A Photosynthetic Bacterium Suitable for Treating High-Salinity Sea Cucumber Boiling Broth
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Medium
2.2. Composition Analysis of SCBB
2.3. The Isolation and Characterization of PSB
2.4. Optimization of the COD Loading Concentrations of SCBB
2.5. Optimization of Growth Conditions
2.6. Treatment Effect of PSB on SCBB Under Batch Culture
2.7. Treatment Effect of PSB on SCBB in Fed-Batch Fermenter
2.8. Data Processing and Analysis
3. Results
3.1. Analysis of the SCBB Composition
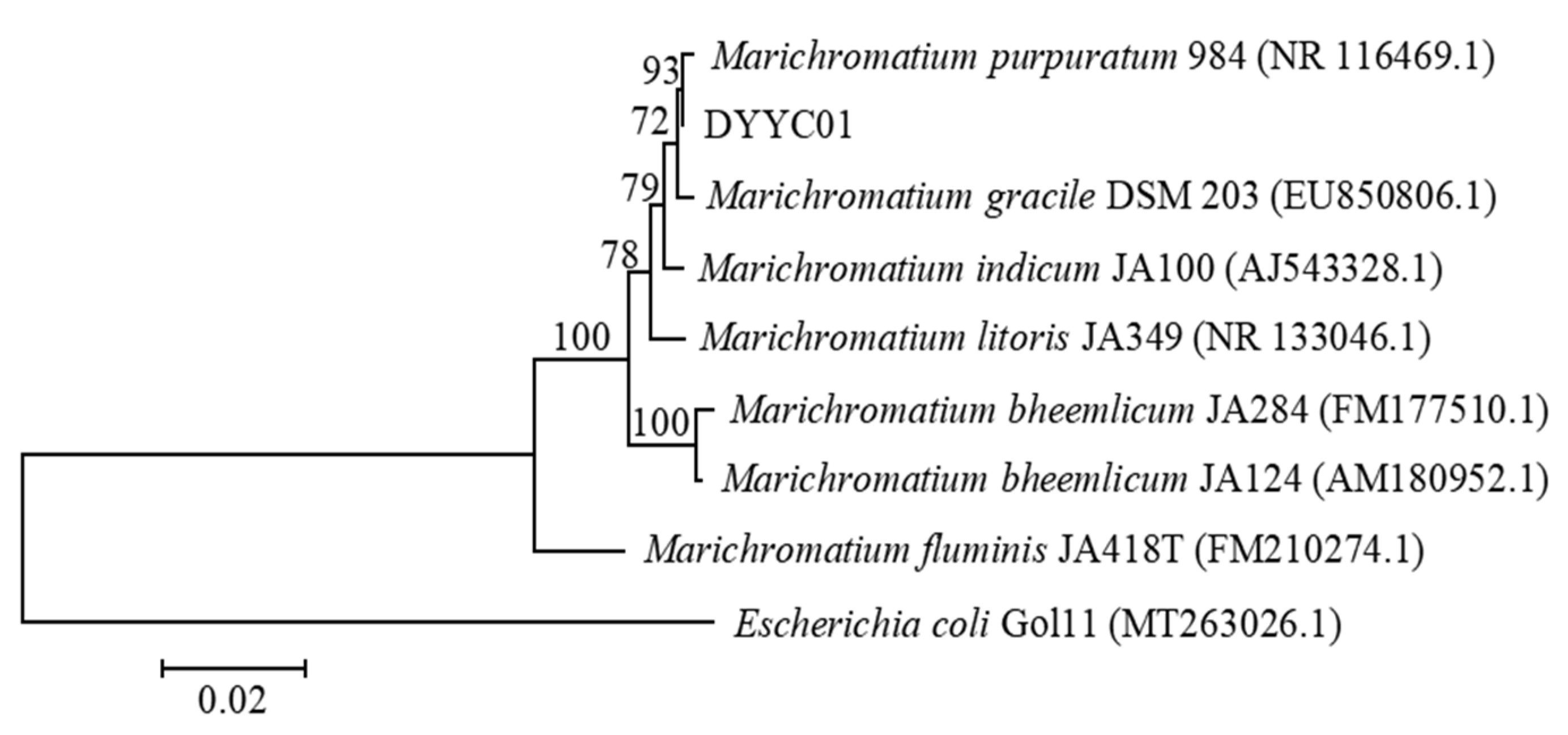
3.2. Identification of Photosynthetic Bacteria
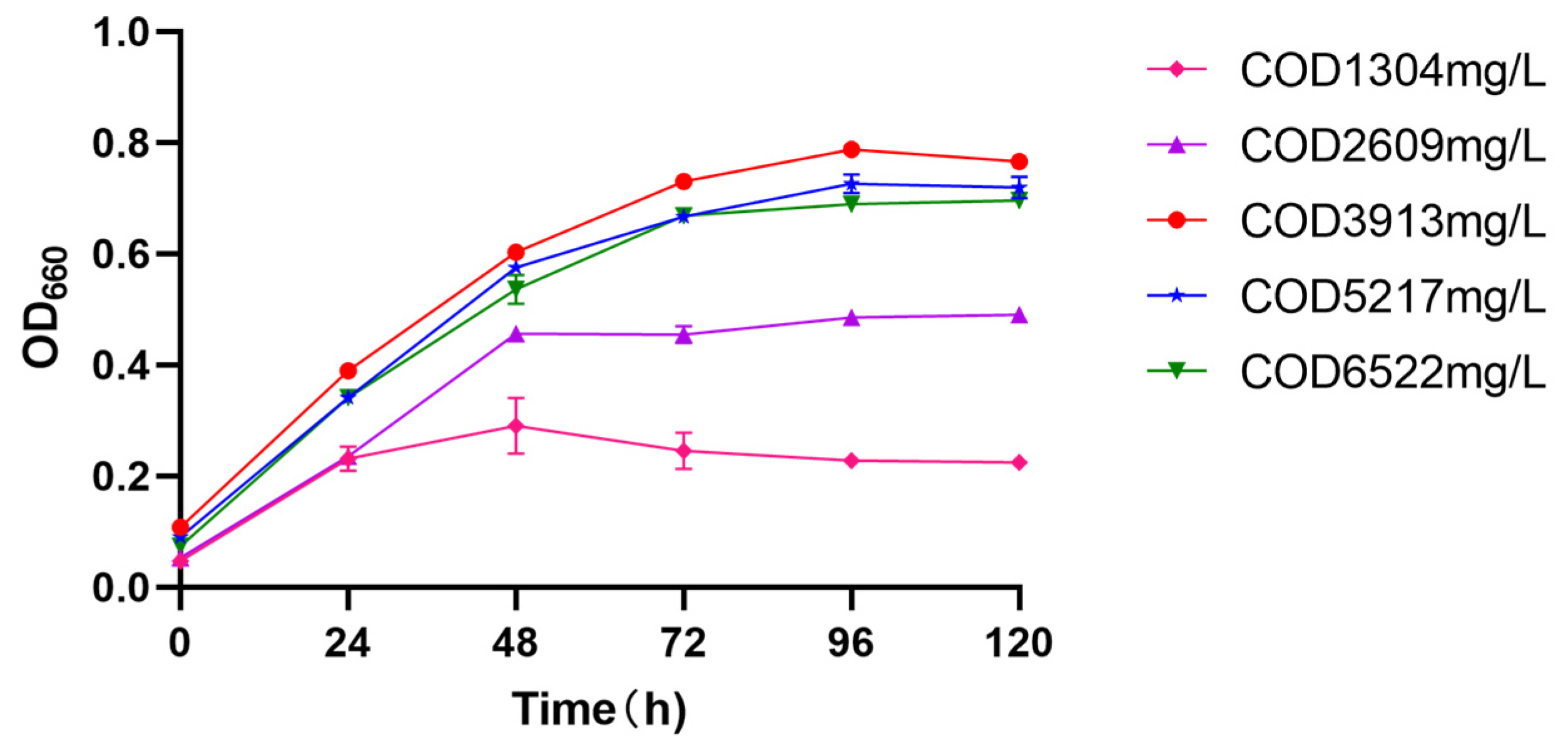
3.3. COD Loading Concentration Optimization
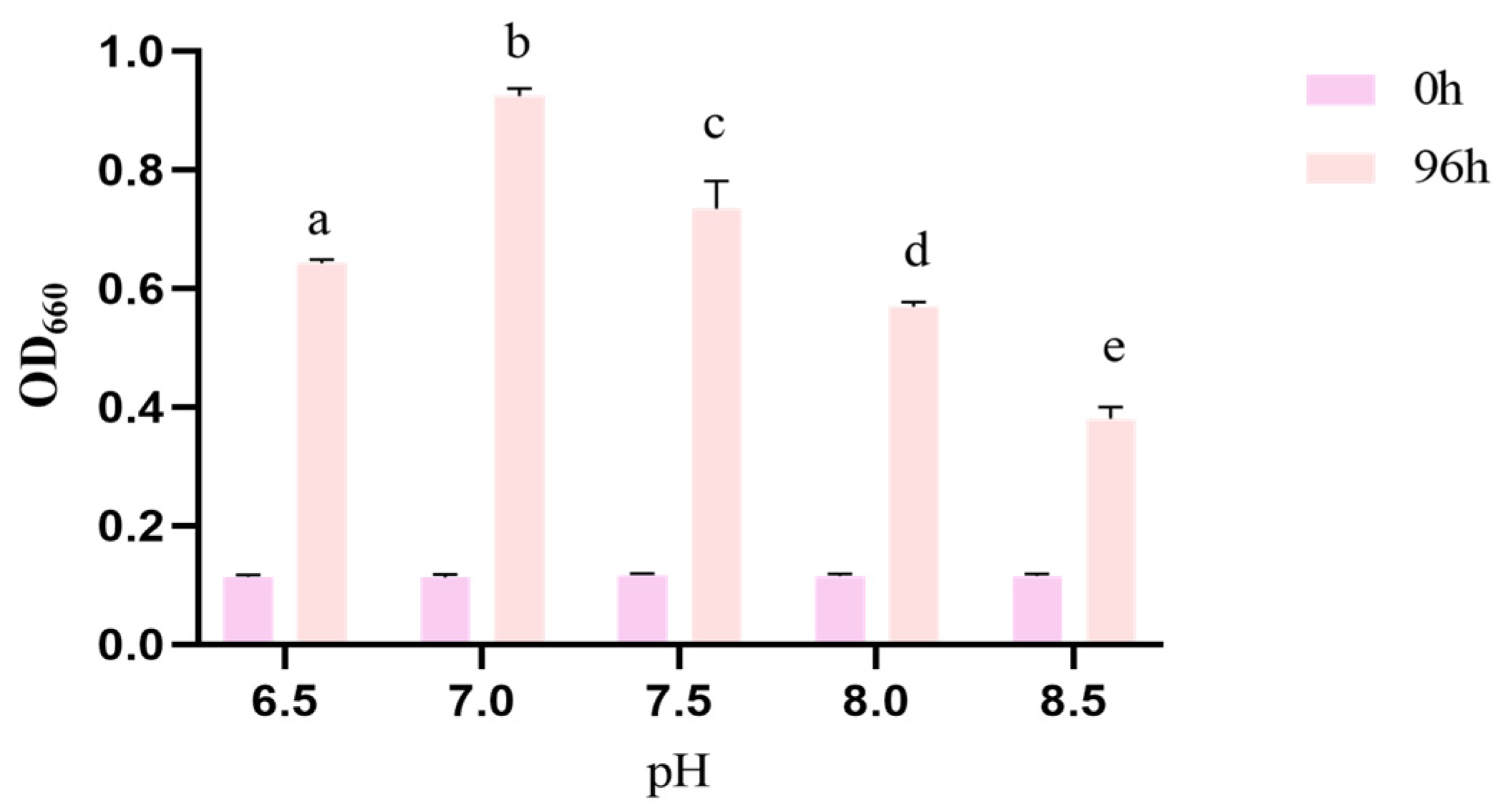
3.4. Growth Conditions Optimization
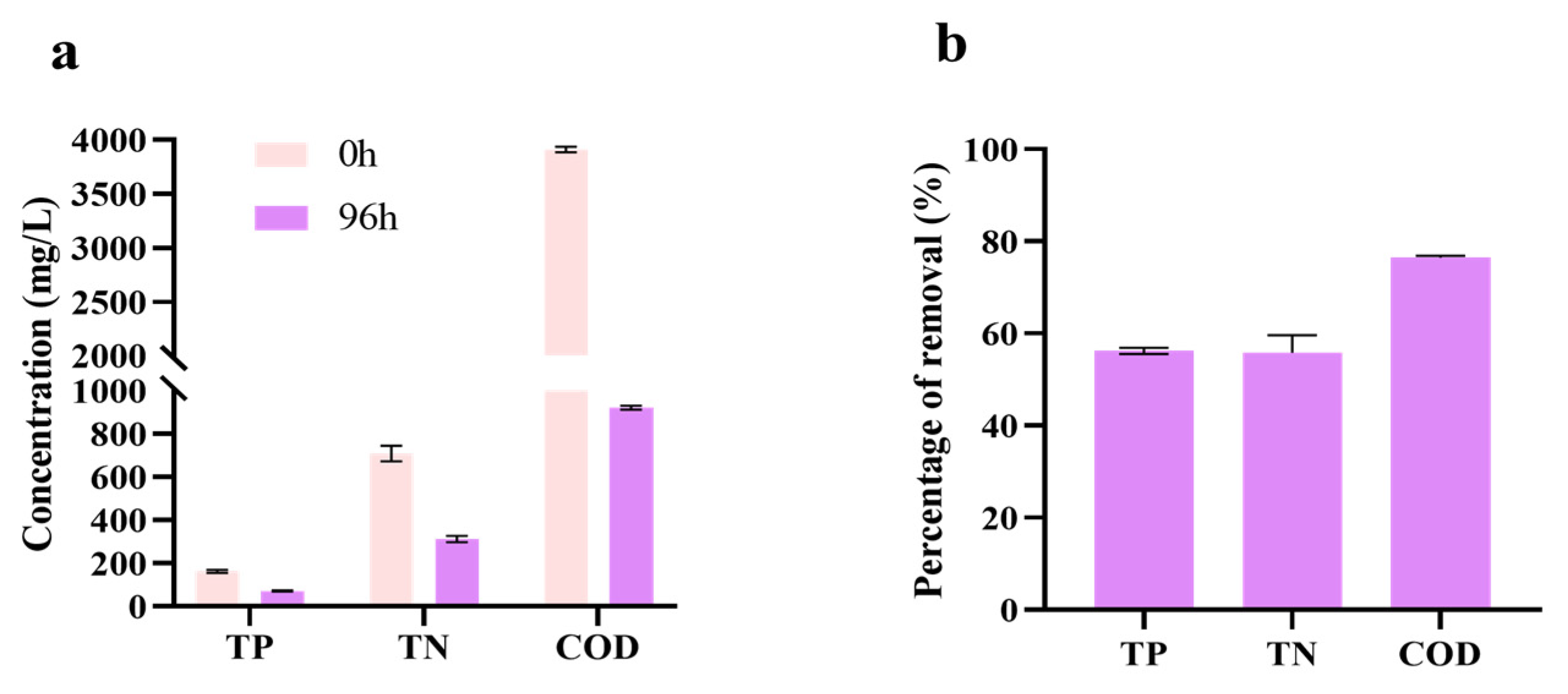
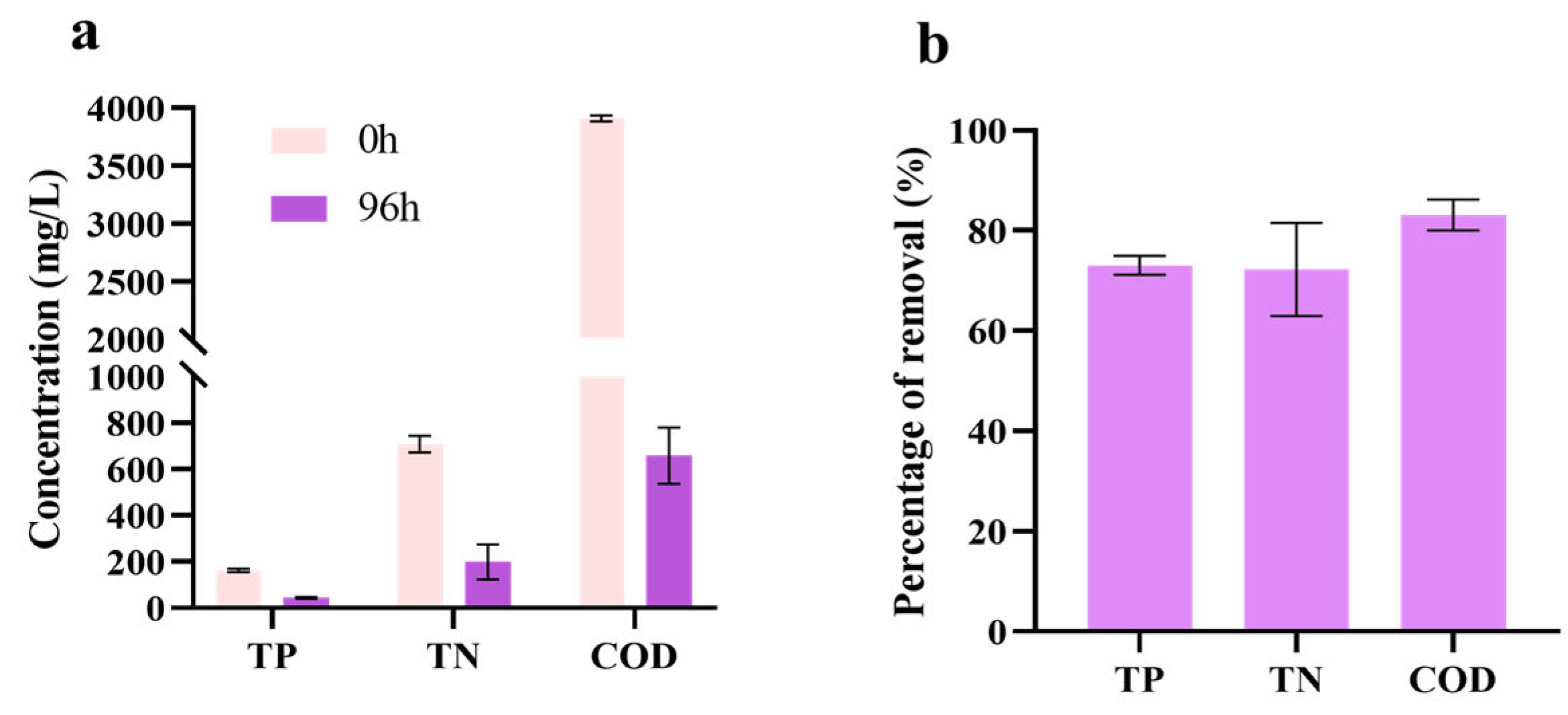
3.5. Treatment Effect on SCBB Under Batch Culture
3.6. Treatment Effect on SCBB in Fed-Batch Fermenter
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Madigan, M.T.; Jung, D.O. An Overview of Purple Bacteria: Systematics, Physiology, and Habitats. In The Purple Phototrophic Bacteria; Hunter, C.N., Daldal, F., Thurnauer, M.C., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 1–15. [Google Scholar] [CrossRef]
- Zhang, X.; Shu, M.; Wang, Y.; Fu, L.; Li, W.; Deng, B.; Liang, Q.; Shen, W. Effect of photosynthetic bacteria on water quality and microbiota in grass carp culture. World J. Microbiol. Biotechnol. 2014, 30, 2523–2531. [Google Scholar] [CrossRef] [PubMed]
- Sund, J.L.; Evenson, C.J.; Strevett, K.A.; Nairn, R.W.; Athay, D.; Trawinski, E. Nutrient conversions by photosynthetic bacteria in a concentrated animal feeding operation lagoon system. J. Environ. Qual. 2001, 30, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Sun, J.; Zheng, J.; Yang, J. An analysis combining proteomics and transcriptomics revealed a regulation target of sea cucumber autolysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2024, 52, 101274. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Ye, F.; Wu, Z.; Wang, Q.; Yuan, Y.; Ye, D. Sustainable Domestic Sewage Reclamation: Insights from Small Villages and Towns in Eastern China. Processes 2025, 13, 435. [Google Scholar] [CrossRef]
- Li, J.; Dong, S.; Gao, Q.; Zhu, C. Nitrogen and phosphorus budget of a polyculture system of sea cucumber (Apostichopus japonicus), jellyfish (Rhopilema esculenta) and shrimp (Fenneropenaeus chinensis). J. Ocean Univ. China 2014, 13, 503–508. [Google Scholar] [CrossRef]
- Fan, X.; Wu, K.; Tian, X.; Benjakul, S.; Li, Y.; Sang, X.; Zhao, Q.; Zhang, J. Endogenous Proteases in Sea Cucumber (Apostichopus japonicas): Deterioration and Prevention during Handling, Processing, and Preservation. Foods 2024, 13, 2153. [Google Scholar] [CrossRef]
- Tang, K.H.; Tang, Y.J.; Blankenship, R.E. Carbon metabolic pathways in phototrophic bacteria and their broader evolutionary implications. Front. Microbiol. 2011, 2, 165. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, M.; Li, X.; Wu, F.; Liu, Y. Nitrogen and phosphorus budget of a Haliotis discus hannai and Apostichopus japonicus polyculture system. Aquac. Res. 2019, 50, 1005–1019. [Google Scholar] [CrossRef]
- Talaiekhozani, A.; Rezania, S. Application of photosynthetic bacteria for removal of heavy metals, macro-pollutants and dye from wastewater: A review. J. Water Process. Eng. 2017, 19, 312–321. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, G.; Zheng, Z.; Meng, F.; Du, T.; He, S. Bio-conversion of photosynthetic bacteria from non-toxic wastewater to realize wastewater treatment and bioresource recovery: A review. Bioresour. Technol. 2019, 278, 383–399. [Google Scholar] [CrossRef]
- Rivera, I.; Buitrón, G.; Bakonyi, P.; Nemestóthy, N.; Bélafi-Bakó, K. Hydrogen production in a microbial electrolysis cell fed with a dark fermentation effluent. J. Appl. Electrochem. 2015, 45, 1223–1229. [Google Scholar] [CrossRef]
- Singh, H.; Paritosh, K.; Vivekanand, V. Microorganism assisted biohydrogen production and bioreactors: An overview. Chem. Eng. Technol. 2021, 46, 204–217. [Google Scholar] [CrossRef]
- Das, D.; Veziroglu, T.N. Advances in biological hydrogen production processes. Int. J. Hydrogen Energy 2008, 33, 6046–6057. [Google Scholar] [CrossRef]
- Akhlaghi, N.; Najafpour-Darzi, G. A comprehensive review on biological hydrogen production. Int. J. Hydrogen Energy 2020, 45, 22492–22512. [Google Scholar] [CrossRef]
- Ogbonna, J.C.; Yoshizawa, H.; Tanaka, H. Treatment of high strength organic wastewater by a mixed culture of photosynthetic microorganisms. J. Appl. Phycol. 2000, 12, 277–284. [Google Scholar] [CrossRef]
- Sreela-or, C.; Imai, T.; Plangklang, P.; Reungsang, A. Optimization of key factors affecting hydrogen production from food waste by anaerobic mixed cultures. Int. J. Hydrogen Energy 2011, 36, 14120–14133. [Google Scholar] [CrossRef]
- Sang, X.; Guan, X.; Tong, Y.; Wang, F.; Zhou, B.; Li, Y.; Zhao, Q. Sulfated Polysaccharides from Sea Cucumber Cooking Liquid Prevents Obesity by Modulating Gut Microbiome, Transcriptome, and Metabolite Profiles in Mice Fed a High-Fat Diet. Foods 2024, 13, 2017. [Google Scholar] [CrossRef]
- Sreela-or, C.; Plangklang, P.; Imai, T.; Reungsang, A. Co-digestion of food waste and sludge for hydrogen production by anaerobic mixed cultures: Statistical key factors optimization. Int. J. Hydrogen Energy 2011, 36, 14227–14237. [Google Scholar] [CrossRef]
- Wang, N.; Xie, L.; Zuo, Y.; Wang, S. Determination of total phosphorus concentration in water by using visible-near-infrared spectroscopy with machine learning algorithm. Environ. Sci. Pollut. Res. Int. 2023, 30, 58243–58252. [Google Scholar] [CrossRef]
- Jaana, K.; Mervi, S.; Kristian, S. Total Nitrogen Determination by a Spectrophotometric Method. Methods Mol. Biol. 2020, 1980, 81–86. [Google Scholar] [CrossRef]
- Santos, A.P.; Santos, M.P. Green analytical method for COD determination using UV–Vis spectroscopy combined with machine learning. Chem. Pap. 2025, 79, 1–8. [Google Scholar] [CrossRef]
- Shivali, K.; Sasikala, C.; Ramana, C.V. MLSA barcoding of Marichromatium spp. and reclassification of Marichromatium fluminis (Sucharita et al.; 2010) as Phaeochromatium fluminis gen. nov. comb. nov. Syst. Appl. Microbiol. 2012, 35, 221–225. [Google Scholar] [CrossRef]
- Kyndt, J.A.; Dubey, S.; Frazier, N.; Meyer, T.E.; Cameron Thrash, J. Genome Sequences of Marichromatium gracile HOL-1 and Its Purple Photosynthetic Coisolate, Afifella sp. H1R. Microbiol. Resour. Announc. 2022, 11, e0003322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, J.; Liu, H.; Liu, S.; Lv, L.; Liang, J. Treatment of high salinity organic wastewater by photosynthetic bacteria to produce value-added products: Performance, community and metabolic characteristics. J. Water Process Eng. 2024, 64, 105593. [Google Scholar] [CrossRef]
- Hou, P.; Liu, S.; Hu, D.; Zhang, J.; Liang, J.; Liu, H.; Zhang, J.; Zhang, G. Predicting biomass conversion and COD removal in wastewater treatment by phototrophic bacteria with interpretable machine learning. J. Environ. Manag. 2025, 375, 124282. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Zhi, R.; Cao, K.-F.; Zhang, G. Bio-stimulation with Bacillus subtilis to realize sucrose wastewater one-step treatment and resource recovery by Aerobic Anoxygenic Phototrophic Bacteria. Desalination Water Treat. 2024, 318, 100408. [Google Scholar] [CrossRef]
- Man, Q.; Zhang, P.; Huang, W.; Zhu, Q.; He, X.; Wei, D. A heterotrophic nitrification-aerobic denitrification bacterium Halomonas venusta TJPU05 suitable for nitrogen removal from high-salinity wastewater. Front. Environ. Sci. Eng. 2021, 16, 69. [Google Scholar] [CrossRef]
- Hong, X.; Chen, Z.; Zhao, C.; Yang, S. Nitrogen transformation under different dissolved oxygen levels by the anoxygenic phototrophic bacterium Marichromatium gracile. World J. Microbiol. Biotechnol. 2017, 33, 113. [Google Scholar] [CrossRef]
- Yang, A.; Zhang, G.; Meng, F.; Zhi, R.; Zhang, P.; Zhu, Y. Nitrogen metabolism in photosynthetic bacteria wastewater treatment: A novel nitrogen transformation pathway. Bioresour. Technol. 2019, 294, 122162. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Teiba, I.; Yoshikawa, T.; Okunishi, S.; Ikenaga, M.; Basuini, M.E.; Maeda, H. Diversity of the Photosynthetic Bacterial Communities in Highly Eutrophicated Yamagawa Bay Sediments. Biocontrol Sci. 2020, 25, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zhi, R.; Zhang, G. Photosynthetic bacteria wastewater treatment with the production of value-added products: A review. Bioresour. Technol. 2020, 299, 122648. [Google Scholar] [CrossRef]
- Zahra, N.; Al Hinai, M.S.; Hafeez, M.B.; Rehman, A.; Wahid, A.; Siddique, K.H.M.; Farooq, M. Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol. Biochem. 2022, 178, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhang, K.; Shi, S.; Jian, L.; Chen, B.; Quadri, S.R.; Tian, X. Alterisphingorhabdus coralli gen. nov. sp. nov.; A novel aerobic anoxygenic phototrophic bacteria isolated from reef-building coral. Int. J. Syst. Evol. Microbiol. 2024, 74, 006577. [Google Scholar] [CrossRef]
- Frigaard, N.-U.; Dahl, C. Sulfur Metabolism in Phototrophic Sulfur Bacteria. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2008; pp. 103–200. [Google Scholar] [CrossRef]
- Laocharoen, S.; Reungsang, A. Isolation, characterization and optimization of photo-hydrogen production conditions by newly isolated Rhodobacter sphaeroides KKU-PS5. Int. J. Hydrogen Energy 2014, 39, 10870–10882. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, S.; Zhang, Z.; Zhang, H.; Xia, C. Enhancement strategies for photo-fermentative biohydrogen production: A review. Bioresour. Technol. 2021, 340, 125601. [Google Scholar] [CrossRef]
- Bibi, F.; Ullah, I.; Alvi, S.A.; Bakhsh, S.A.; Yasir, M.; Al-Ghamdi, A.A.K.; Azhar, E. Isolation, diversity, and biotechnological potential of rhizo- and endophytic bacteria associated with mangrove plants from Saudi Arabia. Genet. Mol. Res. 2017, 16, 10-4238. [Google Scholar] [CrossRef]
- Grattieri, M.; Labarile, R.; Buscemi, G.; Trotta, M. The periodic table of photosynthetic purple non-sulfur bacteria: Intact cell-metal ions interactions. Photochem. Photobiol. Sci. 2022, 21, 101–111. [Google Scholar] [CrossRef]



| Parameter | Salinity | pH | TN | TP | COD |
|---|---|---|---|---|---|
| Concentration | 2–9% | 6.6–6.8 | 278–1100 mg/L | 80–203 mg/L | 1420–15,600 mg/L |
| Amino Acid | Organic Acid | Monosaccharide | |||
|---|---|---|---|---|---|
| Item | Concentration (mg/mL) | Item | Concentration (mg/mL) | Item | Concentration (μg/mg) |
| aspartic acid | 0.235 | oxalic acid | 0.350 | fucose | 2.259 |
| threonine | 0.129 | tartaric acid | 0.395 | rhamnose | N.D |
| serine | 0.085 | formic acid | N.D | galactosamine hydrochloride | 0.244 |
| glutamic acid | 0.322 | malic acid | 0.427 | arabinose | N.D |
| glycine | 0.144 | lactic acid | 1.530 | glucosamine hydrochloride | 2.674 |
| alanine | 0.129 | acetic acid | 0.571 | galactose | 0.770 |
| cystine | 0.052 | citric acid | 0.959 | glucose | 3.725 |
| valine | 0.128 | succinic acid | N.D | mannose | 17.688 |
| methionine | 0.057 | propionic acid | N.D | xylose | N.D |
| isoleucine | 0.095 | - | - | fructose | 29.282 |
| leucine | 0.146 | - | - | ribose | 1.423 |
| tyrosine | 0.061 | - | - | galacturonic acid | N.D |
| phenylalanine | 0.141 | - | - | guluronic acid | N.D |
| histidine | 0.048 | - | - | glucuronic acid | N.D |
| lysine | 0.145 | - | - | mannoturonic acid | N.D |
| arginine | 0.071 | - | - | - | - |
| proline | 0.082 | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, S.; Guo, Y.; Ji, J.; Song, P.; Ma, N.; Qiao, H.; Cai, J. A Photosynthetic Bacterium Suitable for Treating High-Salinity Sea Cucumber Boiling Broth. Fermentation 2025, 11, 284. https://doi.org/10.3390/fermentation11050284
Dong S, Guo Y, Ji J, Song P, Ma N, Qiao H, Cai J. A Photosynthetic Bacterium Suitable for Treating High-Salinity Sea Cucumber Boiling Broth. Fermentation. 2025; 11(5):284. https://doi.org/10.3390/fermentation11050284
Chicago/Turabian StyleDong, Shaokun, Yusi Guo, Jinrui Ji, Pu Song, Ning Ma, Hongjin Qiao, and Jinling Cai. 2025. "A Photosynthetic Bacterium Suitable for Treating High-Salinity Sea Cucumber Boiling Broth" Fermentation 11, no. 5: 284. https://doi.org/10.3390/fermentation11050284
APA StyleDong, S., Guo, Y., Ji, J., Song, P., Ma, N., Qiao, H., & Cai, J. (2025). A Photosynthetic Bacterium Suitable for Treating High-Salinity Sea Cucumber Boiling Broth. Fermentation, 11(5), 284. https://doi.org/10.3390/fermentation11050284






