1. Introduction
Ensiling is a widely adopted method for preserving high-moisture forages, allowing long-term storage while maintaining their nutritional quality for the feeding of ruminants. In tropical and subtropical regions, demand for livestock feed has driven rapid expansion of high-yield grasses such as king grass, sorghum–Sudangrass hybrids, and elephant grass. However, these forages vary greatly in dry matter (DM), water-soluble carbohydrates (WSCs), and fiber structure, leading to inconsistent fermentation performance and stability under tropical conditions [
1,
2].
Despite advances in silage technology, many tropical grasses still exhibit delayed pH decline, excessive ammonia-N (NH
3-N), and formation of undesirable compounds like butyric acid under high-temperature, high-humidity environments [
3,
4]. These issues often arise from insufficient lactic acid bacteria (LAB) populations and proliferation of undesirable microbes such as Clostridium, yeasts, and spoilage fungi. While LAB inoculants improve fermentation in temperate crops like maize and alfalfa [
5,
6], limited research compares microbial dynamics across diverse tropical forages under uniform conditions.
Recent advances in high-throughput sequencing have provided powerful tools to explore bacterial and fungal succession during ensiling. Studies have shown microbial dominance shifts from aerobic epiphytes such as
Enterobacter and
Weissella to LAB species like
Lactobacillus plantarum and
L. buchneri during anaerobic fermentation [
7,
8]. However, most studies focus on a narrow forage range or single-species systems. Moreover, fungal communities—including both beneficial yeasts and spoilage molds—remain understudied, despite their roles in aerobic stability and nutrient loss [
9,
10]. Comprehensive investigations integrating bacterial and fungal community dynamics across multiple forage types are still lacking.
Parallel to microbiome studies, metabolomics—particularly untargeted metabolomics—has emerged as a promising approach for understanding fermentation-driven biochemical changes. This approach has identified key fermentation products such as organic acids, polyphenols, peptides, and amino acids that contribute to silages’ quality, aroma, and potential health benefits [
11]. For instance, chlorogenic acid, lactic acid, and γ-aminobutyric acid (GABA) have been associated with silages dominated by specific LAB species [
12]. However, systematic metabolomic comparisons across different tropical forages and their associations with microbial communities remain scarce.
In practice, even under identical ensiling conditions and using the same inoculants, some forage crops ferment well, while others spoil or exhibit visible fungal contamination, rendering them unsuitable for feeding. This variability suggests that the key determinant lies in the forage material itself rather than fermentation conditions. Intrinsic differences among forage species—such as composition of carbon sources, moisture retention, and epiphytic microbiota—likely play critical roles in shaping microbial succession and fermentation outcomes. To address this, we selected eight representative tropical forages—Sorghum bicolor (sweet sorghum), Sorghum × drummondii (sorghum–Sudangrass hybrid), Sorghum sudanense (Sudangrass), Pennisetum giganteum (giant Napier grass), Pennisetum purpureum cv. Purple (purple elephant grass), Pennisetum sinese (king grass), Leymus chinensis (sheep grass), and Zea mexicana (Mexican teosinte)—and conducted ensiling under standardized conditions. We evaluated fermentation characteristics, bacterial and fungal dynamics (16S rRNA and ITS sequencing), and metabolite profiles using untargeted metabolomics. This study introduces an integrative “forage–microbiota–metabolite” framework to elucidate how plant traits shape microbial ecology and metabolic activity during ensiling. By comparing microecological and metabolic features across forage types, we aim to identify species with superior ensiling potential and provide theoretical support for targeted inoculant strategies and predictive models for silage quality.
2. Materials and Methods
All eight forage species—sorghum–Sudangrass hybrid, purple elephant grass, Mexican teosinte, Sudan grass, giant Napier grass, sweet sorghum, sheep grass, and king grass—were cultivated and harvested from the experimental field of Xichang University, located in Anning Town, Liangshan, China (30°34′ N, 104°04′ E; elevation 1574 m). The region has a subtropical monsoon climate characterized by an average annual temperature of 16.8 °C, annual precipitation of 1000–1200 mm, and relative humidity above 75%. The soil at the experimental site is classified as sandy loam with moderate fertility (pH 6.5, organic matter 28.6 g/kg, total N 1.75 g/kg, available P 25.4 mg/kg, and available K 180 mg/kg). No chemical fertilizers or pesticides were applied during the growth period to avoid external influences on microbial communities.
All forages were harvested at the early heading stage or 55–65 days after sowing, a growth window commonly recognized as suitable for ensiling tropical grasses and consistent with local agronomic practices. At harvest, plant heights ranged from 0.8 m to 1.4 m. Aboveground parts, including stems, nodes, and leaves, were collected at a stubble height of 10–12 cm. Fresh forage was immediately chopped into lengths of approximately 10–20 mm using a high-speed forage chopper (Taizy Agro-Machinery Co., Ltd., Zhengzhou, China). No additives were applied to the silage. For each forage species, approximately 2 kg of fresh chopped material was packed into vacuum-sealable polyethylene bags (30 cm × 40 cm) and sealed with a commercial vacuum sealer (Zhejiang Dongfeng Packing Machine Co., Ltd., Wenzhou, China). Each treatment was performed in triplicate (n = 3), resulting in a total of 24 silage bags. All silage bags were stored at ambient room temperature (~25–28 °C) in a dark and dry environment for a fermentation period of 60 days. After the ensiling period, the bags were opened for evaluation of fermentation characteristics, composition of microbial community (bacterial and fungal), and metabolomic profiles.
The chemical compositions of both fresh and ensiled samples were analyzed, including moisture content, dry matter (DM), crude protein (CP), ether extract (EE), crude fiber (CF), neutral detergent fiber (NDF), acid detergent fiber (ADF), crude starch, crude ash, and water-soluble carbohydrates (WSCs). Moisture content was calculated by subtracting DM from 100%. DM was determined by oven-drying at 105 °C until constant weight (AOAC, 2016; Method 934.01) [
13]. CP was determined using the Kjeldahl method (AOAC, 2016; Method 984.13) [
13]. EE and crude ash were also analyzed following AOAC guidelines (AOAC, 2016; methods 920.39 and 942.05, respectively) [
13]. Crude starch was quantified using the anthrone–sulfuric acid method as described in a prior study [
14]. NDF and ADF were determined using the Van Soest method with heat-stable amylase and sodium sulfite, following the procedure described by He et al. [
15]. Water-soluble carbohydrate (WSC) content was analyzed using the phenol–sulfuric acid method as described by Dubois et al. [
16]. Fermentation quality was evaluated using aqueous extracts of silage. Briefly, 50 g of silage sample was homogenized with 180 mL of sterile distilled water in a blender for 1 min, incubated at 4 °C for 24 h, and filtered through four layers of medical gauze. The pH of the extract was measured using a pH meter (PHSJ-5; LEICI, Shanghai, China). Ammonia nitrogen (NH
4+-N) was determined by the phenol–hypochlorite method [
17]. Organic acids, including lactic acid (LA), acetic acid (AA), propionic acid (PA), and butyric acid (BA), were quantified using a high-performance liquid chromatographic apparatus (HPLC, Agilent Technologies Inc., Santa Clara, CA, USA) equipped with a UV detector and a C18 column, according to the procedure of Zeng et al. [
18].
Microbial DNA was extracted from silage samples using the E.Z.N.A.
® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA), following the manufacturer’s protocol, with minor modifications to accommodate plant-rich, acidic silage substrates. For each silage bag (2 kg), subsamples were collected from five evenly distributed points (top, bottom, left, right, and center), homogenized, and 0.5 g of the composite sample was used for extraction. Briefly, samples were mixed with lysis buffer and mechanically disrupted by bead beating for 10 min to ensure sufficient cell breakage. DNA purification was then carried out using silica spin columns and elution in 50 μL of elution buffer. The concentration and purity of extracted DNA were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Samples with a concentration ≥ 20 ng/μL, OD
60/
280 ratio of 1.8–2.0, and OD
60/
230 > 2.0 were considered qualified. Extracted DNA was diluted to 3.5 ng/μL and stored at −20 °C prior to PCR amplification. Full-length bacterial 16S rRNA genes and fungal ITS regions were amplified using barcode-tagged primers for PacBio SMRT sequencing. Amplicons were purified, quantified, normalized, and used to construct SMRTbell libraries. The library quality was validated before sequencing on the PacBio Sequel platform. Offline BAM files were processed using SMRT Link v13.1 software to generate CCS (circular consensus sequencing) files. Samples were demultiplexed using barcode sequences, and the resulting data were exported in FastQ format. Raw CCS reads were preprocessed using Lima v1.7.0 (
https://github.com/PacificBiosciences/barcoding (accessed on 16 April 2025)) for barcode identification and CCS read extraction. Cutadapt v1.9.1 [
19] was used to trim primer sequences and filter reads by length. Chimeric sequences were removed using UCHIME v4.2 [
20], yielding high-quality CCS reads. Amplicon sequence variants (ASVs) were generated using the DADA2 plugin in QIIME2 [
21], with reads observed fewer than two times across all samples being removed.
Alpha diversity metrics (Shannon and Chao1 indices) were calculated using QIIME2 v2020.6 and visualized with the ggplot2 package in R v4.2.2, evaluating species’ richness and evenness. Rarefaction curves were generated using the core_diversity_analyses.py script in QIIME2. Kruskal–Wallis tests were used to compare alpha diversity across groups, followed by Wilcoxon rank-sum tests for pairwise comparisons with false discovery rate correction. Beta diversity was calculated based on Bray–Curtis distance and visualized by principal coordinate analysis (PCoA) using the capscale function from the vegan package in R (model: capscale(log2(RA) ~ 1)). PERMANOVA was conducted using the adonis function with 999 permutations to test for differences in community composition. ANOSIM was also applied using the anosim function to evaluate group separation, with significance defined as
p < 0.05 [
22]. To further characterize community structure, genus-level abundance bar plots and UPGMA clustering were employed [
23]. One-way ANOVA was used to test the significance of taxonomic differences across groups.
Differentially abundant taxa across eight groups were identified using Linear Discriminant Analysis Effect Size (LEfSe, v1.1.1) [
24]. LEfSe analysis was conducted using the online Galaxy module (
https://huttenhower.sph.harvard.edu/galaxy/, accessed on 25 April 2025), with relative abundances of bacterial taxa as input. Taxonomic features were filtered at a minimum relative abundance of 0.1% across all samples. The Kruskal–Wallis test was used to detect significant differences between groups, followed by pairwise Wilcoxon tests. A logarithmic LDA score threshold of 4.0 was applied to select discriminative features. Spearman’s rank correlation analysis was performed, and the data with a correlation greater than 0.6 and
p-value < 0.05 were selected to construct the correlation network. The coexistence relationship of a species in environmental samples was obtained based on the analysis of network graphs.
Metabolites were extracted using a modified methanol/acetonitrile/water solution (2:2:1, v/v/v). Briefly, 50 mg of freeze-dried sample was mixed with 1000 μL of extraction solvent containing 2 μL of L-2-chlorophenylalanine (internal standard, Aladdin Reagent Co., Ltd., Shanghai, China), followed by vortexing. The mixture was homogenized using ceramic beads at 45 Hz for 10 min, then subjected to ultrasonication on ice for 10 min and incubated at −20 °C for 1 h. After centrifugation at 12,000 rpm for 15 min at 4 °C, 500 μL of the supernatant was diluted with LC–MS-grade water to adjust the final methanol concentration to 60%. The diluted extract was transferred to a new Eppendorf tube, filtered through a 0.22 μm membrane, and centrifuged again under the same conditions. Finally, 120 μL of the clarified supernatant was collected and transferred into 2 mL LC–MS vials for metabolomic analysis.
Metabolomic profiling was performed using a Waters UPLC I-Class PLUS system coupled to a Xevo G2-XS QTOF high-resolution mass spectrometer (Waters Corporation, Milford, MA, USA). Chromatographic separation was carried out on a Waters Acquity UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm). The mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B), used for both positive and negative ionization modes. A 2 μL injection volume was used under a programmed gradient elution [
25]. Mass spectrometry data were acquired in MSE mode using MassLynx V4.2 (Waters), enabling simultaneous collection of low- and high-energy fragment spectra. The collision energy was set to 2 eV for low-energy and ramped from 10 to 40 eV for high-energy acquisition. The scanning frequency was 0.2 s/spectrum. Electrospray ionization (ESI) parameters were as follows: capillary voltage, +2000 V (positive) or −1500 V (negative); cone voltage, 30 V; source temperature, 150 °C; desolvation temperature, 500 °C; cone gas flow, 50 L/h; and desolvation gas flow, 800 L/h.
Raw LC–MS data were processed using Progenesis QI V2.3 (Nonlinear Dynamics, UK) for peak detection, alignment, retention time correction, baseline filtering, and feature extraction. Parameters included precursor tolerance 5 ppm, product ion tolerance 10 ppm, and product ion threshold 5%. Features missing in >50% of samples were removed; zero values were replaced by half of the minimum positive value. Features with identification scores below 36/60 were excluded [
26]. Metabolite annotation was based on matching MS and MS/MS spectra against the METLIN database [
27] and an in-house spectral library (Biomarker Biotech, Beijing). Identification criteria included accurate
m/
z, secondary fragmentation patterns, and isotopic distribution, with tolerances of <100 ppm for precursor ions and <50 ppm for fragment ions. Prior to statistical analysis, data were log-transformed and Pareto-scaled. Differential metabolites were identified using a combination of fold change (FC ≥ 2 or ≤0.5),
t-test (
p < 0.05), and VIP > 1 from OPLS-DA modeling [
28]. Metabolites’ annotation was enriched via the KEGG database [
29], and pathway enrichment was conducted using MetaboAnalyst V5.0 [
30]. Multivariate analyses, including PCA and Spearman correlation, were used to assess metabolic variation and sample quality. For pathway-level inference, hypergeometric distribution tests were applied to evaluate the statistical significance of the KEGG pathway enrichment.
3. Results
3.1. Systematic Evaluation of Fermentation Quality and Nutrient Preservation in Silage from Eight Tropical Forage Crops
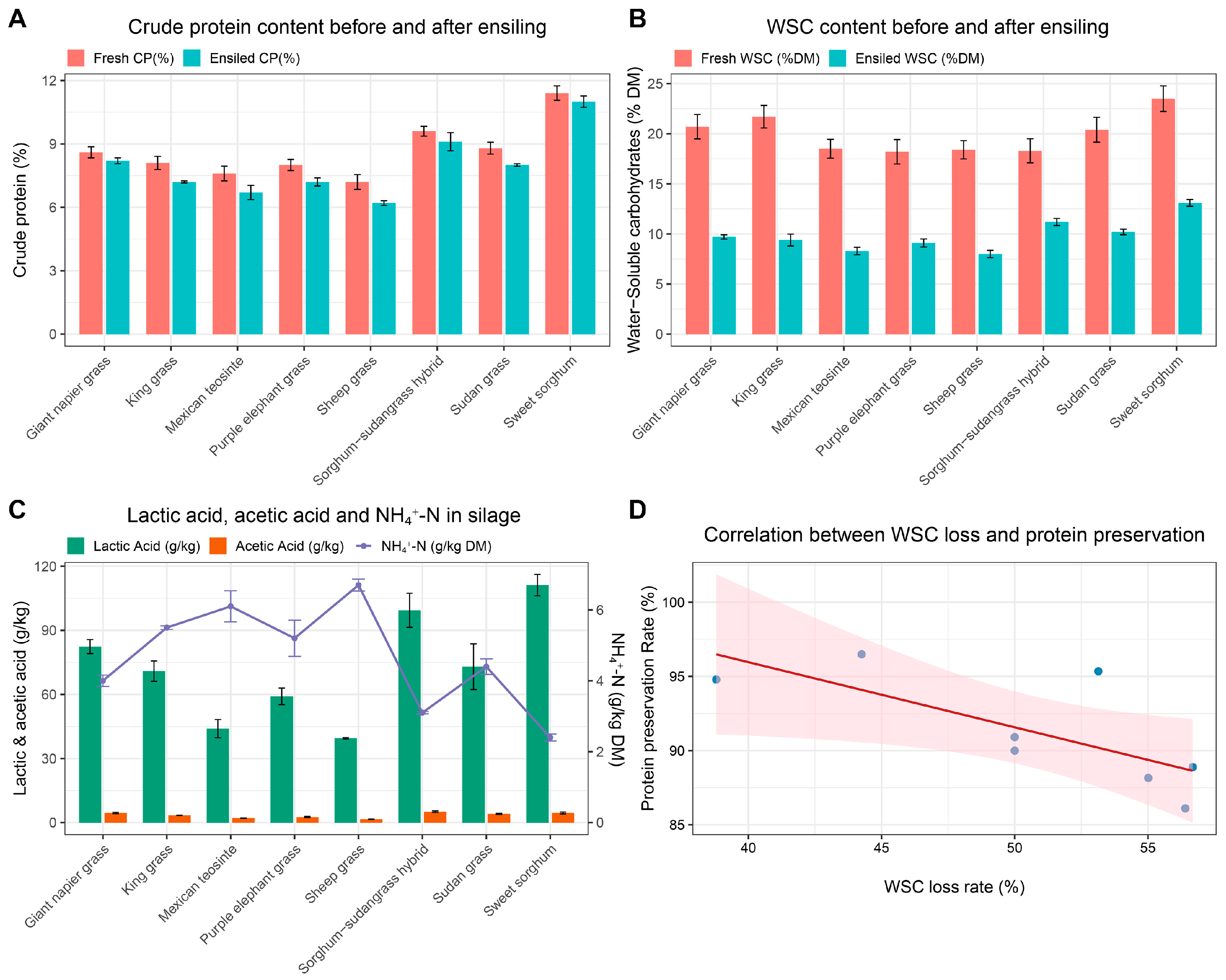
To evaluate the fermentation quality and nutrient preservation of eight tropical forages, we analyzed their physicochemical composition pre- and post-ensiling, along with their production of organic acid and nutrient conversion rates (
Table 1 and
Table 2). Prior to ensiling, notable differences were observed in moisture, crude protein (CP), water-soluble carbohydrates (WSCs), neutral detergent fiber (NDF), and acid detergent fiber (ADF). Sweet sorghum had the highest WSC content (23.5% DM) and a relatively high CP (11.4%). Giant Napier grass also showed a high CP (9.6%) but lower WSCs (18.3%). In contrast, sheep grass had the lowest WSC (18.4%) and CP (7.2%). Over half of the forages had a moisture content exceeding 80%, with sheep grass reaching 82%.
Post-ensiling, compositional shifts reflected forage-specific adaptability to anaerobic conditions (
Figure 1A,B). Sweet sorghum showed the best performance, retaining 11.0% CP and 13.1% WSC, with high lactic acid (111.2 g/kg) and the lowest NH
4+ level (2.4 g/kg DM). Giant Napier grass also retained good nutritional value (9.1% CP, 11.2% WSC) and exhibited low butyric acid (0.6 g/kg). Sheep grass, by contrast, showed the poorest fermentation: CP declined to 6.2%, WSC to 8.0%, with the highest NH
4+ (6.7 g/kg) and butyric acid (4.0 g/kg). Purple elephant grass and king grass showed moderate fermentation performance, with CP around 7.2% and WSC 9.1–9.4%. Although lactic acid levels were moderate and butyric acid remained low, the forages maintained a high fiber content (mainly CF and ADF values, e.g., ADF in purple elephant grass at 38.1%). Sorghum–Sudangrass hybrid and Sudan grass performed favorably, with good lactic acid production (82.4 g/kg and 73 g/kg), WSC retention (9.7% and 10.2%), and controlled NH
4+ levels (4.0 g/kg and 4.4 g/kg). Mexican teosinte showed the weakest fermentation capacity, with low WSC (initial 18.5%, residual 8.3%), minimal lactic acid (44.0 g/kg), high NH
4+ (6.1 g/kg), and substantial propionic acid (30.6 g/kg).
In the analysis of silage fermentation products, clear differences in fermentation patterns and nitrogen preservation emerged among the eight tropical forages based on the interrelationship between lactic acid, acetic acid, and ammonium nitrogen (NH
4+-N) content (
Figure 1C). Forages with high lactic acid concentrations, such as sweet sorghum (111.2 g/kg) and giant Napier grass (99.4 g/kg), exhibited lower NH
4+-N levels (2.4 and 3.1 g/kg DM), reflecting a pattern associated with reduced proteolysis and improved nitrogen retention. In contrast, sheep grass and Mexican teosinte produced less lactic acid (39.5 and 44.0 g/kg) and accumulated more NH
4+-N (6.7 and 6.1 g/kg DM), suggesting significant protein degradation. Moderate acetic acid levels were observed in sweet sorghum (5.1 g/kg) and giant Napier grass (4.5 g/kg). Conversely, sheep grass (1.6 g/kg) and Mexican teosinte (2.1 g/kg) showed a lower acetic acid content compared to other forages.
To further evaluate nutrient preservation, WSC loss and protein preservation rates were analyzed (
Figure 1D,
Supplementary Table S1). Sweet sorghum and giant Napier grass exhibited the lowest WSC losses (44.26% and 38.80%) and the highest protein preservation (96.49% and 94.79%), confirming superior preservation of both energy and nitrogen. By contrast, sheep grass and Mexican teosinte had greater WSC losses (56.40% and 55.01%) and lower protein preservation (86.11% and 88.16%). A significant negative correlation between WSC loss and protein retention (Pearson’s r = −0.730,
p = 0.040) further supports the role of WSC not only as a fermentation substrate but also as a key factor in nitrogen stabilization.
In summary, the eight forages differed markedly in fermentation quality and nutrient preservation. Sweet sorghum and giant Napier grass showed the most favorable profiles, while sheep grass and Mexican teosinte were more prone to fermentation failure and nutrient degradation. The remaining species exhibited an intermediate performance, with outcomes influenced by WSC availability, protein stability, and fiber characteristics.
3.2. Bacterial Diversity Profiling in Silages of Eight Tropical Forages
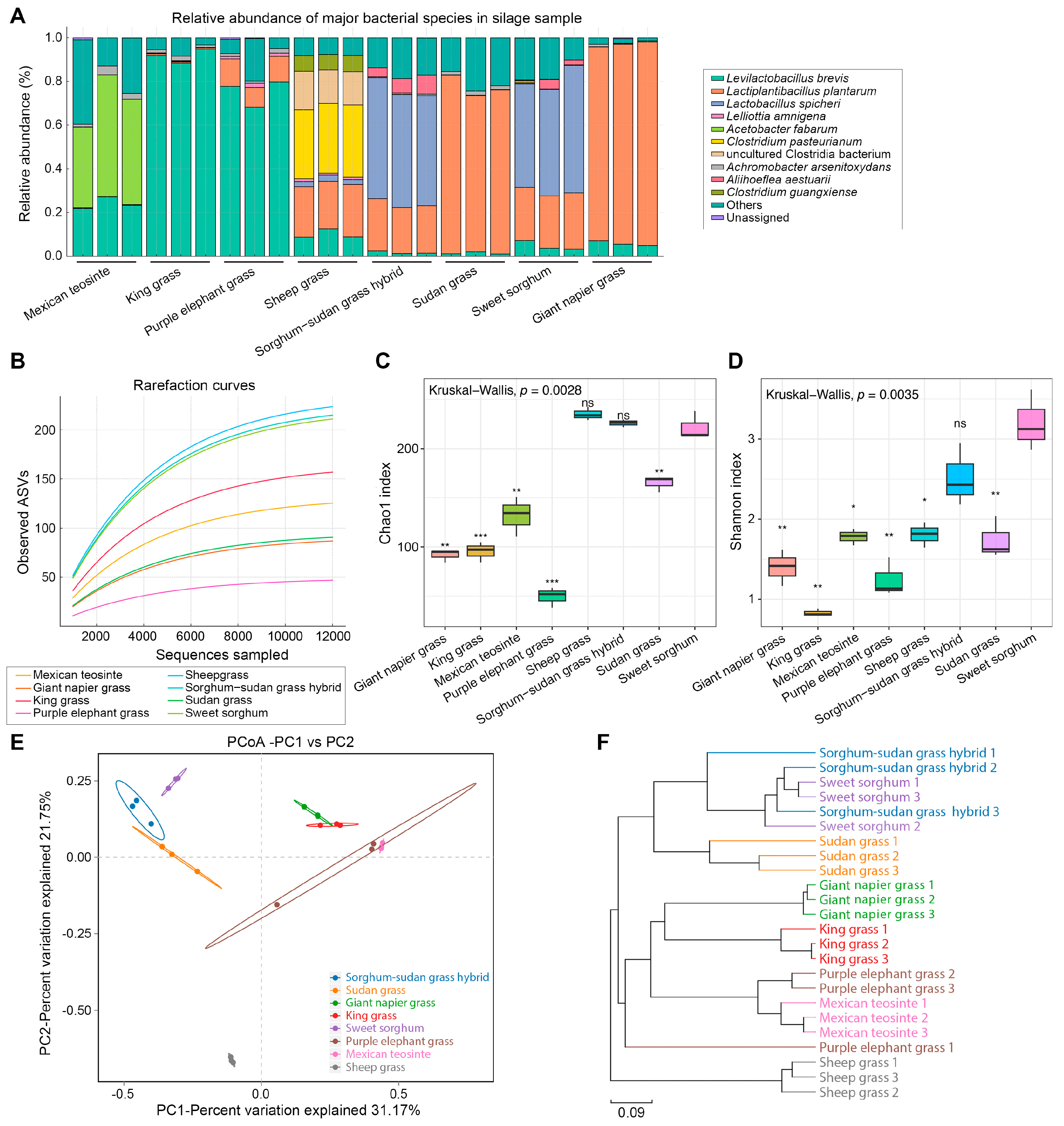
Full-length 16S rRNA gene sequencing was performed on 24 silage samples (triplicates from eight tropical forages) using PacBio circular consensus sequencing (CCS). A total of 287,621 high-quality CCS reads were obtained, with each sample yielding at least 8204 reads (average: 11,984), ensuring sufficient depth and coverage for robust microbial profiling. After DADA2 denoising, ASVs ranged from 31 to 208 per sample. Sweet sorghum exhibited the highest ASV richness (189 ASVs; 174 annotated species), followed by sorghum–Sudangrass hybrid (188 ASVs; 172 species) and Sudan grass (159 ASVs; 132 species). In contrast, giant Napier grass, king grass, and purple elephant grass showed lower ASV counts, with 83, 73, and 38 ASVs (80, 71, and 38 species), respectively.
Bacterial community composition varied markedly among silage types (
Figure 2A). While
lactic acid bacteria (LAB) dominated most samples, their dominant species differed by forage. Giant Napier grass and Sudan grass were strongly dominated by
Lactiplantibacillus plantarum (91.2%, 76.1%), whereas sweet sorghum and sorghum–Sudangrass hybrid were co-dominated by
Lactobacillus spicheri (51.4%, 52.4%) and
L. plantarum (24.7%, 22.3%). In king grass and purple elephant grass,
Levilactobacillus brevis was predominant (91.7%, 75.3%). In contrast, Mexican teosinte and sheep grass silages were dominated by non-LAB taxa, especially
Acetobacter fabarum (47.0% in Mexican teosinte), with LAB comprising < 30% of the communities. These silages also contained high levels of unclassified or potentially contaminant taxa (“Others”). Notably,
Clostridium pasteurianum (32.1%) and
uncultured Clostridia (16.0%) were exclusively found in sheep grass, suggesting clostridial contamination.
To evaluate bacterial richness and evenness across silage types, α-diversity was evaluated using Chao1, Shannon indices, and rarefaction curves (
Figure 2B–D). Both rarefaction and Chao1 analyses revealed significantly higher species richness in sheep grass, sorghum–Sudangrass hybrid, and sweet sorghum, each plateauing above 200 ASVs (Kruskal–Wallis,
p = 0.0028), indicating more complex bacterial communities. In contrast, purple elephant grass, giant Napier grass, and king grass showed a lower richness, with curves flattening below 160 ASVs. Shannon index results mirrored this trend (
p = 0.0035): sweet sorghum (3.20) and the hybrid (2.52) exhibited the highest diversity, while purple elephant grass (1.24), giant Napier grass (1.39), and king grass (1.74) showed the lowest, reflecting imbalanced communities dominated by few taxa.
To explore compositional variation among silages, β-diversity was assessed via Bray–Curtis distances and visualized using PCoA and UPGMA clustering. PCoA revealed a clear separation among silage types (
Figure 2E), with PC1 and PC2 explaining 31.17% and 21.75% of the variance, respectively. Sweet sorghum and the hybrid clustered closely, suggesting similar microbial communities likely driven by comparable substrates and fermentation outcomes. In contrast, sheep grass formed a distinct cluster, reflecting its uniquely diverse and unbalanced microbiota. UPGMA clustering (
Figure 2F) corroborated the PCoA results, identifying three major groups: (1) sweet sorghum, sorghum–Sudangrass hybrid, and Sudan grass; (2) Mexican teosinte, giant Napier grass, king grass, and purple elephant grass; and (3) sheep grass as a separate branch. This pattern aligned with the α-diversity results, indicating an association between bacterial diversity and community structure among forage types.
3.3. Identification of Representative Bacterial Taxa in Different Forage Silages
To identify forage-specific bacterial indicators, LEfSe analysis was conducted using an LDA threshold of 4.0 (
Figure 3A), revealing distinct microbial signatures across silage types. In Mexican teosinte silage, facultative aerobes and environmental bacteria such as Acetobacter fabarum, Carnobacteriaceae, Oceanobacillus, and Halomonas were identified as biomarkers. In contrast,
L. plantarum was the representative taxon in giant Napier grass silage. Both king grass and purple elephant grass silages were dominated by
L. brevis. Notably, in purple elephant grass, LEfSe analysis also detected
Paenibacillus amylolyticus,
Snodgrassella alvi and
Lelliottia amnigena—members of
Enterobacteriaceae—as biomarkers. Sheep grass silage was characterized by an enrichment of
Clostridium species, including
C. pasteurianum,
C. tyrobutyricum, and
Clostridium sensu stricto 12. In sorghum–Sudangrass hybrid silage, although
L. plantarum and
L. spicheri dominated in abundance,
Aliihoeflea aestuarii was identified as a biomarker due to its exclusive occurrence.
L. spicheri also appeared as a biomarker in this group. Overall, LEfSe analysis demonstrated that forage species strongly influence bacterial community composition. Several low-abundance taxa were detected across different silage types, showing forage-specific distribution patterns.
3.4. Co-Occurrence Network Reveals Core Microbial Modules and Functional Assemblages in Silage Fermentation
To unravel symbiotic patterns within silage microbiomes, we constructed a global co-occurrence network based on bacterial diversity data from eight forage silages (
Figure 3B). The network included 43 genera and 92 edges, capturing both positive (red) and negative (green) correlations. Positive associations were observed among Firmicutes. Notably,
Terribacillus (node 1),
Atopostipes (node 2),
Alkalibacterium (node 7),
Bacillus (node 12), and
Oceanobacillus (node 34) formed a central alliance. Conversely, Proteobacteria such as
Achromobacter and
Acinetobacter showed negative or peripheral connections with LAB. Beyond structural relationships, the network highlighted functionally relevant taxa. Although low in abundance, genera like
Aliihoeflea and
Oceanobacillus maintained stable interactions with multiple nodes. A tight subnetwork comprising
Clostridium_sensu_stricto_12,
Clostridium_sensu_stricto_11, and
Caproiciproducens (nodes 23, 35, and 11) was observed.
Using greedy modularity optimization, we identified four distinct microbial modules with unique topological and functional features (
Supplementary Figure S1). Module 1 (14 nodes) was dominated by
Pseudomonas,
Escherichia,
Acetobacter, and
Blautia, which were frequently associated with silage regions where LAB were less dominant. Module 2 (12 nodes) formed the core fermentation cluster, composed of acid- and salt-tolerant taxa such as
Alkalibacterium,
Atopostipes,
Terribacillus, and
Gracilibacillus. Module 3 was enriched in spoilage-related taxa
Clostridium,
Caproiciproducens, and
Akkermansia. Module 4 comprised fiber-degrading and intestinal bacteria such as
Bacteroides,
Ruminococcus, and
Faecalibacterium.
Network centrality analysis identified module-specific keystone taxa
Pseudomonas and
Blautia (Module 1),
Gracilibacillus and
Terribacillus (Module 2), and
Clostridium and
Akkermansia (Module 3), based on degree and betweenness centrality (
Supplementary Table S2). Altogether, the modular network structure reflects associations among microbial taxa that correspond to their functional traits.
3.5. Variation in Fungal Diversity Among Naturally Ensiled Forages
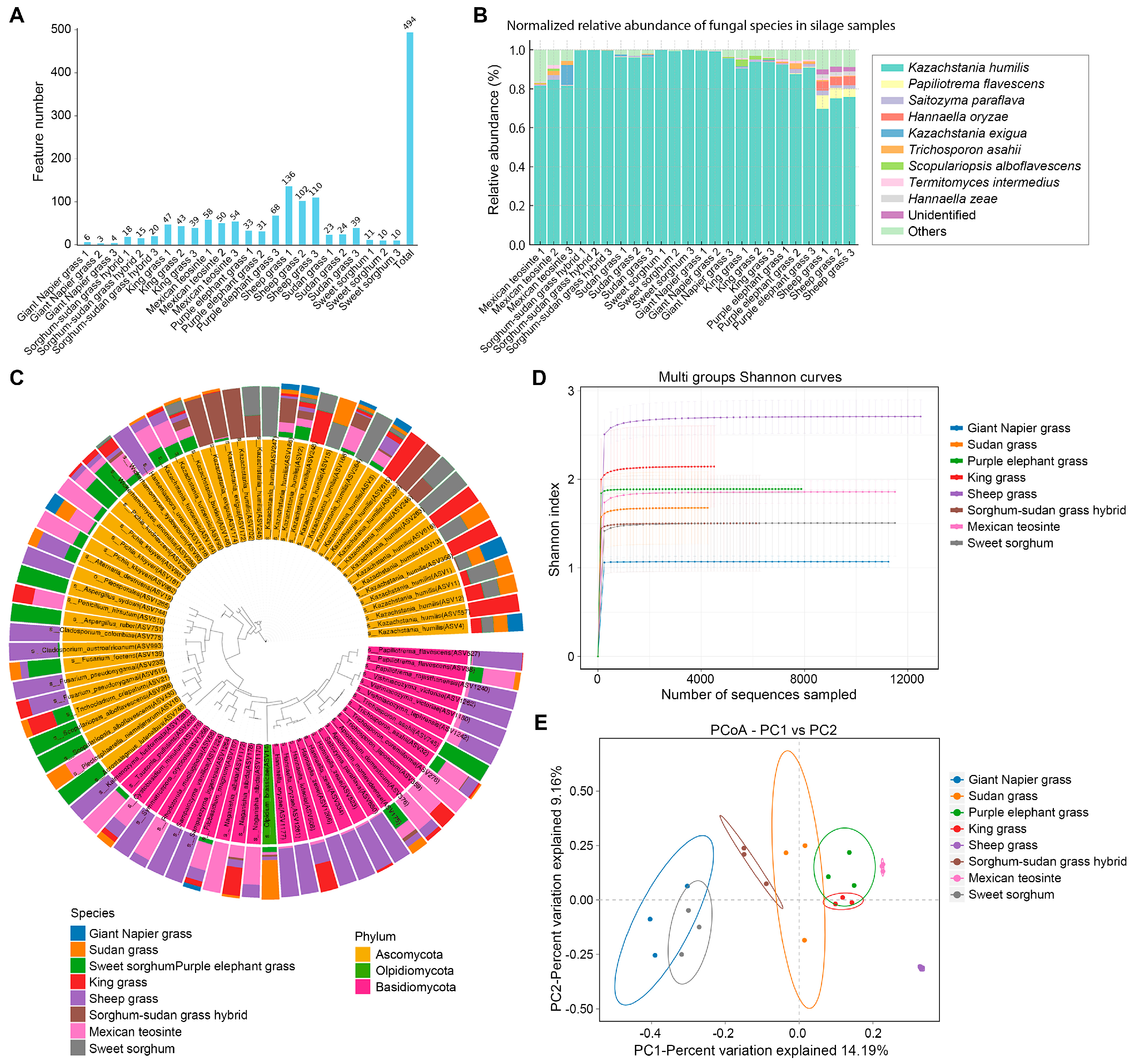
To profile fungal communities in naturally ensiled forages, high-throughput sequencing was performed on 24 silage samples, yielding 263,081 high-quality CCS reads. Each sample had ≥ 5005 reads (avg. 10,962), ensuring adequate depth for diversity analysis. In total, 494 fungal ASVs were identified, with marked differences across forage types (
Figure 4A). Sheep grass exhibited a highly diverse fungal community with the highest fungal richness (avg. 116 ASVs). In contrast, sweet sorghum and its hybrid had only 10 and 4 ASVs, respectively, reflecting simplified fungal communities.
Compared to bacteria, fungal communities showed relatively minor compositional variation across silages (
Figure 4B). Kazachstania humilis dominated most samples (>95% relative abundance), especially in sweet sorghum and sorghum–Sudangrass hybrid, where it was nearly exclusive. In contrast, sheep grass, Mexican teosinte, and purple elephant grass showed reduced
K. humilis and increased representation of
Saitozyma paraflava,
Termitomyces intermedius, and
Papiliotrema flavescens. Hannaella and Trichosporon, occasionally linked to proteolysis and fermentation failure, were also detected.
A circular phylogenetic tree of the top 80 fungal ASVs, annotated with abundance profiles (
Figure 4C), showed that most belonged to Ascomycota (39 ASVs) or Basidiomycota (33 ASVs). Multiple ASVs of
K. humilis formed distinct clades with host-specific distributions. For instance, ASV557 was enriched in king grass, while ASV263 and ASV264 dominated in sweet sorghum. ASV3 and ASV168 were ubiquitous across all samples, demonstrating their broad occurrence in the analyzed silages. Other genera also exhibited forage-specific enrichment:
Cladosporium and
Fusarium in sheep grass;
Aspergillus and
Scopulariopsis in purple elephant grass. Basidiomycota-affiliated ASVs were mostly confined to sheep grass and Mexican teosinte, reflecting host-driven niche specificity.
Fungal α- and β-diversity analyses further underscored forage-specific community patterns. Shannon rarefaction curves (
Figure 4D) showed sheep grass had the highest α-diversity (avg. > 2.5), suggesting either high initial fungal loads or limited inhibition during ensiling. King grass also exhibited a relatively high diversity. In contrast, the hybrid silage showed an extremely low α-diversity (Shannon ~1.0), consistent with reduced ASV richness and rapid fungal suppression. PCoA based on binary Jaccard distance revealed a clear separation among forage types (
Figure 4E), with PC1 explaining 34.19% of the variance. Samples clustered by species, with king grass, Mexican teosinte, and purple elephant grass forming tight clusters, while sweet sorghum, Sudan grass, and the hybrid clustered separately—indicating a strong forage-dependent assembly of fungal communities. This clustering mirrored the α-diversity results and highlighted the dominant influence of host forage type on shaping fungal composition during ensiling.
3.6. Selection Strategy for Pairwise Metabolomic Comparison Based on Fermentation Quality and Microbial Profiles
To support biologically meaningful metabolomic comparisons, we classified the eight tropical forage silages into three types based on fermentation performance and microbial community structure. Classification criteria included lactic acid production, NH4+-N’s accumulation, WSC retention, and microbial composition. Type I (efficient LAB-dominated) included sweet sorghum, sorghum–Sudangrass hybrid, and Sudan grass, and they showed high lactic acid levels, a low NH4+-N, and dominance of L. plantarum and L. spicheri. Their bacterial communities were diverse, while fungal diversity was low and mainly comprised K. humilis. Type II (moderate LAB-dominated) included king grass and purple elephant grass, and they showed an intermediate fermentation performance, moderate WSC loss, and near-monodominance of L. brevis, accompanied by a narrow but acid-tolerant LAB population. Type III (spoilage-prone), comprising sheep grass and Mexican teosinte, characterized by low lactic acid, high NH4+-N, and high fungal and bacterial diversity, with taxa such as A. fabarum and Clostridium spp. Giant Napier grass, although sharing fermentation traits with Type I, showed a uniquely simplified microbiome dominated solely by L. plantarum, and it was therefore considered a functional but microbiologically distinct Type I outlier.
Based on these classifications, we selected three forage pairings that maximized biological and mechanistic contrast for subsequent differential metabolomic analysis. The first comparison, between sweet sorghum (Type I) and sheep grass (Type III), reflects the most extreme divergence in fermentation quality and microbial composition, enabling the identification of key metabolites associated with successful versus failed ensiling. The second pairing, between giant Napier grass (Type I outlier) and king grass (Type II), compares two LAB-dominant silages differing markedly in microbial diversity and fermentation efficiency. The third pairing, between sorghum–Sudangrass hybrid (Type I) and Mexican teosinte (Type III), focuses on the metabolic shifts occurring at the boundary between moderately successful and poorly fermented silages, particularly regarding organic acid production pathways.
3.7. Distinct Metabolic Profiles Between Silages Reveal Divergent Fermentation Pathways and Stress Responses
Metabolomic profiling of 24 silage samples from eight tropical forage species (three replicates each) using LC-QTOF identified 15,827 metabolic features, including 4560 annotated metabolites. Principal component analysis (PCA) revealed clear inter- and intra-group metabolic variations (
Supplementary Figure S2), with PC1 and PC2 explaining 31.31% and 19.43% of the total variance, respectively—capturing 50.74% of the overall metabolic diversity. Type I forages—sweet sorghum, sorghum–Sudangrass hybrid, Sudan grass, and giant Napier grass—clustered closely along PC1, indicating similar metabolic profiles and strong intra-group coherence. Type II forages, king grass and purple elephant grass, also clustered together, reflecting shared fermentation traits. In contrast, Type III forages—sheep grass and Mexican teosinte—were clearly separated along both axes, indicating distinct and heterogeneous metabolomes. The elevated PC2 scores in Mexican teosinte and purple elephant grass suggest notable shifts in key metabolites associated with secondary metabolism.
Integrated annotations from KEGG, HMDB, and LipidMaps databases indicated that the metabolites were primarily enriched in four categories: amino acid metabolism, plant-derived secondary metabolites, bioactive lipids, and flavonoid-related compounds (
Supplementary Figure S3). KEGG enrichment pointed to active amino acid pathways—tryptophan, arginine, tyrosine—as well as biosynthesis of phenylpropanoids, flavonoids, and isoquinoline alkaloids. HMDB classification underscored the abundance of aromatic compounds, organic acid derivatives, and lipid-like molecules—especially phenols, benzenoids, and carboxylic acids. LipidMaps data emphasized the accumulation of fatty acyls, prenol lipids (e.g., terpenoids), and polyphenols. Notably, 224 flavonoid derivatives were identified, highlighting the role of plant polyphenols and bioactive lipids as key drivers of metabolic divergence across forage silages under varying microbial and fermentation conditions.
To elucidate the metabolic distinctions between forage types, we performed differential metabolite analyses across three key silage pairings using OPLS-DA, log
2 fold change, and VIP screening (
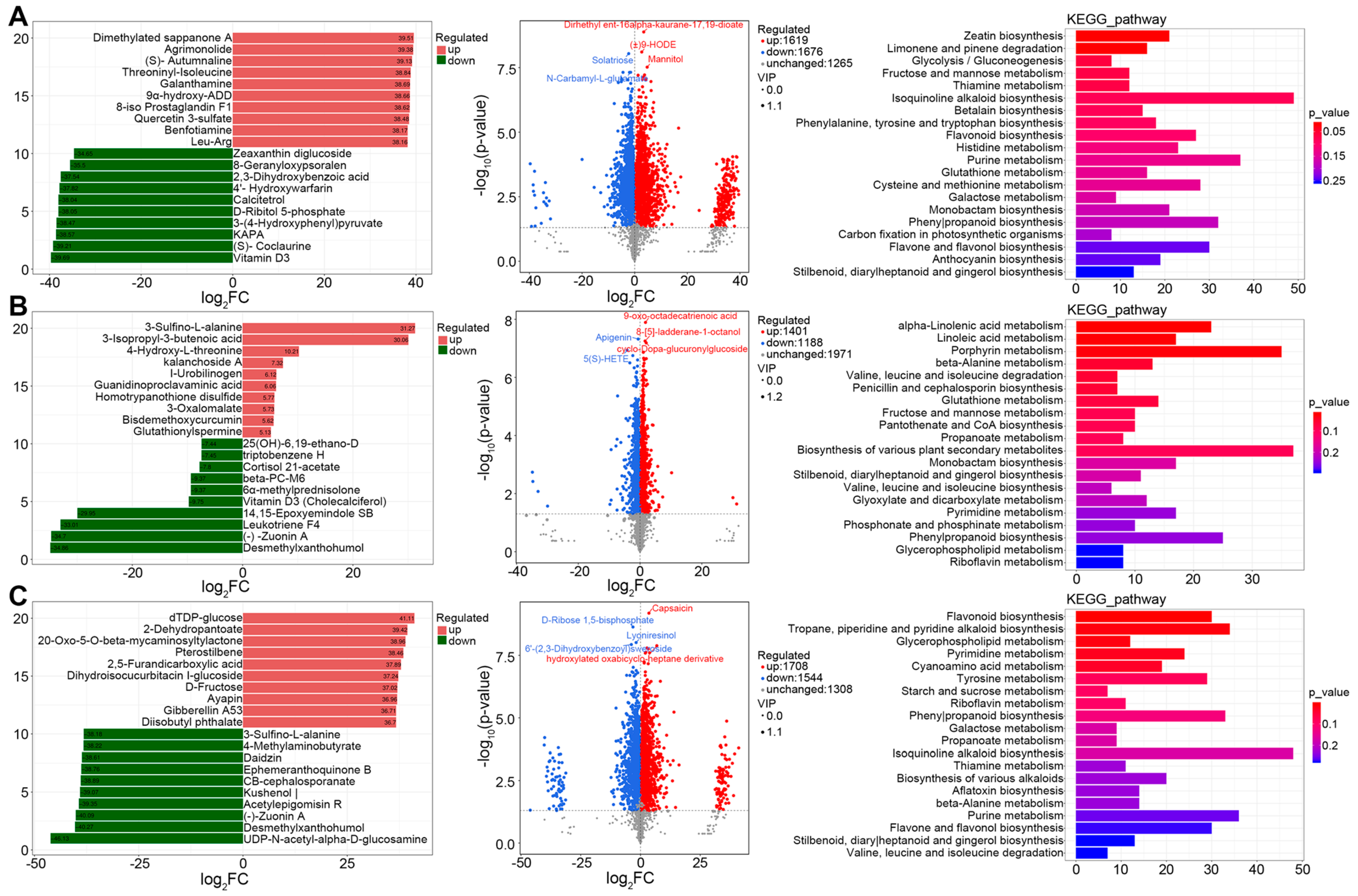
Supplementary Figure S4). Out of 4560 annotated metabolites, the sheep grass vs. sweet sorghum comparison yielded 3295 differential metabolites, reflecting broad shifts in metabolic regulation (
Figure 5A). Sheep grass exhibited elevated levels of oxidative stress markers and natural products (e.g., Agrimonolide, (±) 9-HODE), while antioxidant and lipid-derived compounds like Vitamin D3 were reduced. KEGG enrichment revealed upregulation of isoquinoline alkaloid biosynthesis and glutathione metabolism in sheep grass. Correlation analysis identified tightly linked metabolite modules, including Mannitol and (±) 9-HODE, and an oxidative lipid–sugar alcohol cluster indicative of microecological imbalance and disrupted fermentation homeostasis in sheep grass (
Supplementary Figure S5A).
In the king grass vs. giant Napier grass comparison, 2589 differential metabolites were detected (
Figure 5B). King grass showed elevated nitrogenous metabolites (e.g., N-Acetyl-DL-glutamic acid), pointing to stronger proteolysis and amino acid turnover. In contrast, giant Napier grass accumulated a suite of flavonoids and antimicrobial compounds (e.g., Apigenin, 5(S)-HETE), suggesting enhanced metabolic resilience. KEGG enrichment revealed divergence in flavonoid biosynthesis, lipid oxidation, and nucleotide metabolism, highlighting a more robust stress-buffered fermentation system in giant Napier grass. Correlation networks supported these findings, with distinct metabolite modules linked to plant defense and antioxidant regulation (
Supplementary Figure S5B).
The sorghum–Sudangrass hybrid vs. Mexican teosinte comparison displayed the greatest metabolic divergence, with 3252 differential metabolites (
Figure 5C). Mexican teosinte accumulated hormone-like and stress-associated compounds (e.g., Pterostilbene, Gibberellin A53). In contrast, the hybrid silage was enriched in antimicrobial flavonoids and lignans (e.g., Daidzin, Lyoniresinol). KEGG pathways were enriched for flavonoid biosynthesis and alkaloid metabolism, while correlation analysis identified tightly clustered antioxidant and hormone-linked metabolites, further reinforcing the hypothesis of fermentation instability and functional disarray in Mexican teosinte (
Supplementary Figure S5C).
In summary, these comparisons suggested that metabolic outcomes during silage fermentation are strongly forage-dependent. Superior silages exhibited coordinated enrichment of flavonoids, lipid antioxidants, and amino acid derivatives, while poorly fermented types showed markers of oxidative stress, proteolysis, and microbial imbalance. These findings revealed differences in metabolite profiles among forage types, including antioxidants, flavonoids, and stress-related compounds.
4. Discussion
4.1. Integrated Interpretation of Key Findings with Comparison to Previous Studies
This study presents the first multi-omics evaluation of silages from eight tropical forages, integrating physicochemical traits, bacterial–fungal dynamics, and untargeted metabolomics. We observed differences in fermentation efficiency, microbial assembly, and metabolite profiles, reflecting distinct biochemical trajectories during ensiling. Sweet sorghum and giant Napier grass showed the most favorable fermentation, retaining a high crude protein and WSC, producing abundant lactic acid, and maintaining minimal NH4+-N and butyric acid, indicative of efficient lactic fermentation and suppressed proteolysis. In contrast, sheep grass and Mexican teosinte exhibited a poor performance, with low lactic acid, high NH4+-N, and increased butyric and propionic acids, suggesting clostridial activity and extensive protein degradation. Other forages, such as king grass and purple elephant grass, displayed an intermediate quality.
Microbial communities were strongly forage-dependent. High-quality silages (Type I) were dominated by LAB—primarily L. plantarum and L. spicheri—while poorly fermented silages (Type III) contained spoilage-associated taxa such as Acetobacter and Clostridium. Fungal diversity was generally low, with K. humilis dominating Type I silages, potentially contributing to aerobic stability. Metabolomics revealed that high-quality silages accumulated flavonoids, lipid antioxidants, and phenolic compounds, whereas poor-quality silages contained oxidative stress markers, hormone-like metabolites, and signatures of proteolysis and microbial imbalance.
These results highlight the interaction between forage composition, microbial ecology, and metabolite dynamics in shaping silages’ quality. The novelty lies in systematically linking fermentation chemistry, microbial networks, and metabolic pathways across diverse tropical species—a gap in prior work, which mainly focused on temperate crops like corn, alfalfa, and ryegrass [
31,
32,
33]. Previous metabolomic studies reported changes in amino acids, phenolics, and organic acids during ensiling [
10,
34,
35] but typically examined single species or additive effects rather than contrasting diverse substrates. Our findings extend this knowledge base by demonstrating that tropical forages exhibit even greater variability in fermentation outcomes and metabolite patterns than previously documented for temperate species. To place these findings in context, we compared our results with previous studies that integrated microbial and metabolomic analyses in silage systems.
Consistent with earlier reports, high-quality silages were characterized by lactic acid enrichment and the accumulation of flavonoids, similar to observations in corn silage [
36]. Likewise, strong correlations between LAB dominance and organic acid production, widely reported for barley silage [
37], align with the association we observed between
L. plantarum‘s abundance and high lactic acid in sweet sorghum and giant Napier grass. Unlike these studies, our data captured contrasts: Type III silages accumulated oxidative stress markers and hormone-like compounds rarely addressed in silage research, indicating unique biochemical stress responses. This suggests that successful fermentation is governed by more than WSC availability or buffering capacity [
31,
38]. Our integrated approach reveals that microbial networks’ integrity and secondary metabolite fluxes are equally critical. For example,
Clostridium‘s proliferation in sheep grass coincided with high NH
4+-N and butyric acid—classic indicators of proteolysis [
39,
40] —yet we also detected isoquinoline alkaloid biosynthesis and lipid peroxidation, signaling oxidative and stress-driven pathways seldom quantified before. This metabolomic evidence complements microbial data, implying that proteolytic spoilage in tropical silages involves complex responses beyond nitrogen catabolism.
Beyond bacterial and metabolite dynamics, our fungal analysis provides another key contribution. While fungi are often treated only as spoilage agents [
5,
9], our data confirm
K. humilis‘s dominance in successful fermentations, consistent with its tolerance of acidic, anaerobic conditions [
41], suggesting a stabilizing role during storage.
4.2. Mechanistic Insights into LAB Dominance and Efficient Fermentation
Differences in fermentation performance between forage types can be mechanistically linked to microbial succession and network stability. Type I silages, dominated by LAB such as
L. plantarum and
L. spicheri, likely achieve rapid acidification through intense lactic acid production, creating conditions unfavorable for spoilage organisms. Reducing pH to around 4.2 during early ensiling is widely recognized as critical for inhibiting clostridia, limiting proteolysis, and preventing amino acid deamination [
38,
42]. Our findings support this mechanism, as indicated by the strong negative correlation between WSC loss and protein degradation (r = –0.730,
p = 0.040), highlighting WSC’s dual role as a fermentation substrate and a regulator of nitrogen preservation. Similar correlations were observed in corn silage, where LAB inoculation improved proteomes’ stability by suppressing clostridial activity [
32].
Giant Napier grass, despite near-monoculture dominance by
L. plantarum, exhibited a fermentation quality comparable to sweet sorghum, suggesting that community diversity is less critical than the functional capacity of keystone species.
L. plantarum employs the homofermentative Embden–Meyerhof–Parnas (EMP) pathway to convert sugars primarily into lactic acid, ensuring a rapid pH decline and efficient energy recovery [
31,
43]. This metabolic efficiency explains why giant Napier grass, despite reduced microbial richness, achieved lactic acid concentrations of ~99 g/kg and maintained strong protein preservation. This case exemplifies “functional redundancy minimization,” where dominance of a metabolically superior species offsets low community complexity [
44,
45].
In contrast, Type II silages dominated by
L. brevis exhibited a moderate quality, consistent with its heterofermentative metabolism via the phosphoketolase pathway, which produces acetic acid, ethanol, and CO
2 alongside lactic acid [
46]. While acetic acid improves aerobic stability, its weaker acidification capacity slows pH reduction, prolonging the window for proteolysis and clostridial activity [
41]. These differences underscore the influence of fermentation product profiles on silage outcomes.
Metabolomic integration reinforces these interpretations. Type I silages not only accumulated lactic acid but were also enriched in flavonoids and antioxidant lipids—metabolites linked to plant defense and oxidative stress buffering—suggesting synergistic plant–microbe interactions. Rapid acidification likely mitigates ROS’ accumulation, preserving polyphenolic antioxidants that enhance storage stability. Conversely, Type III silages, where LAB abundance was low, showed signatures of isoquinoline alkaloid biosynthesis and lipid peroxidation, indicating stress-induced pathways triggered by uncontrolled microbial succession. Collectively, these results highlight how LAB dominance shapes primary fermentation chemistry and secondary metabolite trajectories, stabilizing the silage microecosystem.
4.3. Fungal Community Roles in Aerobic Stability and Fermentation Balance
Fungi in silage were long considered primarily agents of aerobic spoilage, yet emerging evidence suggests broader roles in fermentation dynamics [
37,
47]. Our data show fungal communities were generally simplified under anaerobic conditions, with
K. humilis dominating most silages, particularly Type I. This acid-tolerant, fermentative yeast thrives in low-pH environments and may aid residual sugar metabolism [
41]. Its dominance in well-preserved silages suggests a commensal or even stabilizing role, possibly through redox regulation or suppression of filamentous fungi [
48,
49].
In contrast, Type III silages exhibited greater fungal diversity alongside markers of fermentation failure, including high NH
4+-N and oxidative stress metabolites. Genera such as
Hannaella and
Trichosporon—found in sheep grass and Mexican teosinte—are linked to proteolysis and aerobic deterioration [
50]. Their presence implies that inadequate acidification allows opportunistic colonization, exacerbating nutrient loss and generating undesirable metabolites [
51]. The co-occurrence of
Saitozyma and
Termitomyces, both associated with lignocellulose degradation, suggests that fungal-mediated fiber breakdown may occur in poorly fermented systems, altering bacterial substrates and downstream metabolite fluxes [
52,
53].
The predominance of
K. humilis in high-quality silages may also influence aerobic stability. While yeasts metabolizing lactic acid drive heating and spoilage after oxygen exposure [
9], K. humilis appears less problematic, producing less ethanol than
Candida spp. [
41]. Nevertheless, its functional role under feedout conditions merits further study.
4.4. Functional Implications of Key Metabolites in Silage Ecosystems
Untargeted metabolomics revealed a marked biochemical divergence between silages, complementing fermentation and microbial data. Four metabolite classes—amino acid derivatives, plant secondary metabolites, bioactive lipids, and flavonoid-related compounds—strongly correlated with fermentation efficiency and microbiota.
Type I silages accumulated flavonoids (e.g., Apigenin, Daidzin), phenolic acids, and antioxidant lipids, likely persisting or enriched under LAB dominance. These compounds provide the dual benefits of antioxidant protection and antimicrobial activity, particularly against clostridia and enterobacteria [
54], consistent with reduced NH
4+-N levels. Lipid oxidation products such as 5(S)-HETE in giant Napier grass point to active oxylipin pathways, possibly mediating stress signaling during ensiling [
55].
Conversely, Type III silages exhibited signatures of stress and proteolysis. Elevated isoquinoline alkaloids and oxidative markers (e.g., (±)9-HODE) suggest redox imbalance, likely exacerbated by oxygen’s ingress or incomplete acidification. Isoquinoline alkaloids, often linked to plant defense, may also originate from microbial degradation of aromatic amino acids under stress [
56]. Hormone-like metabolites such as Gibberellin A53 and Pterostilbene in Mexican teosinte silage imply persistence of plant metabolism or microbial synthesis of phytohormone analogs during fermentation failure.
Comparisons with prior metabolomic studies reveal both shared and novel features. Accumulation of flavonoids and antioxidant metabolites in high-quality silages mirrors findings in corn silage [
36]. However, the detection of hormone-like compounds and polyphenol derivatives in poor-quality silages is novel, indicating that incomplete fermentation may sustain plant metabolic activity or induce alternative microbial pathways [
10,
34]. These metabolites hold promise as early indicators of instability, supporting their use in quality monitoring.
4.5. Practical Applications and Future Research Based on Mechanistic Insights
Integrating fermentation quality, microbial ecology, and metabolomics provides a strong mechanistic foundation for improving tropical silage management. Our results highlight metabolite signatures as potential predictive biomarkers of fermentation’s success. Rapid detection platforms such as portable LC-MS or spectroscopy could enable on-site monitoring and early intervention. The consistent association between LAB dominance and favorable metabolite profiles supports a microbiome-informed inoculant design. Targeted inoculation with L. plantarum or mixed consortia optimized for functional complementarity can enhance fermentation efficiency and aerobic stability. For forages with low WSC or a high buffering capacity, combining LAB with carbohydrate-hydrolyzing bacteria or enzymes may further improve outcomes.
Beyond inoculant strategies, metabolomics could underpin advanced decision-support systems. Coupled with machine learning, metabolomic fingerprints offer predictive modeling and process optimization under tropical conditions.
However, critical gaps remain. Absence of pre-ensiling microbiome data limits attribution of performance to initial epiphytes versus fermentation-driven succession. Static endpoint sampling precludes insights into metabolite dynamics, and many detected compounds lack annotation, hindering functional interpretation. Future research should incorporate longitudinal metabolomics, metatranscriptomics, and isotope tracing to elucidate microbial–metabolite linkages and validate causal mechanisms.