Cutinase Production in Komagataella phaffii (Pichia pastoris): Performance Differences Between Host Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Construction of Plasmids, Transformation, and Yeast Selection
2.3. Production of Recombinant Cutinases ANCUT1 and ANCUT3 in K. phaffii in Shake Flasks
2.4. Enzymatic Assays
2.5. Protein Assay
2.6. SDS-PAGE and Zymograms
2.7. Statistical Analysis
3. Results
3.1. Effect of Methanol on Growth and Cutinase Production in K. phaffii KM71H Strains
3.2. Effect of Inoculum Concentration on Cutinase Production in KM71H Strains
3.3. Comparing Cutinase Production in K. phaffi MutS and Mut+ Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamada, Y.; Matsuda, M.; Maeda, K.; Mikata, K. The phylogenetic relationships of methanol-assimilating yeasts based on the partial sequences of 18S and 26S ribosomal RNAs: The proposal of Komagataella gen. nov. (Saccharomycetaceae). Biosci. Biotechnol. Biochem. 1995, 59, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Khlebodarova, T.M.; Bogacheva, N.V.; Zadorozhny, A.V.; Bryanskaya, A.V.; Vasilieva, A.R.; Chesnokov, D.O.; Pavlova, E.I.; Peltek, S.E. Komagataella phaffii as a platform for heterologous expression of enzymes used for Industry. Microorganisms 2024, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Chen, Z.; Chen, Y.; Wang, X.; Zhang, Y.; Li, X.; Wang, F. Multiple strategies for high-efficiency expression of Thermomyces lanuginosus lipase in Pichia pastoris and production of biodiesel in solvent-free system. Fuel 2023, 333, 126246. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Heterologous protein expression in Pichia pastoris: Latest research progress and applications. ChemBioChem 2018, 19, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Pal, M.; Sharma, R.K. Pichia as yeast cell factory for production of industrially important bio-products: Current trends, challenges, and future prospects. J. Bioresour. Bioprod. 2022, 7, 209–218. [Google Scholar] [CrossRef]
- Barone, G.D.; Emmerstorfer-Augustin, A.; Biundo, A.; Pisano, I.; Coccetti, P.; Mapelli, V.; Camattari, A. Industrial production of proteins with Pichia pastoris—Komagataella phaffii. Biomolecules 2023, 13, 1085. [Google Scholar] [CrossRef]
- Unver, Y.; Dagci, I. Komagataella phaffii (Pichia pastoris) as a powerful yeast expression system for biologics broduction. Front. Biosci. (Elite Ed) 2024, 16, 19. [Google Scholar] [CrossRef]
- Lv, W.; Cai, M. Advancing recombinant protein expression in Komagataella phaffii: Opportunities and challenges. FEMS Yeast Res. 2025, 25, foaf010. [Google Scholar] [CrossRef]
- Looser, V.; Bruhlmann, B.; Bumbak, F.; Stenger, C.; Costa, M.; Camattari, A.; Fotiadis, D.; Kover, K. Cultivation strategies to enhance productivity of Pichia pastoris: A review. Biotechnol. Adv. 2015, 33, 1177–1193. [Google Scholar] [CrossRef]
- Liu, W.C.; Zhou, F.; Xia, D.; Shiloach, J. Expression of multidrug transporter P-glycoprotein in Pichia pastoris affects the host’s methanol metabolism. Microb. Biotechnol. 2019, 12, 1226–1236. [Google Scholar] [CrossRef]
- Cregg, J.M. Expression in the methylotrophic yeast Pichia pastoris. In Gene expression systems: Using nature e for the art of expression; Fernandez, J., Hoeffler, J., Eds.; Academic Press: San Diego, CA, USA, 1998; pp. 157–191. [Google Scholar]
- Ruo, P.A.; Leong, S.T.; Mooi, K.C.; Noorizan, M.; Boon, Y. Comparing the expression of human DNA topoisomerase I in KM71H and X33 strains of Pichia pastoris. Electron. J. Biotechnol. 2016, 21, 9–17. [Google Scholar] [CrossRef]
- Mayson, B.E.; Kilburn, D.G.; Zamost, B.L.; Raymond, C.K.; Lesnicki, G.J. Effects of methanol concentration on expression levels of recombinant protein in fed-batch cultures of Pichia methanolica. Biotechnol. Bioeng. 2003, 81, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Pedro, A.Q.; Oppolzer, D.; Bonifácio, M.J.; Maia, C.J.; Queiroz, J.A.; Passarinha, L.A. Evaluation of MutS and Mut+ Pichia pastoris strains for membrane-bound catechol-O-methyltransferase biosynthesis. Appl. Biochem. Biotechnol. 2015, 18, 3840–3855. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Gao, M.; Yan, J.; Chen, S.; Sun, J.; Hua, Q.; Ding, J.; Shi, Z. Evaluation of the sub-optimal induction strategies for heterologous proteins production by Pichia pastoris Mut+/MutS strains and related transcriptional and metabolic analysis. World J. Microbiol. Biotechnol. 2018, 34, 180. [Google Scholar] [CrossRef]
- Castro-Rodríguez, J.A.; Rodríguez-Sotres, R.; Farres, A. Determinants for an efficient enzymatic catalysis in Poly(Ethylene Terephthalate) degradation. Catalysts 2023, 13, 591. [Google Scholar] [CrossRef]
- Pio, T.F.; Macedo, G.A. Chapter 4 Cutinases: Properties and industrial applications. Adv. Appl. Microbiol. 2009, 66, 77–95. [Google Scholar] [CrossRef]
- Oliveira de Torres, C.; de Assis, M.A.; Mazutti, M.A.; Guimarães Pereira, G.C.; de Oliveira, D. Production of recombinant cutinases and their potential applications in polymer hydrolysis: The current status. Process Biochem. 2023, 134, 30–46. [Google Scholar] [CrossRef]
- Arnau, C.; Ramon, R.; Casas, C.; Valero, F. Optimization of the heterologous production of a Rhizopus oryzae lipase in Pichia pastoris system using mixed substrates on controlled fed-batch bioprocess. Enzym. Microb. Technol. 2010, 46, 494–500. [Google Scholar] [CrossRef]
- Wu, D.; Yu, X.W.; Wang, T.C.; Wang, R.; Xu, Y. High yield Rhizopus chinenisis prolipase production in Pichia pastoris: Impact of methanol concentration. Biotechnol. Bioprocess Eng. 2011, 16, 305–311. [Google Scholar] [CrossRef]
- Dutta, K.; Sen, S.; Veeranki, V.D. Production, characterization and applications of microbial cutinases. Process Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Nimchua, T.; Punnapayak, H.; Zimmermann, W. Comparison of the hydrolysis of polyethylene terephthalate fibers by a hydrolase from Fusarium oxysporum LCH I and Fusarium solani f. sp. pisi. Biotechnol. J. 2007, 2, 361–364. [Google Scholar] [CrossRef]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase structure, function and biocatalytic applications. Electron. J. Biotechnol. 1998, 1, 160–173. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hooks, D.; Brightwell, G. Current understanding on the heterogenous expression of plastic depolymerising enzymes in Pichia pastoris. Bioengineering 2025, 12, 68. [Google Scholar] [CrossRef] [PubMed]
- Koschorreck, K.; Liu, D.; Kazenwadel, C.; Schmid, R.D.; Hauer, B. Heterologous expression, characterization and site-directed mutagenesis of cutinase CUTAB1 from Alternaria brassicicola. Appl. Microbiol. Biotechnol. 2010, 87, 991–997. [Google Scholar] [CrossRef]
- Farrés, A.; Peña-Montes, C.; Hernández-Domínguez, E.E.; Morales-García, S.L.; Sánchez-Sánchez, M.; Solís-Báez, I. Cutinasas Recombinantes de Aspergillus Nidulans Para Degradación de Poliésteres. WO-2017/204615 A2, 30 November 2017. [Google Scholar]
- Alvarado, E.; Castro, R.; Castro-Rodríguez, J.A.; Navarro, A.; Farres, A. Poly(lacticacid) degradation by recombinant cutinases from Aspergillus nidulans. Polymers 2024, 16, 1994. [Google Scholar] [CrossRef]
- Ahmed Al-Tammar, K.; Omar, O.; Abdul Murad, A.M.; Abu Bakar, F.D. Expression and characterization of a cutinase (AnCUT2) from Aspergillus niger. Open Life Sci. 2016, 11, 29–38. [Google Scholar] [CrossRef]
- Nyyssölä, A.; Pihlajaniemi, V.; Järvinen, R.; Mikander, S.; Kontkanen, H.; Kruus, K.; Kallio, H.; Buchert, J. Screening of microbes for novel acidic cutinases and cloning and expression of an acidic cutinase from Aspergillus niger CBS 513.88. Enzym. Microb. Technol. 2013, 52, 272–278. [Google Scholar] [CrossRef]
- Shirke, A.N.; Basore, D.; Butterfoss, G.L.; Bonneau, R.; Bystroff, C.; Gross, R.A. Toward rational thermostabilization of Aspergillus oryzae cutinase: Insights into catalytic and structural stability. PROTEINS: Struct. Funct. Bioinform. 2016, 84, 60–72. [Google Scholar] [CrossRef]
- Murguiondo, C.; Barriuso, J.; Prieto, A. Optimized enzymatic PLA hydrolysis by a recombinant fungal cutinase: A step toward a closed PLA cycle. Int. J. Biol. Macromol. 2025, 301, 140482. [Google Scholar] [CrossRef]
- Hu, X.; Zhaoying, G.; Zhanyong, W.; Tingting, S.; Lei, Y.; Ping, L. Enzymatic degradation of poly(butylene succinate) by cutinase cloned from Fusarium solani. Polym. Degrad. Stab. 2016, 134, 211–219. [Google Scholar] [CrossRef]
- Kwon, M.A.; Kim, H.S.; Yang, T.H.; Song, B.K.; Song, J.K. High-level expression and characterization of Fusarium solani cutinase in Pichia pastoris. Protein Expr. Purif. 2009, 68, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Lienqueo, M.E.; Salazar, O.; Calado, C.R.C.; Fonseca, L.P.; Cabral, J.M.S. Influence of tryptophan tags on the purification of cutinase, secreted by a recombinant Saccharomyces cerevisiae, using cationic expanded bed adsorption and hydrophobic interaction chromatography. Biotechnol. Lett. 2008, 30, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Liu, C.; Zhang, W.; Qu, M.; Li, Y.; Zang, Y.; Xiong, X.; Pan, K.; Zhao, X. Characteristics of a recombinant Fusarium verticillioides cutinase and its effects on enzymatic hydrolysis of rice straw. Int. J. Biol. Macromol. 2021, 171, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Seman, W.M.K.W.; Bakar, S.A.; Bukhari, N.A.; Gaspar, S.M.; Othman, R.; Nathan, S.; Mahadi, N.M.; Jahim, J.; Murad, A.M.A.; Bakar, F.D.A. High level expression of Glomerella cingulata cutinase in dense cultures of Pichia pastoris grown under fed-batch conditions. J. Biotechnol. 2014, 184, 219–228. [Google Scholar] [CrossRef]
- Hong, R.; Sun, Y.; Su, L.; Gu, L.; Wang, F.; Wu, J. High-level expression of Humicola insolens cutinase in Pichia pastoris without carbon starvation and its use in cotton fabric bioscouring. J. Biotechnol. 2019, 304, 10–15. [Google Scholar] [CrossRef]
- Duan, X.; Liu, Y.; You, X.; Jiang, Z.; Yang, S.; Yang, S. High-level expression and characterization of a novel cutinase from Malbranchea cinnamomea suitable for butyl butyrate production. Biotechnol. Biofuels 2017, 10, 223. [Google Scholar] [CrossRef]
- Wang, G.Y.; Michailides, T.J.; Hammock, B.D.; Lee, Y.-M.; Bostock, R.M. Molecular cloning, characterization, and expression of a redox-responsive cutinase from Monilinia fructicola (Wint.) Honey. Fungal Genet. Biol. 2002, 35, 261–276. [Google Scholar] [CrossRef]
- Vázquez-Alcántara, L.; Oliart-Ros, R.M.; García-Bórquez, A.; Peña-Montes, C. Expression of a Cutinase of Moniliophthora roreri with Polyester and PET-Plastic Residues Degradation Activity. Microbiol. Spectr. 2021, 9, e0097621. [Google Scholar] [CrossRef]
- Nyyssölä, A.; Pihlajaniemi, V.; Häkkinen, M.; Kontkanen, H.; Saloheimo, M.; Nakari-Setälä, T. Cloning and characterization of a novel acidic cutinase from Sirococcus conigenus. Appl. Microbiol. Biotechnol. 2014, 98, 3639–3650. [Google Scholar] [CrossRef]
- Gamerith, C.; Vastano, M.; Ghorbanpour, S.M.; Zitzenbacher, S.; Ribitsch, D.; Zumstein, M.T.; Sander, M.; Acero, E.H.; Pellis, A.; Guebitz, G.M. Enzymatic degradation of aromatic and aliphatic polyesters by P. pastoris expressed cutinase 1 from Thermobifida cellulosilytica. Front. Microbiol. 2017, 8, 938. [Google Scholar] [CrossRef] [PubMed]
- Carbios, S.B. Method for Recycling Plastic Products. U.S. Patent 10, 512 B2, 28 March 2019. [Google Scholar]
- Ulaganathan, N.; Dhodduraj, K.; Chandukishore, T.; Prabhu, A.A. Production of thermostable recombinant cutinase using mixed food waste: A sustainable approach toward environmental remediation. Chem. Eng. J. 2024, 496, 153892. [Google Scholar] [CrossRef]
- Su, L.; Hong, R.; Wu, J. Enhanced extracellular expression of gene-optimized Thermobifida fusca cutinase in Escherichia coli by optimization of induction strategy. Process Biochem. 2015, 50, 1039–1046. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Z.; Chen, J.; Wu, J. Study on improvement of extracellular production of recombinant Thermobifida fusca cutinase by Escherichia coli. Appl. Biochem. Biotechnol. 2011, 165, 666–675. [Google Scholar] [CrossRef]
- Shirke, A.N.; Su, A.; Jones, J.A.; Butterfoss, G.L.; Koffas, M.A.G.; Kim, J.R.; Gross, R.A. Comparative thermal inactivation analysis of Aspergillus oryzae and Thiellavia terrestris cutinase: Role of glycosylation. Biotechnol. Bioeng. 2016, 114, 63–73. [Google Scholar] [CrossRef]
- Liu, M.; Yang, S.; Long, L.; Cao, Y.; Ding, S. Engineering a chimeric lipase-cutinase (Lip-Cut) for efficient enzymatic deinking of waste paper. BioResources 2018, 13, 981–996. [Google Scholar] [CrossRef]
- Duan, X.; Jiang, Z.; Liu, Y.; Yan, Q.; Xiang, M.; Yang, S. High-level expression of codon-optimized Thielavia terrestris cutinase suitable for ester biosynthesis and biodegradation. Int. J. Biol. Macromol. 2019, 135, 768–775. [Google Scholar] [CrossRef]
- Castro-Rodríguez, J.A.; Ramírez-Gonzalez, K.F.; Franco-Guerrero, F.; Sabido-Ramos, A.; Abundio-Sanchez, I.F.; Rodríguez-Sotres, R.; Rodríguez-Romero, A.; Farres, A. ANCUT1, a fungal cutinase MgCl2 -activated by a non-essential activation mechanism for Poly(ethylene terephthalate) hydrolysis. Catalysts 2025, 15, 757. [Google Scholar] [CrossRef]
- Li, X.; Shi, B.; Huang, J.W.; Zeng, Z.; Yang, Y.; Zhang, L.; Min, J.; Chen, C.C.; Guo, R.T. Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris. Bioresour. Bioprocess. 2023, 10, 26. [Google Scholar] [CrossRef]
- Fritzsche, S.; Hübner, H.; Oldiges, M.; Castiglione, K. Comparative evaluation of the extracellular production of a polyethylene terephthalate degrading cutinase by Corynebacterium glutamicum and leaky Escherichia coli in batch and fed-batch processes. Microb. Cell Fact. 2024, 23, 274. [Google Scholar] [CrossRef]
- Ma, H.-N.; Hsiang, C.-C.; Ng, I.-S. Tailored expression of ICCM cutinase in engineered Escherichia coli for efficient polyethylene terephthalate hydrolysis. Enzym. Microb. Technol. 2024, 179, 110476. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Sulaiman, S.; You, D.J.; Kanaya, E.; Koga, Y.; Kanaya, S. Crystal structure and thermodynamic and kinetic stability of metagenome- derived LC-cutinase. Biochemistry 2014, 53, 1858–1869. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-R.-; Jang, Y.A.; Song, J.K.; Eom, G.T. Secretory production of an engineered cutinase in Bacillus subtilis for efficient biocatalytic depolymerization of polyethylene terephthalate. Bioprocess. Biosyst. Eng. 2022, 45, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Shirke, A.N.; White, C.; Englaender, J.A.; Zwarycz, A.; Butterfoss, G.L.; Linhardt, R.J.; Gross, R.A. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: Mechanism and effect on PET hydrolysis. Biochemistry 2018, 57, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Krishnamoorthy, H.; Dasu, V.V. Novel cutinase from Pseudomonas cepacia NRRL B 2320: Purification, characterization and identification of cutinase encoding genes. J. Gen. Appl. Microbiol. 2013, 59, 171–184. [Google Scholar] [CrossRef]
- Bermúdez-García, E.; Peña-Montes, C.; Castro-Rodríguez, J.A.; González-Canto, A.; Navarro-Ocaña, A.; Farrés, A. ANCUT2, a Thermo-alkaline cutinase from Aspergillus nidulans and its potential applications. Appl. Biochem. Biotechnol. 2017, 182, 1014–1036. [Google Scholar] [CrossRef]
- Bermúdez-García, E.; Peña-Montes, C.; Martins, I.; Pais, J.; Pereira, C.; Sánchez, S.; Farrés, A. Regulation of the cutinases expressed by Aspergillus nidulans and evaluation of their role in cutin degradation. Appl. Microbiol. Biotechnol. 2019, 103, 3863–3874. [Google Scholar] [CrossRef]
- Peña-Montes, C.; Bermúdez-García, E.; Castro-Ochoa, D.; Vega-Pérez, F.; Esqueda-Domínguez, K.; Castro-Rodríguez, J.A.; González-Canto, A.; Segoviano-Reyes, L.; Navarro-Ocaña, A.; Farrés, A. ANCUT1, a novel thermoalkaline cutinase from Aspergillus nidulans and its application on hydroxycinnamic acids lipophilization. Biotechnol. Lett. 2024, 46, 409–430. [Google Scholar] [CrossRef]
- Llanos-Reyes, A.C. Aplicación de las Cutinasas Recombinantes Ancut3 y Ancut4 Provenientes de Aspergillus nidulans en la Degradación de Poliésteres. Master’s Thesis, Universidad Nacional Autónoma de México, Ciudad de México, México, 2018. [Google Scholar]
- Lovera-Espinosa, D.; Torres-Jiménez, M.K. Optimización de Condiciones Para Mejorar la Producción y Degradación del Tereftalato de Polietileno (PET) por la Cutinasa ANCUT3 Expresada en Pichia pastoris. Bachelor’s Thesis, Universidad Nacional Autónoma de México, Ciudad de México, México, 2019. [Google Scholar]
- Castro-Ochoa, D.; Peña-Montes, C.; González-Canto, A.; Alva- Gasca, A.; Esquivel-Bautista, R.; Navarro-Ocaña, A.; Farrés, A. ANCUT2, an extracellular cutinase from Aspergillus nidulans induced by olive oil. Appl. Biochem. Biotechnol. 2012, 166, 1275–1290. [Google Scholar] [CrossRef] [PubMed]
- Invitrogen. pPICZα A, B, and C Pichia Expression Vectors for Selection on ZeocinTM and Purification of Secreted, Recombinant Proteins; Catalog, (V195-20): Carlsbad, CA, USA, 2010; pp. 1–38. Available online: https://tools.thermofisher.com/content/sfs/manuals/ppiczalpha_man.pdf (accessed on 10 July 2025).
- Esqueda-Domínguez, K.L. Producción, Identificación y Caracterización de una Cutinasa de Aspergillus nidulans PWI. Master’s Thesis, Universidad Nacional Autónoma de México, Ciudad de México, México, 2012. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Inan, M.; Meagher, M.M. Fermentation strategies for recombinant protein expression in the methylotrophic yeast Pichia pastoris. Biotechnol. Bioprocess. Eng. 2000, 5, 275–287. [Google Scholar] [CrossRef]
- Chahinian, H.; Sarda, L. Distinction between esterases and lipases: Comparative biochemical properties of sequence-related carboxylesterases. Protein Pept. Lett. 2009, 16, 1149–1161. [Google Scholar] [CrossRef]
- Higgins, D.R.; Cregg, J.M. Methods in Molecular Biology: Pichia Protocols; Humana Press: Totowa, NJ, USA, 1998; pp. 1–15. [Google Scholar]
- Viader-Salvado, J.M.; Castillo-Galvan, M.; Fuentes-Garibay, J.A.; Iracheta-Cardenas, M.M.; Guerrero- Olazaran, M. Optimization of five environmental factors to increase beta-propeller phytase production in Pichia pastoris and impact on the physiological response of the host. Biotechnol. Prog. 2013, 29, 1377–1385. [Google Scholar] [CrossRef]
- Orman, M.A.; Çalık, P.; Özdamar, T.H. The influence of carbon sources on recombinant-human- growth-hormone production by Pichia pastoris is dependent on phenotype: A comparison of Muts and Mut+ strains. Biotechnol. Appl. Biochem. 2009, 52, 245–255. [Google Scholar] [CrossRef]
- Pla, I.A.; Damasceno, L.M.; Vannelli, T.; Ritter, G.; Batt, C.; Shuler, M.L. Evaluation of Mut+ and MutS Pichia pastoris phenotypes for high level extracellular scFv expression under feedback control of the methanol concentration. Biotechnol. Prog. 2006, 22, 881–888. [Google Scholar] [CrossRef]
- Yang, J.; Cai, H.; Liu, J.; Zeng, M.; Chen, J.; Cheng, Q.; Zhang, L. Controlling AOX1 promoter strength in Pichia pastoris by manipulating poly (dA:dT) tracts. Sci. Rep. 2018, 8, 1401. [Google Scholar] [CrossRef]

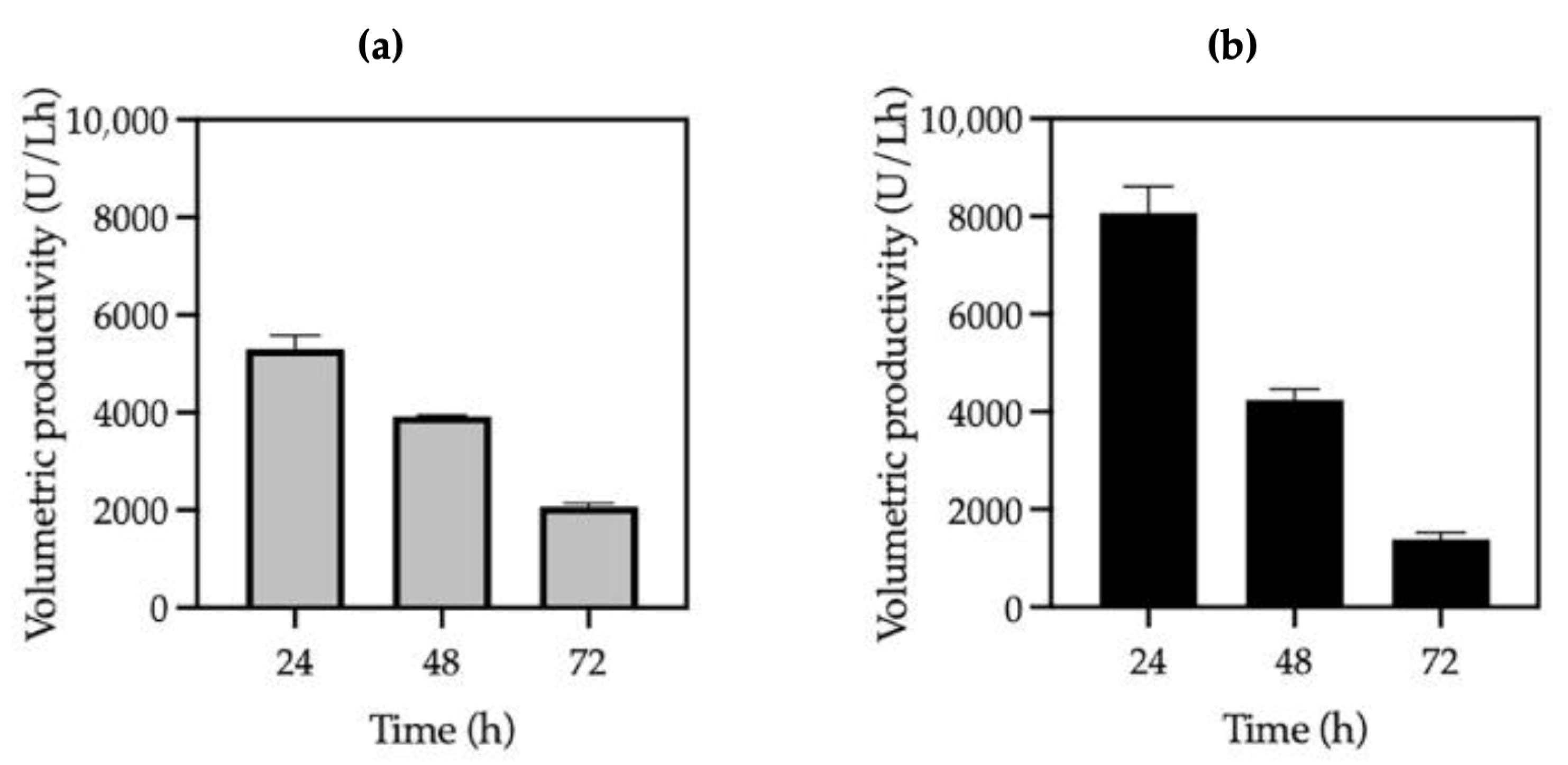
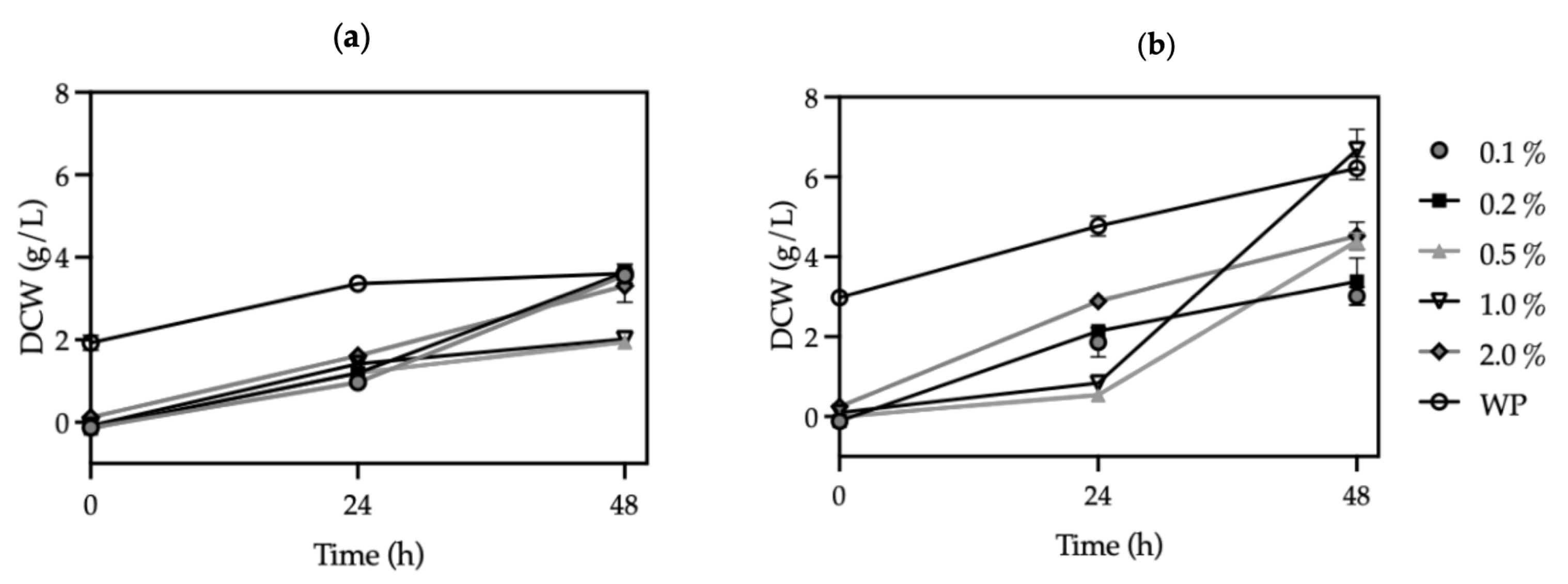
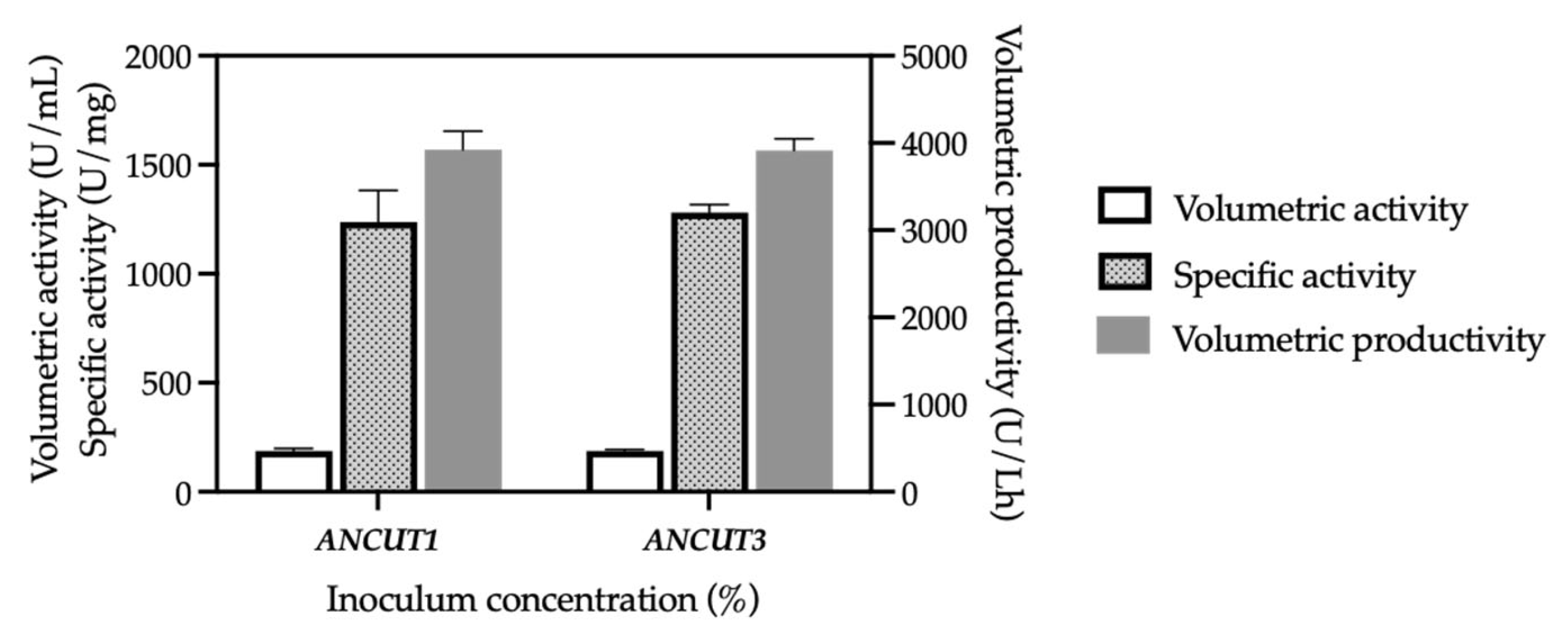

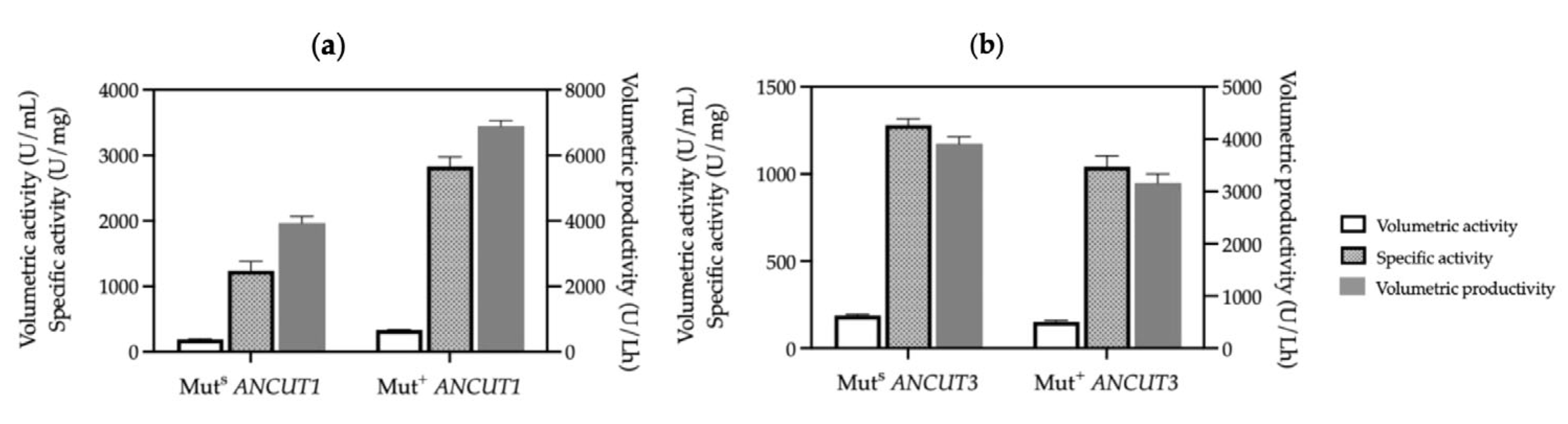
| Strain | Methanol (%) | Optical Density (600 nm) | Biomass (g/L) | Protein Concentration (mg/mL) | Volumetric Activity (U/mL) | Specific Activity (U/mg) | Productivity (U/Lh) |
|---|---|---|---|---|---|---|---|
| KM71H aox1::pPICZα-A-ANCUT1 | 0.5 | 7.78 ± 0.24 | 3.89 ± 0.13 | 0.18 ± 0.00 | 188 ± 1 | 1037 ± 33 | 3917 ± 30 |
| 1.0 | 8.46 ± 0.52 | 4.11 ± 0.27 | 0.17 ± 0.01 | 182 ± 7 | 1077 ± 87 | 3793 ± 148 | |
| 1.5 | 8.19 ± 1.35 | 4.10 ± 0.70 | 0.15 ± 0.00 | 80 ± 2 | 535 ± 41 | 1673 ± 46 | |
| 2.0 | 6.17 ± 0.47 | 3.05 ± 0.24 | 0.17 ± 0.00 | 86 ± 1 | 499 ± 7 | 1792 ± 25 | |
| 3.0 | 5.26 ± 0.17 | 2.58 ± 0.09 | 0.15 ± 0.00 | 80 ± 7 | 523 ± 70 | 1673 ± 141 | |
| KM71H aox1::pPICZα-A-ANCUT3 | 0.5 | 8.97 ± 1.56 | 4.51 ± 0.81 | 0.10 ± 0.00 | 132 ± 8 | 1262 ± 60 | 2729 ± 167 |
| 1.0 | 12.65 ± 2.07 | 6.42 ± 1.08 | 0.10 ± 00 | 158 ± 9 | 1666 ± 223 | 3271 ± 189 | |
| 1.5 | 12.21 ± 0.11 | 6.20 ± 0.06 | 0.09 ± 0.00 | 204 ± 10 | 2184 ± 48 | 4250 ± 208 | |
| 2.0 | 10.91 ± 0.28 | 5.52 ± 0.15 | 0.10 ± 0.00 | 174 ± 12 | 1721 ± 172 | 3625 ± 250 | |
| 3.0 | 6.84 ± 0.23 | 3.40 ± 0.12 | 0.09 ± 0.00 | 113 ± 11 | 1129 ± 145 | 2354 ± 229 |
| Strain | Inoculum (%) | Optical Density (600 nm) | Biomass (g/L) | Protein Concentration (mg/mL) | Volumetric Activity (U/mL) | Specific Activity (U/mg) | Productivity (U/Lh) |
|---|---|---|---|---|---|---|---|
| KM71H aox1::pPICZα-A-ANCUT1 (0.5% methanol) | 0.1 | 7.14 ± 0.25 | 3.56 ± 0.13 | 0.13 ± 0.00 | 99 ± 12 | 779 ± 106 | 2068 ± 247 |
| 0.2 | 7.27 ± 0.29 | 3.63 ± 0.15 | 0.13 ± 0.00 | 140 ± 4 | 1106 ± 71 | 2910 ± 77 | |
| 0.5 | 8.06 ± 0.16 | 4.04 ± 0.08 | 0.15 ± 0.01 | 188 ± 10 | 1237 ± 145 | 3921 ± 213 | |
| 1.0 | 8.32 ± 0.18 | 4.17 ± 0.09 | 0.16 ± 0.02 | 179 ± 9 | 1139 ± 107 | 3720 ± 187 | |
| 2.0 | 13.10 ± 1.47 | 6.66 ± 0.77 | 0.28 ± 0.01 | 99 ± 6 | 357 ± 35 | 2073 ± 131 | |
| WP | 14.23 ± 0.86 | 7.25 ± 0.45 | 0.10 ± 0.01 | 99 ± 6 | 1022 ± 132 | 2073 ± 131 | |
| KM71H aox1::pPICZα-A-ANCUT3 (1.5% methanol) | 0.1 | 6.10 ± 0.44 | 3.02 ± 0.23 | 0.14 ± 0.00 | 114 ± 5 | 839 ± 44 | 2369 ± 109 |
| 0.2 | 6.8 ± 1.13 | 3.38 ± 0.59 | 0.13 ± 0.00 | 90 ± 5 | 688 ± 54 | 1867 ± 106 | |
| 0.5 | 8.76 ± 0.40 | 4.40 ± 0.21 | 0.11 ± 0.00 | 110 ± 8 | 1018 ± 97 | 2294 ± 163 | |
| 1.0 | 13.15 ± 0.98 | 6.68 ± 0.51 | 0.10 ± 0.01 | 116 ± 7 | 1155 ± 125 | 2413 ± 137 | |
| 2.0 | 8.99 ± 0.66 | 4.52 ± 0.35 | 0.15 ± 0.00 | 188 ± 6 | 1281 ± 36 | 3916 ± 131 | |
| WP | 12.25 ± 0.55 | 6.22 ± 0.28 | 0.11 ± 0.00 | 101 ± 7 | 905 ± 47 | 2096 ± 150 |
| Strain | Methanol (%) | Inoculum (%) | Optical Density (600 nm) | Biomass (g/L) | Protein Concentration (mg/mL) | Volumetric Activity (U/mL) | Specific Activity (U/mg) | Productivity (U/Lh) |
|---|---|---|---|---|---|---|---|---|
| KM71H ANCUT1 | 0.5 | 0.5 | 8.06 ± 0.16 | 4.04 ± 0.08 | 0.15 ± 0.01 | 188 ± 10 | 1237 ± 145 | 3921 ± 213 |
| X-33ANCUT1 | 9.42 ± 0.58 | 4.74 ± 0.30 | 0.12 ± 0.00 | 331 ± 8 | 2831 ± 144 | 6887 ± 176 | ||
| KM71H ANCUT3 | 1.5 | 2.0 | 8.99 ± 0.66 | 4.52 ± 0.35 | 0.15 ± 0.00 | 188 ± 6 | 1281 ± 36 | 3916 ± 131 |
| X-33ANCUT3 | 15.19 ± 0.87 | 7.74 ± 0.45 | 0.15 ± 0.00 | 152 ± 8 | 1043 ± 61 | 3159 ± 175 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabido-Ramos, A.; Tagle-Gil, M.; León-Montes, K.E.; Castro-Rodríguez, J.A.; Farrés, A. Cutinase Production in Komagataella phaffii (Pichia pastoris): Performance Differences Between Host Strains. Fermentation 2025, 11, 483. https://doi.org/10.3390/fermentation11080483
Sabido-Ramos A, Tagle-Gil M, León-Montes KE, Castro-Rodríguez JA, Farrés A. Cutinase Production in Komagataella phaffii (Pichia pastoris): Performance Differences Between Host Strains. Fermentation. 2025; 11(8):483. https://doi.org/10.3390/fermentation11080483
Chicago/Turabian StyleSabido-Ramos, Andrea, Montserrat Tagle-Gil, Krystel Estefany León-Montes, José Augusto Castro-Rodríguez, and Amelia Farrés. 2025. "Cutinase Production in Komagataella phaffii (Pichia pastoris): Performance Differences Between Host Strains" Fermentation 11, no. 8: 483. https://doi.org/10.3390/fermentation11080483
APA StyleSabido-Ramos, A., Tagle-Gil, M., León-Montes, K. E., Castro-Rodríguez, J. A., & Farrés, A. (2025). Cutinase Production in Komagataella phaffii (Pichia pastoris): Performance Differences Between Host Strains. Fermentation, 11(8), 483. https://doi.org/10.3390/fermentation11080483







