Metabolic Engineering of Escherichia coli for De Novo Biosynthesis of Mandelic Acid
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Compounds and Standards
2.2. Strains and Plasmid Construction
2.3. Protein Network Analysis
2.4. Cultivation and Induction of Engineered Strains
2.5. Protein Expression and SDS-PAGE Analysis
2.6. Whole-Cell Biocatalysis
2.7. Extraction and Analysis of MA
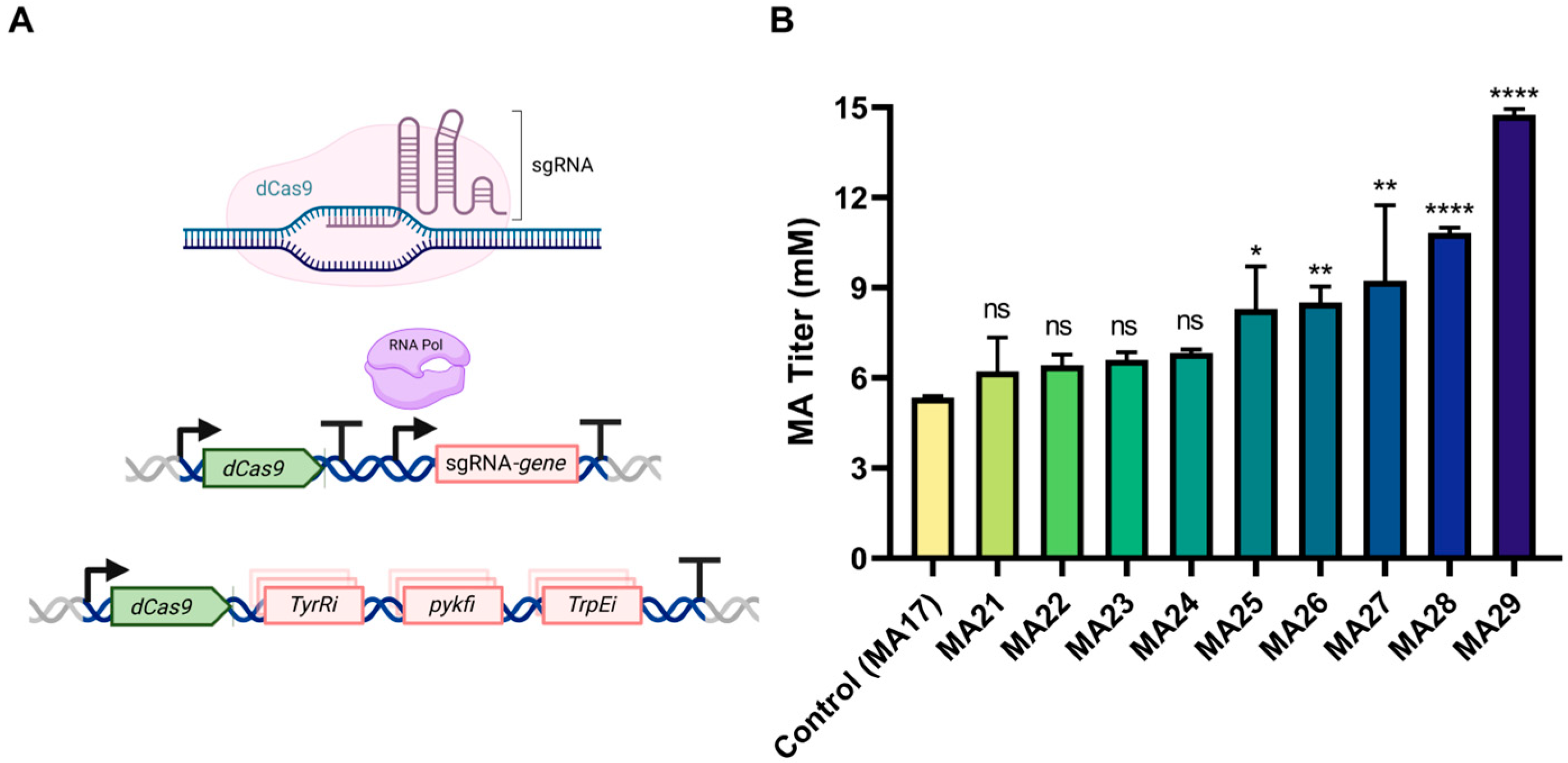
2.8. CRISPRi-Mediated Gene Interference
2.9. HCDC of E. coli
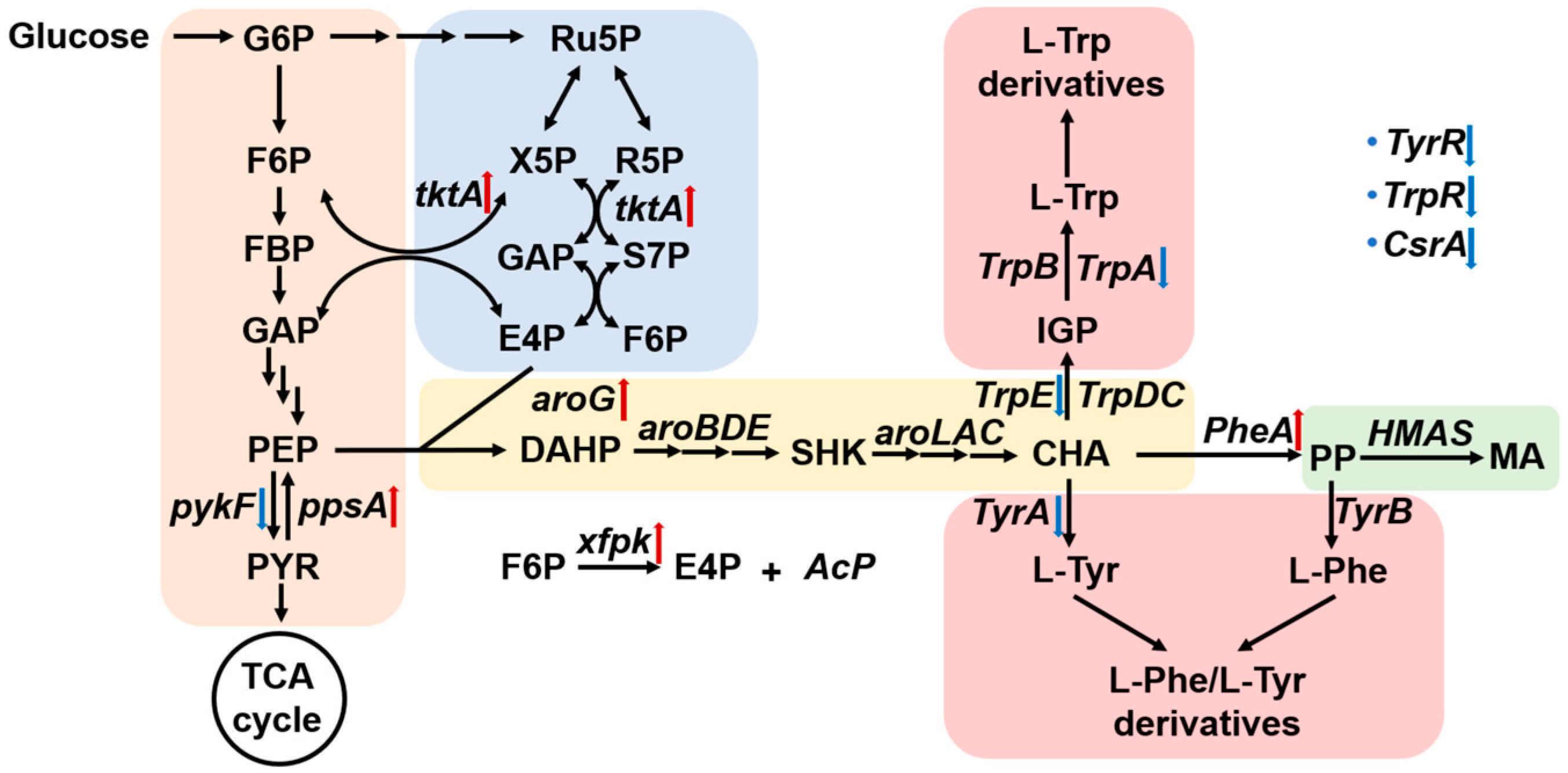
3. Results and Discussion
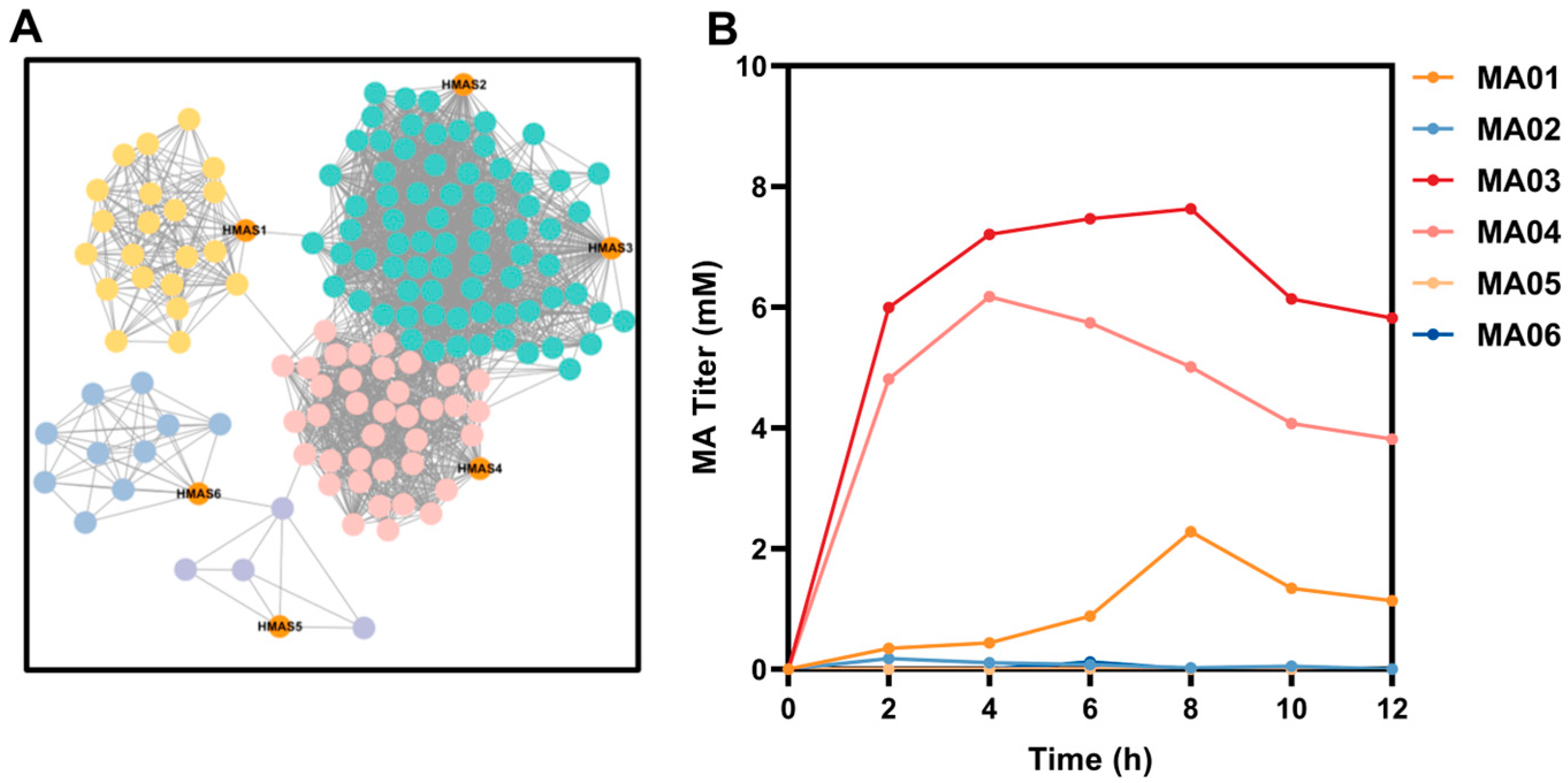
3.1. Selection and Functional Validation of HMAS Homologs
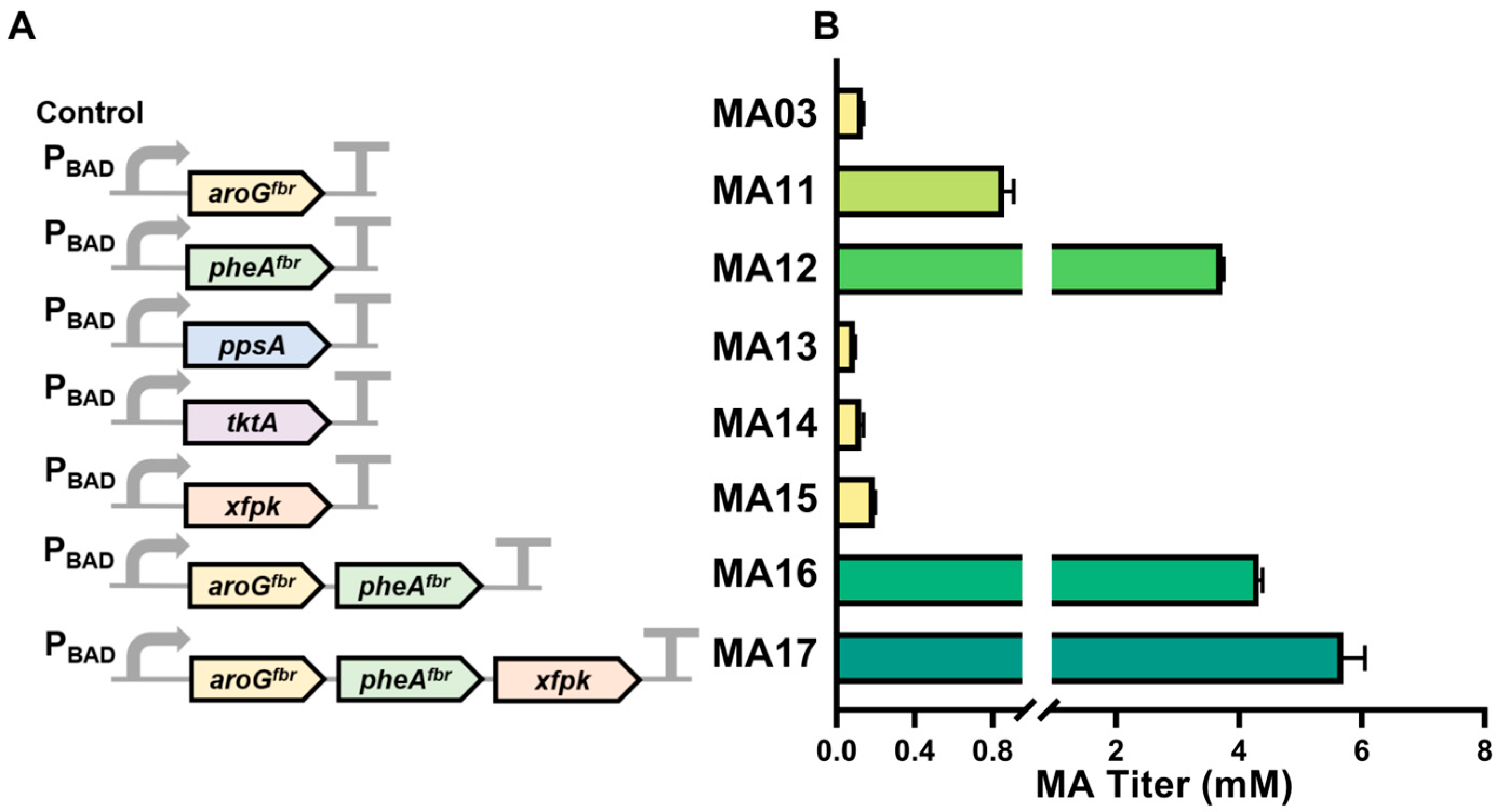
3.2. Optimization of Endogenous Key Gene Overexpression
3.3. CRISPRi-Mediated Inhibition of Competing Pathways to Enhance MA Production
3.4. HCDC of Engineered E. coli in a 5 L Bioreactor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MA | mandelic acid |
| HMAS | hydroxymandelate synthase |
| E4P | erythrose-4-phosphate |
| PEP | phosphoenolpyruvate |
| CRISPRi | CRISPR interference |
| HCDC | high-cell-density cultivation |
| MOFs | metal–organic frameworks |
| CPs | coordination polymers |
| NHase | nitrile hydratases |
| ManDH | D-mandelate dehydrogenase |
| PP | phenylpyruvate |
| PGA | phenylglyoxylic acid |
| E. coli | Escherichia coli |
| S. cerevisiae | Saccharomyces cerevisiae |
| EFI-EST | Enzyme Function Initiative–Enzyme Similarity Tool |
| SSN | sequence similarity network |
| PPP | pentose phosphate pathway |
| L-Tyr | L-tyrosine |
| L-Trp | L-tryptophan |
References
- Reddy, K.L.; Mathew, J.P.; Maniappan, S.; Tom, C.; Shiby, E.; Pujala, R.K.; Kumar, J. Mandelic acid appended chiral gels as efficient templates for multicolour circularly polarized luminescence. Nanoscale 2022, 14, 4946–4956. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Geng, S.; Wang, L.; Wen, Z.; Sun, X.; Huang, H. Bacterial mandelic acid degradation pathway and its application in biotechnology. J. Appl. Microbiol. 2022, 133, 273–286. [Google Scholar] [CrossRef]
- Wójcik, A.; Kubiak, M.; Rotsztejn, H. Influence of azelaic and mandelic acid peels on sebum secretion in ageing women. Adv. Dermatol. Allergol. 2013, 3, 140–145. [Google Scholar] [CrossRef]
- Van Putten, P.L. Mandelic acid and urinary tract infections. Antonie Leeuwenhoek 1979, 45, 622–623. [Google Scholar] [CrossRef]
- Świsłocka, R.; Świderski, G.; Nasiłowska, J.; Sokołowska, B.; Wojtczak, A.; Lewandowski, W. Research on the electron structure and antimicrobial properties of mandelic acid and its alkali metal salts. Int. J. Mol. Sci. 2023, 24, 3078. [Google Scholar] [CrossRef]
- Poterała, M.; Dranka, M.; Borowiecki, P. Chemoenzymatic preparation of enantiomerically enriched (R)-(–)-Mandelic acid derivatives: Application in the synthesis of the active agent pemoline. Eur. J. Org. Chem. 2017, 2017, 2290–2304. [Google Scholar] [CrossRef]
- Xia, M.; Yang, M.; Wang, Y.; Tian, F.; Hu, J.; Yang, W.; Tao, S.; Lu, L.; Ding, X.; Jiang, S.; et al. dl-Mandelic acid exhibits high sperm-immobilizing activity and low vaginal irritation: A potential non-surfactant spermicide for contraception. Biomed. Pharmacother. 2020, 126, 110104. [Google Scholar] [CrossRef] [PubMed]
- Dulęba, J.; Siódmiak, T.; Mastalerz, R.; Kocot, N.; Dębińska, A.; Suchomska, E.; Czeczka, M.; Cała, K.; Marszałł, M. The application of lipase from Burkholderia cepacia in the enantioselective pharmaceutical biocatalysis. Farm. Pol. 2022, 78, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.; Culbertson, E. Effects of topical mandelic acid treatment on facial skin viscoelasticity. Facial Plast. Surg. 2018, 34, 651–656. [Google Scholar] [CrossRef]
- Cai, P.; Gao, Z.; Yin, X.; Luo, Y.; Zhao, X.; Pan, Y. Facile enantioseparation and recognition of mandelic acid and its derivatives in self-assembly interaction with chiral ionic liquids. J. Sep. Sci. 2019, 42, 3589–3598. [Google Scholar] [CrossRef]
- Tay, H.M.; Hua, C. A structural study into halogenated derivatives of mandelic acid as building blocks of chiral coordination polymers. J. Coord. Chem. 2022, 75, 1656–1669. [Google Scholar] [CrossRef]
- Martínková, L.; Křen, V. Biocatalytic production of mandelic acid and analogues: A review and comparison with chemical processes. Appl. Microbiol. Biotechnol. 2018, 102, 3893–3900. [Google Scholar] [CrossRef] [PubMed]
- Monteith, J.J.; Rousseaux, S.A.L. A Dual Ni/Photoredox Cross-Coupling Approach toward Mandelic Acids. Org. Lett. 2024, 26, 4566–4570. [Google Scholar] [CrossRef]
- Singh, R.V.; Sambyal, K. Green synthesis aspects of (R)-(–)-mandelic acid; a potent pharmaceutically active agent and its future prospects. Crit. Rev. Biotechnol. 2022, 43, 1226–1235. [Google Scholar] [CrossRef]
- Stolz, A.; Eppinger, E.; Sosedov, O.; Kiziak, C. Comparative Analysis of the Conversion of Mandelonitrile and 2-Phenylpropionitrile by a Large Set of Variants Generated from a Nitrilase Originating from Pseudomonas fluorescens EBC191. Molecules 2019, 24, 4232. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Xia, Y.; Zhou, Z. Recent advances and promises in nitrile hydratase: From mechanism to industrial applications. Front. Bioeng. Biotechnol. 2020, 8, 352. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Z.; Wang, J. A thermophilic biofunctional multienzyme cascade reaction for Cell-Free synthesis of salvianic acid A and 3,4-Dihydroxymandelic acid. ACS Sustain. Chem. Eng. 2019, 7, 18247–18253. [Google Scholar] [CrossRef]
- Lukito, B.R.; Wang, Z.; Sekar, B.S.; Li, Z. Production of (R)-mandelic acid from styrene, L-phenylalanine, glycerol, or glucose via cascade biotransformations. Bioresour. Bioprocess. 2021, 8, 22. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Wu, B.; Hong, M. High-Enantioselectivity adsorption separation of racemic mandelic acid and methyl mandelate by robust Chiral UIO-68-Type ZR-MOFS. Inorg. Chem. 2023, 63, 381–389. [Google Scholar] [CrossRef]
- Reifenrath, M.; Boles, E. Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae. Metab. Eng. 2018, 45, 246–254. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Bergmans, H.E.; Van Die, I.M.; Hoekstra, W.P. Transformation in Escherichia coli: Stages in the process. J. Bacteriol. 1981, 146, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI web resource for Genomic Enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Oberg, N.; Zallot, R.; Gerlt, J.A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) web resource for genomic enzymology tools. J. Mol. Biol. 2023, 435, 168018. [Google Scholar] [CrossRef]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. BBA Proteins Proteom. 2015, 1854, 1019–1037. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, W.; Shao, S.; Wang, Q. Gene silencing through CRISPR interference in bacteria: Current advances and future prospects. Front. Microbiol. 2021, 12, 635227. [Google Scholar] [CrossRef]
- Hawkins, J.S.; Wong, S.; Peters, J.M.; Almeida, R.; Qi, L.S. Targeted Transcriptional Repression in Bacteria Using CRISPR Interference (CRISPRI); Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; pp. 349–362. [Google Scholar] [CrossRef]
- Blin, K.; Pedersen, L.E.; Weber, T.; Lee, S.Y. CRISPy-web: An online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 2016, 1, 118–121. [Google Scholar] [CrossRef]
- Müller, U.; Van Assema, F.; Gunsior, M.; Orf, S.; Kremer, S.; Schipper, D.; Wagemans, A.; Townsend, C.A.; Sonke, T.; Bovenberg, R.; et al. Metabolic engineering of the E. coli l-phenylalanine pathway for the production of d-phenylglycine (d-Phg). Metab. Eng. 2006, 8, 196–208. [Google Scholar] [CrossRef]
- Liu, P.; Jin, Q.; Li, X.; Zhang, R.; Yuan, H.; Liu, C.; Wang, P. Directed evolution and metabolic engineering generate an Escherichia coli cell factory for de novo production of 4-hydroxymandelate. Bioresour. Technol. 2024, 413, 131497. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Zheng, Y.; Su, T.; Qi, Q. Microbial CRISPRI and CRISPRA systems for metabolic engineering. Biotechnol. Bioprocess Eng. 2019, 24, 579–591. [Google Scholar] [CrossRef]
- Gordon, G.C.; Korosh, T.C.; Cameron, J.C.; Markley, A.L.; Begemann, M.B.; Pfleger, B.F. CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab. Eng. 2016, 38, 170–179. [Google Scholar] [CrossRef]
- Meza, E.; Becker, J.; Bolivar, F.; Gosset, G.; Wittmann, C. Consequences of phosphoenolpyruvate: Sugar phosphotranferase system and pyruvate kinase isozymes inactivation in central carbon metabolism flux distribution in Escherichia coli. Microb. Cell Factories 2012, 11, 127. [Google Scholar] [CrossRef]
- Zhao, C.; Lin, Z.; Dong, H.; Zhang, Y.; Li, Y. Reexamination of the Physiological Role of PykA in Escherichia coli Revealed that It Negatively Regulates the Intracellular ATP Levels under Anaerobic Conditions. Appl. Environ. Microbiol. 2017, 83, e00316-17. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Liu, Z.; He, J.; Wang, J.; Yan, Y.; Wang, X.; Cui, Y.; Bi, Y.; Du, Z.; Song, Y.; et al. TyrR, the regulator of aromatic amino acid metabolism, is required for mice infection of Yersinia pestis. Front. Microbiol. 2015, 6, 110. [Google Scholar] [CrossRef][Green Version]
- Coulson, T.J.D.; Patten, C.L. The TyrR Transcription Factor Regulates the Divergent akr-ipdC Operons of Enterobacter cloacae UW5. PLoS ONE 2015, 10, e0121241. [Google Scholar] [CrossRef]
- Cho, B.; Federowicz, S.; Park, Y.; Zengler, K.; Palsson, B.Ø. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat. Chem. Biol. 2011, 8, 65–71. [Google Scholar] [CrossRef]
- Bashiri, G.; Johnston, J.M.; Evans, G.L.; Bulloch, E.M.M.; Goldstone, D.C.; Jirgis, E.N.M.; Kleinboelting, S.; Castell, A.; Ramsay, R.J.; Manos-Turvey, A.; et al. Structure and inhibition of subunit I of the anthranilate synthase complex of Mycobacterium tuberculosis and expression of the active complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 2297–2308. [Google Scholar] [CrossRef]
- Lynch, J.H.; Dudareva, N. Aromatic amino acids: A complex network ripe for future exploration. Trends Plant Sci. 2020, 25, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Min, B.E.; Hwang, H.G.; Seo, S.W.; Jung, G.Y. Pathway optimization by re-design of untranslated regions for L-tyrosine production in Escherichia coli. Sci. Rep. 2015, 5, srep13853. [Google Scholar] [CrossRef]
- Revelles, O.; Millard, P.; Nougayrède, J.; Dobrindt, U.; Oswald, E.; Létisse, F.; Portais, J. The Carbon Storage Regulator (Csr) System Exerts a Nutrient-Specific Control over Central Metabolism in Escherichia coli Strain Nissle 1917. PLoS ONE 2013, 8, e66386. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Xiao, X.; Xing, W.; Na, R.; Song, Y.; Cao, G.; Wang, P. Metabolic Engineering of Escherichia coli for De Novo Biosynthesis of Mandelic Acid. Fermentation 2025, 11, 331. https://doi.org/10.3390/fermentation11060331
Liu C, Xiao X, Xing W, Na R, Song Y, Cao G, Wang P. Metabolic Engineering of Escherichia coli for De Novo Biosynthesis of Mandelic Acid. Fermentation. 2025; 11(6):331. https://doi.org/10.3390/fermentation11060331
Chicago/Turabian StyleLiu, Chang, Xuefeng Xiao, Wanbin Xing, Rina Na, Yunuo Song, Guoqiang Cao, and Pengchao Wang. 2025. "Metabolic Engineering of Escherichia coli for De Novo Biosynthesis of Mandelic Acid" Fermentation 11, no. 6: 331. https://doi.org/10.3390/fermentation11060331
APA StyleLiu, C., Xiao, X., Xing, W., Na, R., Song, Y., Cao, G., & Wang, P. (2025). Metabolic Engineering of Escherichia coli for De Novo Biosynthesis of Mandelic Acid. Fermentation, 11(6), 331. https://doi.org/10.3390/fermentation11060331





