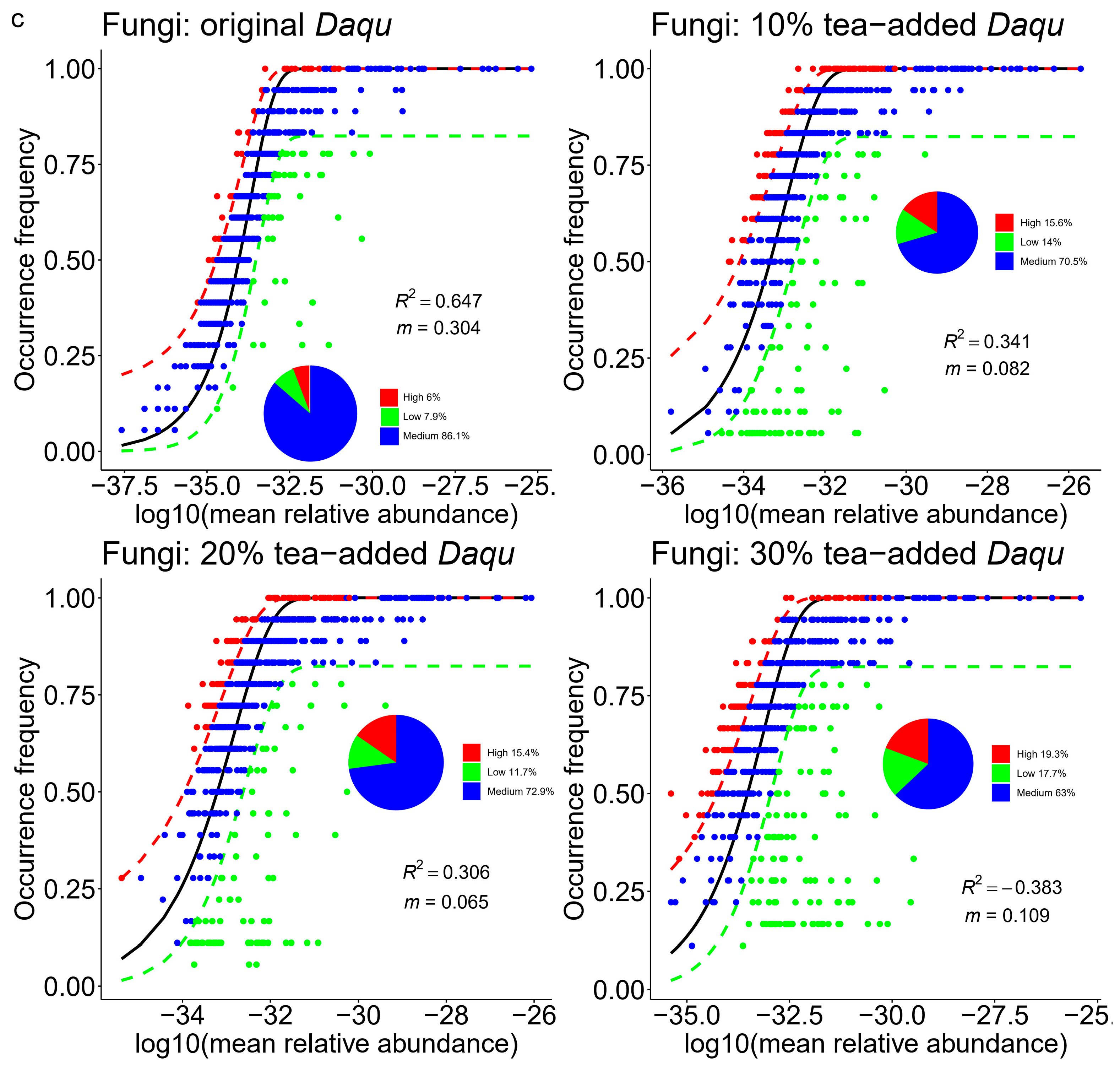
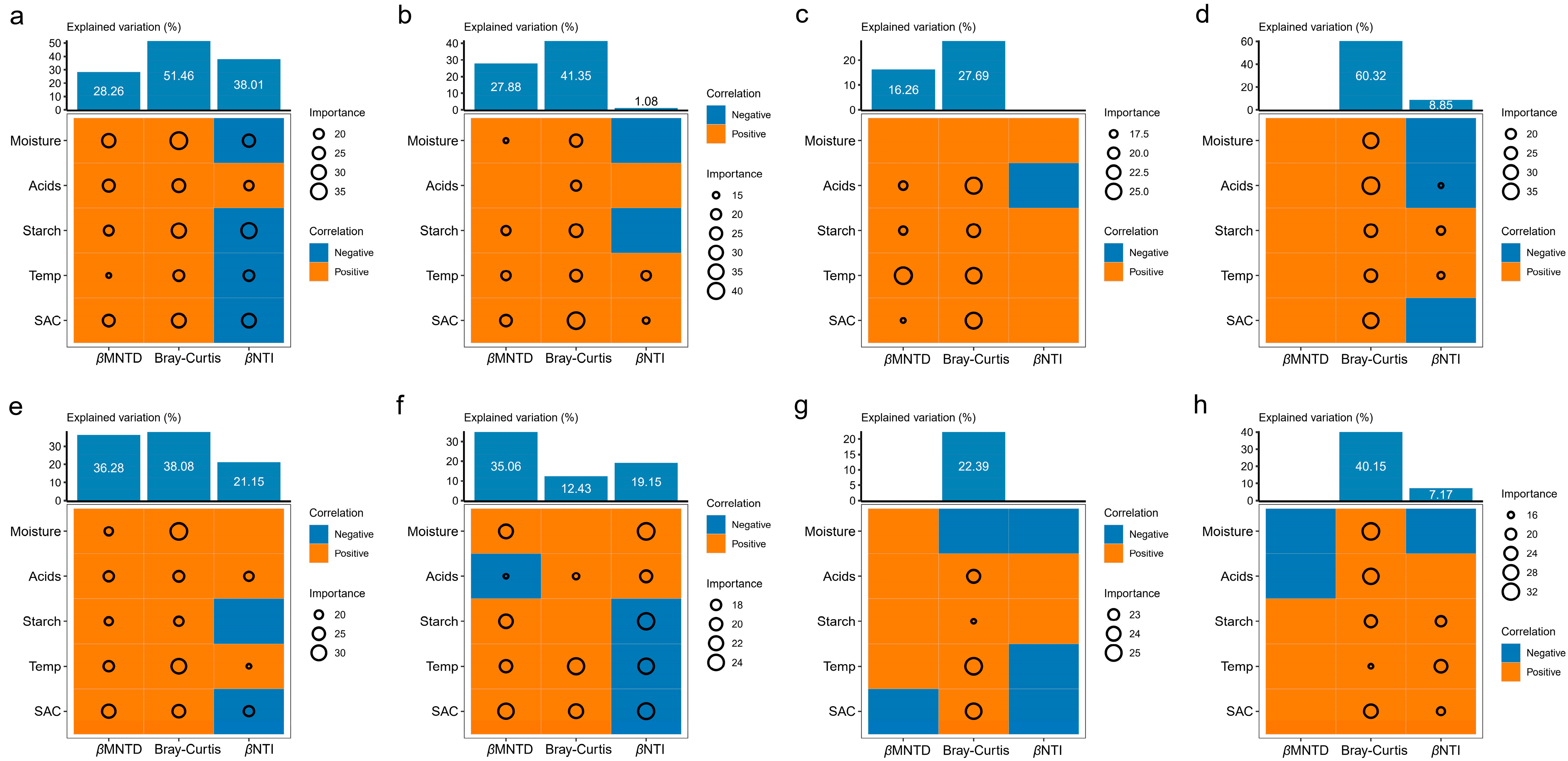
| Daqu Types | Models |
|---|---|
| Bacterial community | |
| Original Daqu | Richness = 443.13 + 24.84ln(T) |
| 10% tea-added Daqu | Richness = 453.56 + 22.56ln(T) |
| 20% tea-added Daqu | Richness = 461.07 + 22.49ln(T) |
| 30% tea-added Daqu | Richness = 466.58 + 20.39ln(T) |
| Fungal community | |
| Original Daqu | Richness = 565.04 + 21.92ln(T) |
| 10% tea-added Daqu | Richness = 634.71 + 9.89ln(T) |
| 20% tea-added Daqu | Richness = 643.44 + 11.08ln(T) |
| 30% tea-added Daqu | Richness = 648.27 + 13.01ln(T) |
| Daqu Types | Models | R2 |
|---|---|---|
| Bacterial community | ||
| Original Daqu | Ss = 0.636 − 0.007T | 0.148 |
| 10% tea-added Daqu | Ss = 0.566 − 0.010T | 0.171 |
| 20% tea-added Daqu | Ss = 0.515 − 0.010T | 0.193 |
| 30% tea-added Daqu | Ss = 0.595 − 0.007T | 0.135 |
| Fungal community | ||
| Original Daqu | Ss = 0.577 − 0.008T | 0.122 |
| 10% tea-added Daqu | Ss = 0.523 − 0.009T | 0.164 |
| 20% tea-added Daqu | Ss = 0.512 − 0.008T | 0.133 |
| 30% tea-added Daqu | Ss = 0.599 − 0.008T | 0.163 |
| Periods | Adonis | Anosim |
|---|---|---|
| Bacterial community | ||
| Day1 | R2 = 0.369 n.s. | R = 0.179 n.s. |
| Day6 | R2 = 0.627 *** | R = 0.702 *** |
| Day12 | R2 = 0.606 *** | R = 0.694 *** |
| Day15 | R2 = 0.402 * | R = 0.287 * |
| Day30 | R2 = 0.333 n.s. | R = 0.157 n.s. |
| Day40 | R2 = 0.549 ** | R = 0.528 ** |
| Fungal community | ||
| Day1 | R2 = 0.374 n.s. | R = 0.296 * |
| Day6 | R2 = 0.546 ** | R = 0.519 ** |
| Day12 | R2 = 0.333 n.s. | R = 0.077 n.s. |
| Day15 | R2 = 0.365 n.s. | R = 0.176 n.s. |
| Day30 | R2 = 0.490 ** | R = 0.472 ** |
| Day40 | R2 = 0.579 *** | R = 0.630 ** |
| Kindom | Daqu Treatments | Pagel’s (1999) λ [34] | Blomberg et al. (2003) K [33] | Fritz & Purvis (2010) D Test [35] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acids | Starch | Moisture | Temp | SAC | Acids | Starch | Moisture | Temp | SAC | Acids | Starch | Moisture | Temp | SAC | ||
| Bacteria | original Daqu | 0.704 | 0.037 | 0.237 | 0.999 *** | 0.167 | 0.231 ** | 0.163 | 0.233 ** | 1.225 * | 0.195 * | 0.783 * | 0.615 * | 0.927 *** | −4.339 | 0.530 |
| 10% tea-added Daqu | 0.475 ** | 0.000 | 0.011 | 0.232 | 0.023 | 0.173 * | 0.117 | 0.120 | 0.146 | 0.140 | 0.250 * | <0.001 | 0.113 | 0.468 * | 0.248 | |
| 20% tea-added Daqu | 0.773 *** | 0.786 *** | 0.268 ** | 0.257 | 0.464 *** | 0.284 *** | 0.177 * | 0.097 | 0.136 | 0.192 ** | 0.713 *** | 0.879 *** | 0.327 * | 0.080 | −0.013 | |
| 30% tea-added Daqu | 0.093 | 0.654 ** | 0.239 | 0.000 | 0.161 | 0.198 | 0.264 | 0.211 | 0.166 | 0.210 | 0.012 | 0.124 | −0.675 | 0.217 | 0.127 | |
| Fungi | original Daqu | 0.861 *** | 0.696 * | 0.759 ** | 0.824 *** | 0.430 | 0.089 * | 0.060 | 0.015 | 0.070 ** | <0.001 | 0.891 *** | 0.253 | 0.391 * | 0.481 *** | 0.214 |
| 10% tea-added Daqu | <0.001 | 0.532 * | 0.813 *** | 0.233 | 0.020 | 0.362 | 0.347 * | 0.571 *** | 0.108 | 0.271 | 0.140 | 0.481 | 0.819 ** | −0.141 | 0.188 | |
| 20% tea-added Daqu | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.510 | 0.812 | 0.771 | 0.575 | 0.490 | −0.975 | 0.958 | −1.256 | −0.791 | −1.279 | |
| 30% tea-added Daqu | <0.001 | <0.001 | <0.001 | 0.059 | 0.314 | 0.244 | 0.246 | 0.179 | 0.218 | 0.212 | 0.521 | 0.869 * | −0.041 | −0.346 | 0.356 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Li, F.; Xiao, H.; Zhao, T.; Zhong, Y.; Hu, Z.; Jiang, L.; Wang, X.; Wang, X. Green Tea Modulates Temporal Dynamics and Environmental Adaptation of Microbial Communities in Daqu Fermentation. Fermentation 2025, 11, 511. https://doi.org/10.3390/fermentation11090511
Zhao L, Li F, Xiao H, Zhao T, Zhong Y, Hu Z, Jiang L, Wang X, Wang X. Green Tea Modulates Temporal Dynamics and Environmental Adaptation of Microbial Communities in Daqu Fermentation. Fermentation. 2025; 11(9):511. https://doi.org/10.3390/fermentation11090511
Chicago/Turabian StyleZhao, Liang, Fangfang Li, Hao Xiao, Tengfei Zhao, Yanxia Zhong, Zhihui Hu, Lu Jiang, Xiangyong Wang, and Xinye Wang. 2025. "Green Tea Modulates Temporal Dynamics and Environmental Adaptation of Microbial Communities in Daqu Fermentation" Fermentation 11, no. 9: 511. https://doi.org/10.3390/fermentation11090511
APA StyleZhao, L., Li, F., Xiao, H., Zhao, T., Zhong, Y., Hu, Z., Jiang, L., Wang, X., & Wang, X. (2025). Green Tea Modulates Temporal Dynamics and Environmental Adaptation of Microbial Communities in Daqu Fermentation. Fermentation, 11(9), 511. https://doi.org/10.3390/fermentation11090511





