Preliminary Genetic and Physiological Characterization of Starmerella magnoliae from Spontaneous Mead Fermentation in Patagonia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Mead Production and Yeast Isolation
2.3. Yeast Identification
2.4. Intraspecific Characterization of Starmerella magnoliae Isolates
2.5. Phylogenetic Analysis
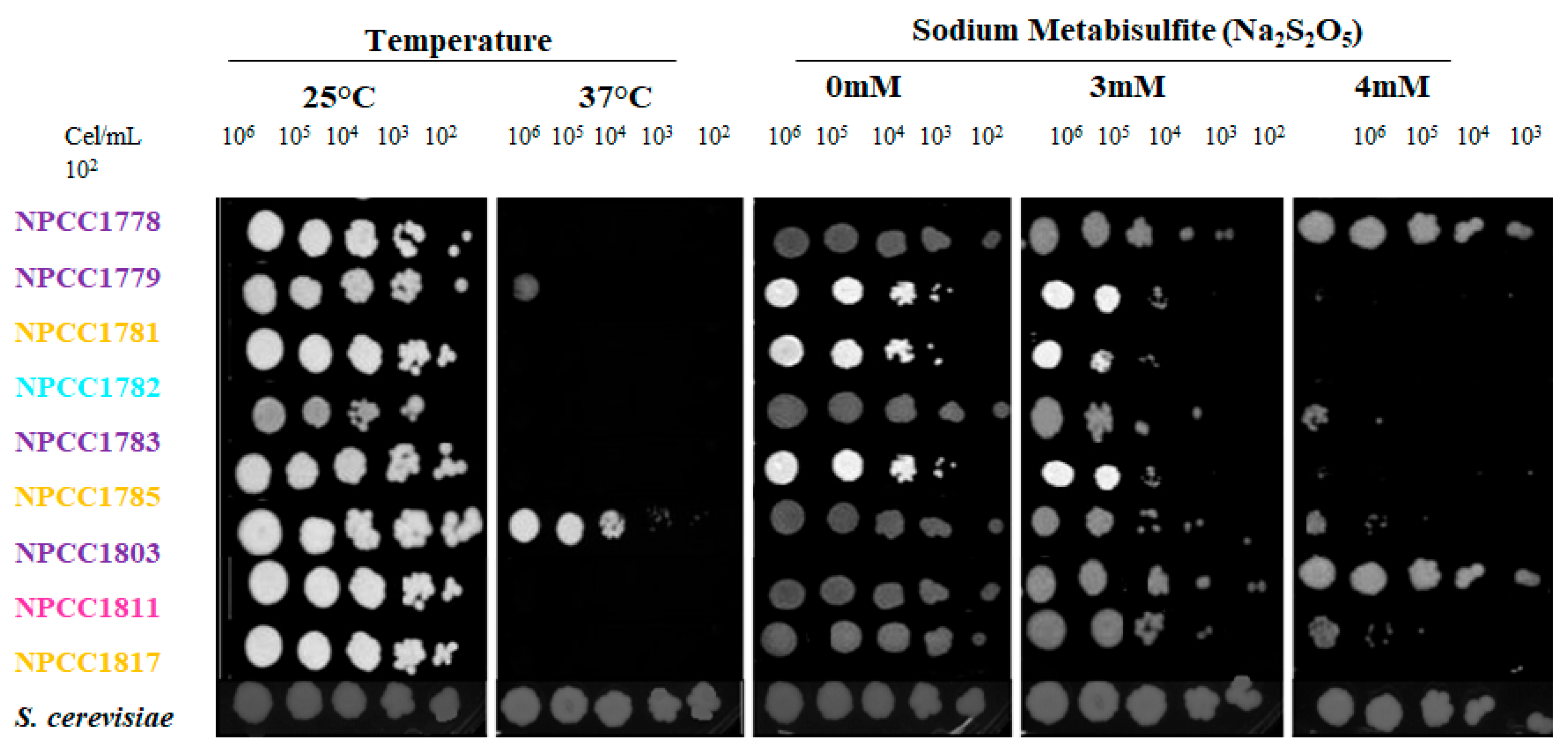
2.6. Stress Tolerance Assays
2.6.1. Sulfite Tolerance
2.6.2. Temperature Tolerance
2.6.3. Ethanol Tolerance
2.7. Microfermentation of Mead with S. magnoliae
2.8. Physicochemical Parameters
2.9. Statistical Analysis
3. Results
3.1. Yeast Diversity in Mead
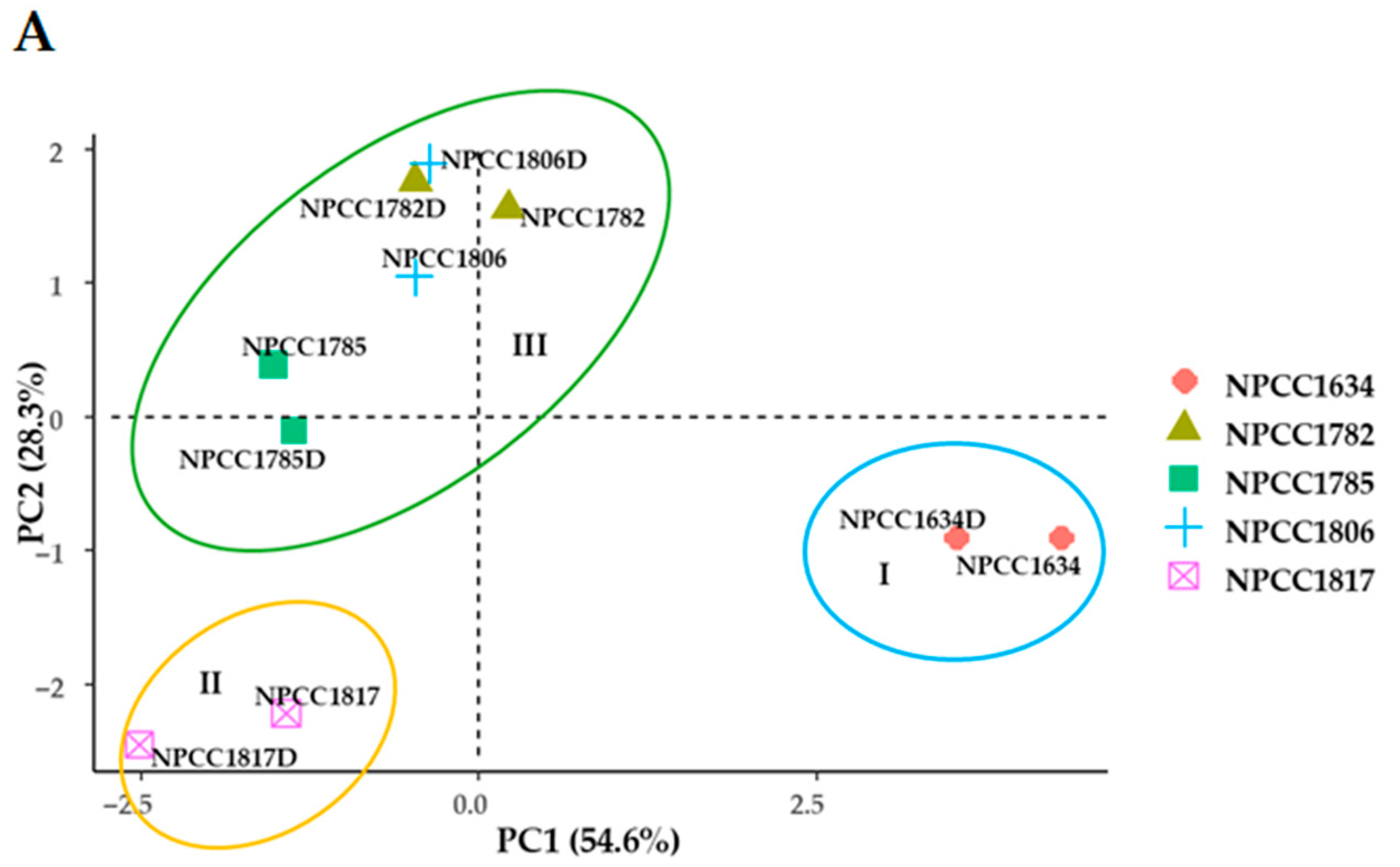
3.2. Intraspecific Characterization of S. magnoliae
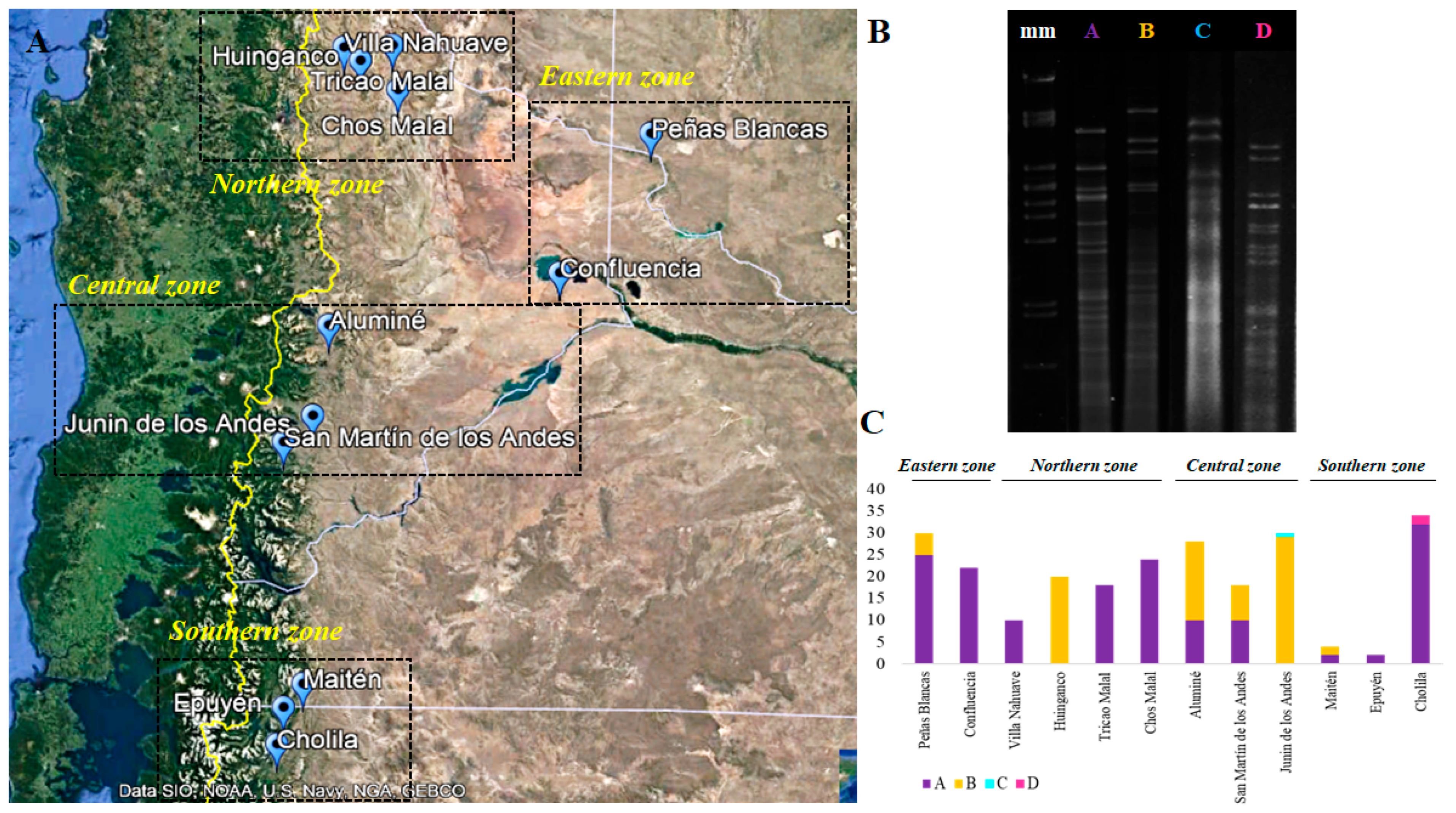
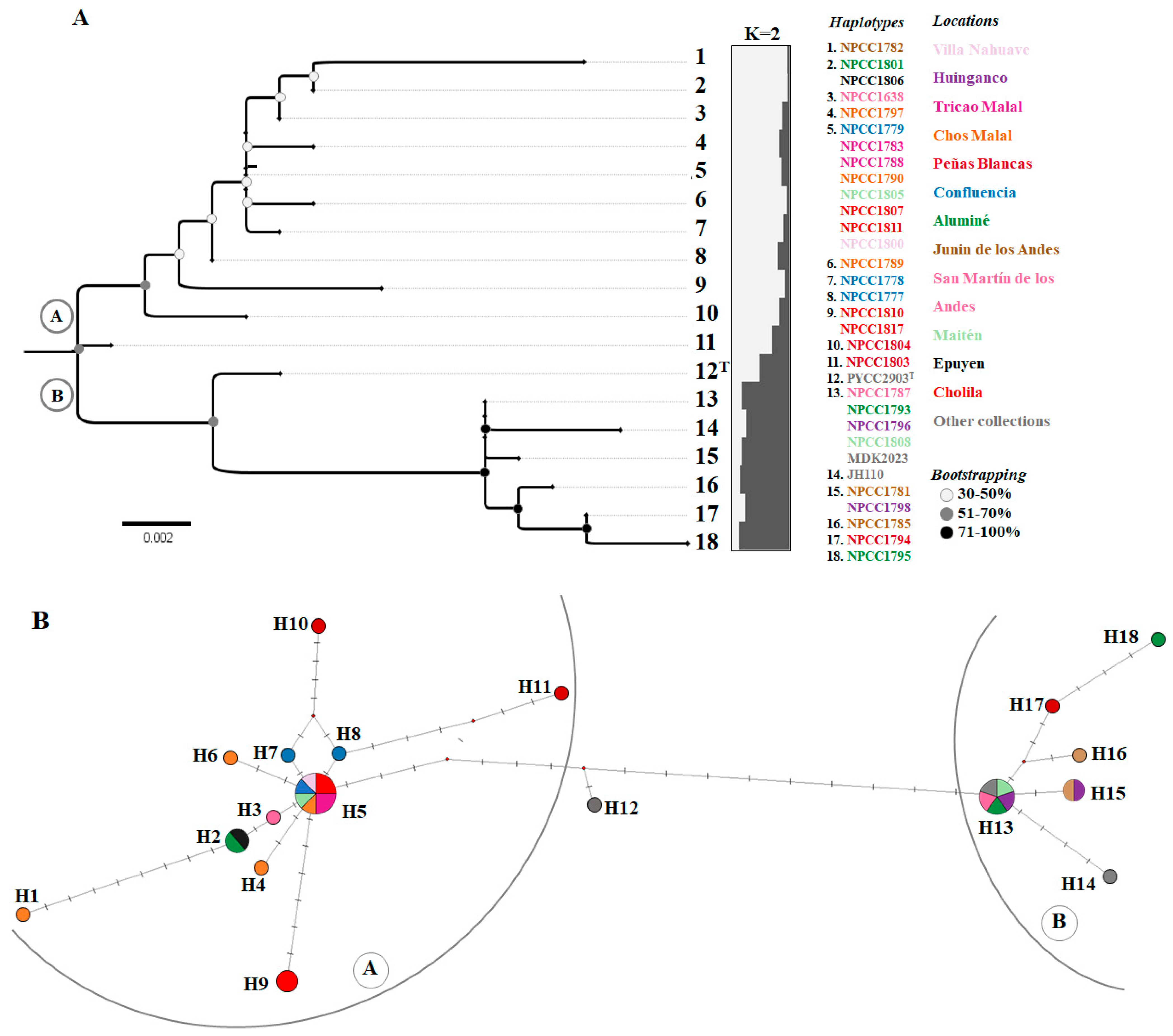
3.3. Phylogenetic and Phylogeographic Analysis of S. magnoliae
3.4. Starmerella Magnoliae Growth Under Different Stress Conditions
3.5. Microfermentation of Mead with S. magnoliae
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| App | μmax (h−1) | λ (h) | ||||||
|---|---|---|---|---|---|---|---|---|
| S.mangoliae | 0% | 3% | 5% | 10% | 0% | 3% | 5% | 10% |
| NPCC1638 | 0.051 ± 0.004 efghi | 0.128 ± 0.007 d | 0.093 ± 0.004 de | 0.098 ± 0.011 ab | 1.437 ± 0.002 efg | 1.264 ± 0.164 fg | 2.389 ± 0.304 hi | 3.525 ± 0.187 e |
| NPCC1777 | 0.049 ± 0.008 fghij | 0.090 ± 0.004 hij | 0.061 ± 0.004 fghi | 0.087 ± 0.004 bc | 1.320 ± 0.054 gh | 3.444 ± 0.295 b | 2.461 ± 0.051 ghi | 3.325 ± 0.291 e |
| NPCC1778 | 0.049 ± 0.003 fghij | 0.114 ± 0.002 def | 0.070 ± 0.002 fg | 0.107 ± 0.013 a | 1.959 ± 0.026 cd | 2.550 ± 0.129 cd | 1.248 ± 0.131 kl | 1.642 ± 0.117 h |
| NPCC1779 | 0.049 ± 0.003 ghijk | 0.109 ± 0.003 efg | 0.065 ± 0.007 fghi | 0.111 ± 0.021 a | 1.962 ± 0.024 cd | 2.545 ± 0.128 cd | 1.560 ± 0.282 jkl | 1.699 ± 0.160 h |
| NPCC1781 | 0.044 ± 0.002 ijk | 0.101 ± 0.011 fghij | 0.055 ± 0.002 ghijk | 0.073 ± 0.011 cd | 3.084 ± 0.013 a | 3.675 ± 0.238 ab | 7.728 ± 0.471 a | 3.942 ± 0.384 cd |
| NPCC1782 | 0.086 ± 0.002 c | 0.094 ± 0.009 ghij | 0.113 ± 0.001 bc | 0.116 ± 0.015 a | 2.873 ± 0.019 b | 2.379 ± 0.151 d | 1.151 ± 0.158 l | 1.534 ± 0.338 h |
| NPCC1783 | 0.048 ± 0.002 hijk | 0.106 ± 0.003 efgh | 0.069 ± 0.002 fgh | 0.104 ± 0.009 ab | 1.886 ± 0.090 cd | 2.536 ± 0.130 cd | 1.481 ± 0.391 jkl | 1.688 ± 0.166 h |
| NPCC1785 | 0.044 ± 0.002 jk | 0.086 ± 0.001 ij | 0.052 ± 0.001 hijk | 0.072 ± 0.011 cd | 3.004 ± 0.098 ab | 3.716 ± 0.043 a | 7.701 ± 0.626 a | 3.624 ± 0.103 de |
| NPCC1787 | 0.055 ± 0.001 ef | 0.186 ± 0.001 c | 0.132 ± 0.042 a | 0.051 ± 0.001 ef | 1.400 ± 0.081 fg | 1.482 ± 0.202 f | 1.595 ± 0.240 jkl | 2.605 ± 0.211 fg |
| NPCC1788 | 0.048 ± 0.001 ghijk | 0.107 ± 0.003 efg | 0.067 ± 0.006 fgh | 0.113 ± 0.020 a | 1.994 ± 0.013 cd | 2.546 ± 0.138 cd | 1.392 ± 0.069 jkl | 1.678 ± 0.153 h |
| NPCC1789 | 0.048 ± 0.001 ghijk | 0.107 ± 0.004 efg | 0.060 ± 0.007 fghi | 0.106 ± 0.021 a | 1.891 ± 0.071 cd | 2.558 ± 0.152 cd | 1.569 ± 0.325 jkl | 1.701 ± 0.151 h |
| NPCC1790 | 0.048 ± 0.002 hijk | 0.106 ± 0.004 efgh | 0.068 ± 0.002 fgh | 0.106 ± 0.011 a | 1.964 ± 0.160 cd | 2.567 ± 0.135 cd | 1.633 ± 0.239 jkl | 1.695 ± 0.143 h |
| NPCC1793 | 0.056 ± 0.001 e | 0.261 ± 0.041 b | 0.098 ± 0.011 cd | 0.115 ± 0.003 a | 1.576 ± 0.071 e | 2.509 ± 0.256 cd | 2.662 ± 0.297 ghi | 6.919 ± 0.166 b |
| NPCC1794 | 0.064 ± 0.002 d | 0.019 ± 0.001 mn | 0.018 ± 0.001 n | 0.023 ± 0.003 h | 1.041 ± 0.092 jk | 0.334 ± 0.068 i | 3.388 ± 0.210 ef | 1.162 ± 0.209 i |
| NPCC1795 | 0.017 ± 0.001 m | 0.043 ± 0.001 kl | 0.038 ± 0.001 klm | 0.040 ± 0.004 efgh | 1.184 ± 0.003 hij | 0.385 ± 0.041 i | 4.486 ± 0.324 c | 2.468 ± 0.382 fg |
| NPCC1796 | 0.050 ± 0.002 fghij | 0.100 ± 0.002 fghij | 0.060 ± 0.002 fghi | 0.079 ± 0.010 c | 0.584 ± 0.069 l | 0.935 ± 0.024 h | 2.907 ± 0.542 fg | 4.248 ± 0.220 c |
| NPCC1797 | 0.049 ± 0.001 fghij | 0.107 ± 0.005 efg | 0.069 ± 0.003 fg | 0.111 ± 0.014 a | 2.030 ± 0.038 c | 2.563 ± 0.130 cd | 1.434 ± 0.256 jkl | 1.657 ± 0.126 h |
| NPCC1798 | 0.042 ± 0.002 k | 0.086 ± 0.001 j | 0.056 ± 0.004 ghij | 0.074 ± 0.009 cd | 3.027 ± 0.112 ab | 3.830 ± 0.129 a | 7.319 ± 0.143 a | 3.628 ± 0.101 de |
| NPCC1800 | 0.051 ± 0.001 efghi | 0.120 ± 0.010 de | 0.076 ± 0.003 ef | 0.101 ± 0.008 ab | 1.466 ± 0.213 efg | 1.166 ± 0.155 gh | 2.744 ± 0.296 ghi | 2.340 ± 0.239 g |
| NPCC1801 | 0.047 ± 0.001 hijk | 0.035 ± 0.010 lm | 0.032 ± 0.002 lmn | 0.042 ± 0.005 efg | 1.040 ± 0.115 jk | 0.311 ± 0.031 i | 3.748 ± 0.516 de | 1.177 ± 0.149 i |
| NPCC1803 | 0.084 ± 0.004 c | 0.054 ± 0.008 k | 0.041 ± 0.002 jkl | 0.038 ± 0.006 fgh | 1.216 ± 0.190 hi | 2.631 ± 0.052 c | 1.806 ± 0.170 j | 0.752 ± 0.098 j |
| NPCC1804 | 0.084 ± 0.001 c | 0.052 ± 0.001 k | 0.049 ± 0.007 ijkl | 0.039 ± 0.009 fgh | 1.585 ± 0.270 e | 2.605 ± 0.065 cd | 1.708 ± 0.076 jk | 0.768 ± 0.068 j |
| NPCC1805 | 0.026 ± 0.013 l | 0.102 ± 0.013 fghi | 0.016 ± 0.003 n | 0.027 ± 0.006 gh | 0.637 ± 0.041 l | 0.306 ± 0.039 i | 2.870 ± 0.241 gh | 1.738 ± 0.115 h |
| NPCC1806 | 0.131 ± 0.010 a | 0.116 ± 0.008 def | 0.115 ± 0.018 ab | 0.113 ± 0.017 a | 0.541 ± 0.108 l | 1.882 ± 0.094 e | 5.862 ± 0.065 b | 2.760 ± 0.144 f |
| NPCC1807 | 0.108 ± 0.004 b | 0.011 ± 0.003 n | 0.113 ± 0.022 bc | 0.044 ± 0.007 efg | 0.537 ± 0.028 l | 0.414 ± 0.007 i | 7.798 ± 0.070 a | 8.267 ± 0.339 a |
| NPCC1808 | 0.055 ± 0.004 efg | 0.298 ± 0.015 a | 0.100 ± 0.012 bcd | 0.044 ± 0.007 efg | 1.557 ± 0.008 ef | 2.508 ± 0.262 cd | 2.705 ± 0.216 ghi | 1.134 ± 0.124 i |
| NPCC1810 | 0.052 ± 0.002 efgh | 0.041 ± 0.004 kl | 0.023 ± 0.007 mn | 0.057 ± 0.008 de | 1.113 ± 0.027 ij | 0.268 ± 0.029 i | 2.373 ± 0.316 i | 1.092 ± 0.151 ij |
| NPCC1811 | 0.048 ± 0.002 hijk | 0.108 ± 0.004 efg | 0.070 ± 0.002 fg | 0.110 ± 0.012 a | 1.864 ± 0.024 d | 2.557 ± 0.150 cd | 1.665 ± 0.281 jk | 1.749 ± 0.178 h |
| NPCC1817 | 0.050 ± 0.002 efghij | 0.043 ± 0.003 kl | 0.065 ± 0.003 fghi | 0.070 ± 0.015 cd | 0.916 ± 0.009 k | 0.307 ± 0.035 i | 3.966 ± 0.352 d | 1.675 ± 0.349 h |
References
- Codex Alimentarius Commission. Standard for Honey (CXS 12-1981) (Adopted in 1981; Revised in 1987, 2001; Amended in 2019, 2022). Food and Agriculture Organization of the United Nations (FAO) & World Health Organization (WHO), 2022. Available online: https://www.fao.org/fao-who-codexalimentarius (accessed on 25 January 2025).
- Piana, G.; D’Albore, R.; Isola, A. La Miel, 1st ed.; Mundi Prensa: Madrid, Spain, 1989; Volume 1, p. 124. [Google Scholar]
- Calderón, D.; Piromalli, P.; Apablaza, O.; Virgillito, O. Guía Para la Elaboración de Hidromiel y Licor de Miel, 1st ed.; Ministerio de Agricultura, Ganadería y Pesca: Buenos Aires, Argentina, 2017; p. 58. [Google Scholar]
- Ministerio de la Producción. PRODUCE: Conoce los Requisitos de Calidad de las Bebidas a Base de Miel. Gobierno del Perú, 2024. Available online: https://www.gob.pe/institucion/produce/noticias/1034225-produce-conoce-los-requisitos-de-calidad-de-las-bebidas-a-base-de-miel (accessed on 5 February 2025).
- Iurlina, M.O.; Fritz, R. Characterization of microorganisms in Argentinean honeys from different sources. Int. J. Food Microbiol. 2005, 105, 297–304. [Google Scholar] [CrossRef]
- Estrada, H.; Gamboa, M.M.; Chaves, C.; Arias, M.L. Evaluación de la actividad antimicrobiana de la miel de abeja contra Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Escherichia coli, Salmonella enteritidis, Listeria monocytogenes y Aspergillus niger. Evaluación de su carga microbiológica. Arch. Latinoam. Nutr. 2005, 55, 167–171. [Google Scholar]
- Dell’Elce, A.; Aguirre, M.S.; Weidmann, C.; Patricelli, P.; Formentini, E. Actividad bactericida in vitro de la miel sobre Escherichia coli y Staphylococcus aureus: Comparación con la actividad de antibióticos betalactámicos. In Proceedings of the V Jornadas de Difusión de la Investigación y Extensión, Esperanza, Argentina, 8 November 2017. [Google Scholar]
- Déak, T. Yeasts in specific types of foods. In Handbook of Food Spoilage Yeasts, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 117–201. [Google Scholar]
- Carvalho, C.M.; Meirinho, S.; Estevinho, M.L.F.; Choupina, A. Yeast species associated with honey: Different identification methods. Arch. Zootec. 2010, 59, 103–113. Available online: http://scielo.isciii.es/scielo.php?script=sci_arttext&pid=S0004-05922010000100011 (accessed on 18 March 2025). [CrossRef]
- Echeverrigaray, S.; Scariot, F.J.; Foresti, L.; Schwarz, L.V.; Rocha, R.K.M.; da Silva, G.P.; Moreira, J.P.; Delamare, A.P.L. Yeast biodiversity in honey produced by stingless bees raised in the highlands of southern Brazil. Int. J. Food Microbiol. 2021, 347, 109200. [Google Scholar] [CrossRef] [PubMed]
- Carrizo Villoldo, A.E.; Carrizo, C.B.; Benítez Ahrendts, M.R.; Carrillo, L. Levaduras aisladas de mieles como antagonistas de mohos patógenos de cultivos. Rev. Fac. Agron. 2020, 119, 054. [Google Scholar] [CrossRef]
- Ziuzia, P.; Janiec, Z.; Wróbel-Kwiatkowska, M.; Lazar, Z.; Rakicka-Pustułka, M. Honey’s yeast—New source of valuable species for industrial applications. Int. J. Mol. Sci. 2023, 24, 7889. [Google Scholar] [CrossRef]
- Barry, J.P.; Metz, M.S.; Hughey, J.; Quirk, A.; Bochman, M.L. Two novel strains of Torulaspora delbrueckii isolated from the honey bee microbiome and their use in honey fermentation. Fermentation 2018, 4, 22. [Google Scholar] [CrossRef]
- Pereira, A.P.; Dias, T.; Andrade, J.; Ramalhosa, E.; Estevinho, L.M. Mead production: Selection and characterization assays of Saccharomyces cerevisiae strains. Food Chem. Toxicol. 2009, 47, 2057–2063. [Google Scholar] [CrossRef]
- Vidrih, R.; Hribar, J. Mead. The oldest alcoholic beverage. In Traditional Foods, 2nd ed.; Kristbergsson, K., Oliveira, J., Eds.; Springer: New York, NY, USA, 2016; Volume 10, pp. 471–479. [Google Scholar] [CrossRef]
- Administración Nacional de Medicamentos, Alimentos y Tecnología Médica (ANMAT). Código Alimentario Argentino. Capítulo XIII Bebidas Fermentadas. 2024. Available online: https://www.argentina.gob.ar/sites/default/files/anmat-capitulo_xiii_beb_fermentadasactualiz_2018-12.pdf (accessed on 13 May 2025).
- Codex Alimentarius Commission. General Standard for Food Additives (CODEX STAN 192-1995, Rev. 2024). 2024. Available online: https://www.fao.org/gsfaonline/docs/CXS_192e.pdf (accessed on 5 May 2025).
- Gurini, L.; Apablaza, O.; Basilio, A.; Ciappini, M.C.; Fagúndez, G.; Gaggiotti, M.; Gutiérrez, A.; Salgado, C.R.; Winter, J. Guía para la Caracterización de Mieles Argentina, 1st ed.; Instituto Nacional de Tecnología Agropecuaria (INTA): Buenos Aires, Argentina, 2019. Available online: https://inta.gob.ar (accessed on 6 March 2025).
- Lopez, V.; Querol, A.; Ramon, D.; Fernandez-Espinar, M.T. A simplified procedure to analyse mitochondrial DNA from industrial yeasts. Int. J. Food Microbiol. 2001, 68, 75–85. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res. 2006, 3, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posadas, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Forster, P.; Rohl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Park, H.; Lopez, N.H.; Bakalinsky, A.T. Use of sulfite resistance in Saccharomyces cerevisiae as a dominant selectable marker. Curr. Genet. 1999, 36, 339–344. [Google Scholar] [CrossRef]
- González Flores, M.; Rodríguez, M.E.; Oteiza, J.M.; Barbagelata, R.J.; Lopes, C.A. Physiological characterization of Saccharomyces uvarum and Saccharomyces eubayanus from Patagonia and their potential for cidermaking. Int. J. Food Microbiol. 2017, 249, 9–17. [Google Scholar] [CrossRef]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 2875–2902. [Google Scholar]
- Detry, R.; Simon-Delso, N.; Bruneau, E.; Daniel, H.-M. Specialisation of yeast genera in different phases of bee bread maturation. Microorganisms 2020, 8, 1789. [Google Scholar] [CrossRef]
- Roxo, I.; Amaral, A.L.; Portugal, A.; Trovão, J. A checklist of fungi isolated from honey (2000–2022). Stud. Fungi 2023, 8, 14. [Google Scholar] [CrossRef]
- Yu, J.-H.; Lee, D.-H.; Oh, Y.-J.; Han, K.-C.; Ryu, Y.-W.; Seo, J.-H. Selective utilization of fructose to glucose by Candida magnoliae, an erythritol producer. Appl. Biochem. Biotechnol. 2006, 132, 870. [Google Scholar] [CrossRef]
- Soares de Medeiros, A.S. Fermentation of Fruit Juices by the Osmotolerant Yeast Candida magnoliae. Master’s Thesis, Universidade Federal de Pernambuco, Recife, Brazil, 2014. [Google Scholar]
- Lee, D.-H.; Kim, S.-J.; Seo, J.-H. Molecular cloning and characterization of two novel fructose-specific transporters from the osmotolerant and fructophilic yeast Candida magnoliae JH110. Appl. Microbiol. Biotechnol. 2014, 98, 3569–3578. [Google Scholar] [CrossRef]
- da Costa Neto, D.J.; de Morais, P.B. The vectoring of Starmerella species and other yeasts by stingless bees in a Neotropical savanna. Fungal Ecol. 2020, 47, 100973. [Google Scholar] [CrossRef]
- Eizaguirre, J.I.; Peris, D.; Rodríguez, M.E.; Lopes, C.A.; De Los Ríos, P.; Hittinger, C.T.; Libkind, D. Phylogeography of the wild Lager-brewing ancestor (Saccharomyces eubayanus) in Patagonia. Environ. Microbiol. 2018, 20, 3732–3743. [Google Scholar] [CrossRef]
- Almeida, P.; Goncalves, C.; Teixeira, S.; Libkind, D.; Bontrager, M.; Masenuf-Pomarede, I.; Albertin, W.; Durrens, P.; Sherman, D.J.; Marullo, P.; et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 2014, 5, 4044. [Google Scholar] [CrossRef]
- Gonzalez Flores, M.; Rodriguez, M.E.; Peris, D.; Barrio, E.; Querol, A.; Lopes, C.A. Human-associated migration of Holarctic Saccharomyces uvarum strains to Patagonia. Fungal Ecol. 2020, 48, 100990. [Google Scholar] [CrossRef]
- Fiore, C.; Arrizon, J.; Gschaedler, A.; Flores, J.; Romano, P. Comparison between yeasts from grape and agave musts for traits of technological interest. World J. Microbiol. Biotechnol. 2005, 21, 1141–1147. [Google Scholar] [CrossRef]
- Englezos, V.; Rantsiou, K.; Torchio, F.; Rolle, L.; Gerbi, V.; Cocolin, L. Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (syn. Candida zemplinina) in wine fermentation: Physiological and molecular characterizations. Int. J. Food Microbiol. 2015, 199, 33–40. [Google Scholar] [CrossRef]
- Instituto Nacional de Alimentación (INAL). Decreto Nº 188/008. IMPO—Diario Oficial, 31 March 2008. Available online: https://www.impo.com.uy/bases/decretos-reglamento/188-2008/1 (accessed on 25 June 2025).
- Lopes, C.A.; Barrio, E.; Querol, A. Natural hybrids of S. cerevisiae × S. kudriavzevii share alleles with European wild populations of Saccharomyces kudriavzevii. FEMS Yeast Res. 2010, 10, 412–421. [Google Scholar] [CrossRef]
- Sampaio, J.P.; Gonçalves, P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 2008, 74, 2144–2152. [Google Scholar] [CrossRef]
- Desai, M.V.; Dubey, K.V.; Vakil, B.V.; Ranade, V.V. Isolation, identification and screening of the yeast flora from Indian cashew apple for sugar and ethanol tolerance. Int. J. Biotechnol. Wellness Ind. 2012, 1, 259–265. [Google Scholar]
- Plesset, J.; Palm, C.; McLaughlin, C.S. Induction of heat shock proteins and thermotolerance by ethanol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1982, 108, 1340–1345. [Google Scholar] [CrossRef]
- Ding, J.; Huang, X.; Zhang, L.; Zhao, N.; Yang, D.; Zhang, K. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2009, 85, 253–263. [Google Scholar] [CrossRef]
- Stanley, D.; Bandara, A.; Fraser, S.; Chambers, P.J.; Stanley, G.A. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 2010, 109, 13–24. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C. Wine Chemistry and Biochemistry; Springer: New York, NY, USA, 2009; p. 879. [Google Scholar]
- Ciani, M.; Ferraro, L.; Fatichenti, F. Influence of glycerol production on the aerobic and anaerobic growth of the wine yeast Candida stellata. Enzyme Microb. Technol. 2000, 27, 698–703. [Google Scholar] [CrossRef]
- Santos, A.R.O.; Leon, M.P.; Barros, K.O.; Freitas, L.F.D.; Hughes, A.F.S.; Morais, P.B.; Lachance, M.A.; Rosa, C.A. Starmerella camargoi f.a., sp. nov., and five other new Starmerella species from flowers and bees. Int. J. Syst. Evol. Microbiol. 2018, 68, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Moro, P. Estudio del Impacto de Levaduras No-Saccharomyces Para Mejorar la Calidad de Vinos Tintos. Master’s Thesis, Universidad de Valladolid, Valladolid, Spain, 2021. [Google Scholar]


| Sampling Site | Sample a N° | Plant Species b | Yeast Species Isolated from Mead c * | |
|---|---|---|---|---|
| Sweet d | Dry e | |||
| Neuquén Province | ||||
| Aluminé | 1 | Centaurea cyanus | Starmerella magnoliae (100%) | Starmerella magnoliae (80%) Zygosaccharomyces rouxii (20%) |
| 2 | Centaurea cyanus | Starmerella magnoliae (100%) | Zygosaccharomyces rouxii (100%) | |
| Chos Malal | 3 | Centaurea solstitialis Centaurea cyanus | Starmerella magnoliae (100%) | Rhodotorula mucilaginosa (40%) Saccharomyces cerevisiae (20%) Starmerella magnoliae (40%) |
| 4 | Centaurea cyanus | Rhodotorula mucilaginosa (100%) | Starmerella magnoliae (100%) | |
| Huinganco | 5 | Unidentified | Starmerella magnoliae (100%) | Saccharomyces cerevisiae (100%) |
| 6 | Unidentified | Starmerella magnoliae (100%) | Saccharomyces cerevisiae (100%) | |
| Junín de los Andes | 7 | Unidentified | Starmerella magnoliae (100%) | Starmerella magnoliae (100%) |
| 8 | Unidentified | Starmerella magnoliae (100%) | Rhodotorula mucilaginosa (80%) Saccharomyces cerevisiae (20%) | |
| Neuquén | 9 | Unidentified | Starmerella magnoliae (100%) | Nakaseomyces glabratus (40%) Saccharomyces uvarum (20%) Starmerella magnoliae (40%) |
| 10 | Nakaseomyces glabratus (100%) | Saccharomyces uvarum (20%) Starmerella magnoliae (80%) | ||
| San Martín de los Andes | 11 | Lomatia hirsuta | Starmerella magnoliae (100%) | Zygosaccharomyces rouxii (20%) Starmerella magnoliae (80%) |
| Tricao Malal | 12 | Centaurea cyanus | Pichia membranifaciens (20%) Starmerella magnoliae (80%) | Starmerella magnoliae (100%) |
| Villa Nahueve | 13 | Azorella prolifera Centaurea cyanus | Starmerella magnoliae (100%) | Zygosaccharomyces rouxii (100%) |
| Río Negro Province | ||||
| Peñas Blancas | 14 | Lotus corniculatus Medicago sativa | Starmerella magnoliae (100%) | Starmerella magnoliae (100%) |
| 15 | Lotus corniculatus Medicago sativa | Starmerella magnoliae (100%) | Zygosaccharomyces rouxii (100%) | |
| Chubut Province | ||||
| Cholila | 16 | Lomatia hirsuta Fabiana imbricata Rhamnus lycioides Salix humboldtiana Brassica rapa Taraxacum officinale | Starmerella magnoliae (60%) Pichia membranifaciens (20%) Zygosaccharomyces rouxii (20%) | Zygosaccharomyces rouxii (20%) Starmerella magnoliae (80%) |
| 17 | Lomatia hirsuta Fabiana imbricata Rhamnus lycioides | Starmerella magnoliae (100%) | Starmerella magnoliae (100%) | |
| Epuyen | 18 | Taraxacum officinale Lomatia hirsuta Discaria trinervis Rhamnus lycioides Trifolium | Starmerella magnoliae (20%) Zygosaccharomyces rouxii (40%) Starmerella bombi (40%) | Pichia membranifaciens (100%) |
| El Maiten | 19 | Carduus spp. Erodium cicutarium Chamaemelum nobile | Pichia membranifaciens (40%) Zygosaccharomyces rouxii (40%) Starmerella magnoliae (20%) | Pichia membranifaciens (40%) Zygosaccharomyces rouxii (40%) Starmerella magnoliae (20%) |
| Sampling Site | mtDNA-RFLP | Representative Strain * |
|---|---|---|
| Peñas Blancas-RN | A (25 S1,S2,D1) | NPCC1803 S1; NPCC1804 D1 |
| B (5 S2) | NPCC1794 S2; | |
| Villa Nahuave-NQ | A (10 S1) | NPCC1800 S1 |
| Huinganco-NQ | B (20 S1,S2) | NPCC1796 S1; NPCC1798 S2 |
| Tricao Malal-NQ | A (18 S1,D1) | NPCC1783 S1; NPCC1788 D1 |
| Chos Malal-NQ | A (24 S1,D1,D2) | NPCC1797 S1; NPCC1790 D1; NPCC1789 D2 |
| Confluencia-NQ | A (22 S1,D1,D2) | NPCC1777 D1; NPCC1778 D2; NPCC1779 S1 |
| Aluminé-NQ | A (10 S1) | NPCC1801 S1 |
| B (18 S2,D1) | NPCC1793 S2; NPCC1795 D1 | |
| Junín de los Andes-NQ | B (29 S1,S2,D1) | NPCC1781 S1; NPCC1785 D1 |
| C (1 S2) | NPCC1782 S2 | |
| San Martín de los Andes-NQ | A (10 S1) | NPCC1638 S1 |
| B (8 D1) | NPCC1787 D1 | |
| Maitén-CH | A (2 S1) | NPCC1805 S1 |
| B (2 D1) | NPCC1808 D1 | |
| Epuyén-CH | A (2 S1) | NPCC1806 S1 |
| Cholila-CH | A (32 S1,S2,D1,D2) | NPCC1807 S1; NPCC1811 D1 |
| D (2 S2,D2) | NPCC1817 S2; NPCC1810 D2 |
| Parameters | S. magnoliae | S. cerevisiae | ||||
|---|---|---|---|---|---|---|
| NPCC1782 | NPCC1785 | NPCC1806 | NPCC1817 | NPCC1634 | ||
| Glycerol (g/L) | 8.26 ± 0.34 B | 8.59 ± 0.11 B | 8.83 ± 0.48 B | 7.47 ± 1.79 AB | 6.52 ± 1.07 A | |
| Citric acid (g/L) | 0.26 ± 0.005 A | 0.26 ± 0.009 A | 0.26 ± 0.007 A | 0.25 ± 0.001 A | 0.46 ± 0.025 B | |
| Malic acid (g/L) | ND | ND | ND | ND | 0.47 ± 0.08 | |
| Acetic acid (g/L) | 1.01 ± 0.30 abC | 0.714 ± 0.11 abBC | 1.16 ± 0.04 bC | 0.57 ± 0.13 aA | 0.68 ± 0.01 B | |
| Lactic acid (g/L) | 1.17 ± 0.18 aB | 1.89 ± 0.13 bcB | 1.35 ± 0.36 abB | 2.22 ± 0.31 cC | 0.17 ± 0.02 A | |
| Succinic acid (g/L) | 0.35 ± 0.07 A | 0.39 ± 0.053 A | 0.27 ± 0.09 A | 0.31 ± 0.07 A | 0.72 ± 0.04 B | |
| Ethanol (% v/v) | 3.99 ± 1.20 A | 4.36 ± 0.32 A | 4.05 ± 0.24 A | 4.39 ± 0.007 A | 9.29 ± 1.32 B | |
| Methanol (% v/v) | ND | ND | ND | ND | ND | |
| Residual sugars (g/L) | Glucose | 125.47 ± 7.56 aB | 160.07 ± 2.06 bB | 124.94 ± 7.58 aB | 145.16 ± 6.12 bB | 1.81 ± 2.08 A |
| Sucrose | 0.01 ± 1 × 10−3 | 0.44 ± 0.63 | 0.79 ± 1.12 | 3.89 ± 0.07 | 0.55 ± 0.78 | |
| Fructose | 9.02 ± 0.06 abB | 17.44 ± 1.38 bC | 5.37 ± 3.45 aB | 34.12 ± 6.61 cC | 1.80 ± 9.4 × 10−3A | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinjan, V.; González Flores, M.; Rodriguez, M.E.; Lopes, C.A. Preliminary Genetic and Physiological Characterization of Starmerella magnoliae from Spontaneous Mead Fermentation in Patagonia. Fermentation 2025, 11, 494. https://doi.org/10.3390/fermentation11090494
Kleinjan V, González Flores M, Rodriguez ME, Lopes CA. Preliminary Genetic and Physiological Characterization of Starmerella magnoliae from Spontaneous Mead Fermentation in Patagonia. Fermentation. 2025; 11(9):494. https://doi.org/10.3390/fermentation11090494
Chicago/Turabian StyleKleinjan, Victoria, Melisa González Flores, María Eugenia Rodriguez, and Christian Ariel Lopes. 2025. "Preliminary Genetic and Physiological Characterization of Starmerella magnoliae from Spontaneous Mead Fermentation in Patagonia" Fermentation 11, no. 9: 494. https://doi.org/10.3390/fermentation11090494
APA StyleKleinjan, V., González Flores, M., Rodriguez, M. E., & Lopes, C. A. (2025). Preliminary Genetic and Physiological Characterization of Starmerella magnoliae from Spontaneous Mead Fermentation in Patagonia. Fermentation, 11(9), 494. https://doi.org/10.3390/fermentation11090494






