Catabolism Mechanism and Growth-Promoting Effect of Xylooligosaccharides in Lactiplantibacillus plantarum Strain B20
Abstract
1. Introduction
2. Material and Methods
2.1. Chemicals and Reagents
2.2. Bacterial Strain and Growth Conditions
2.3. Compare the Growth-Promoting Effect of Different Oligosaccharides on Lb. plantarum B20
2.4. Dose- and Fraction-Dependent Effect of XOS on Lb. plantarum B20
2.5. Quantification of Bacterial Growth and Lactic Acid Production
2.6. Genomic Analysis of XOS Catabolism in Lb. plantarum
2.7. RNA Isolation and Sequencing of Lb. plantarum B20
2.8. Bioinformatic Analysis of Transcriptomic Data
2.9. Quantitative Real-Time Polymerase Chain Reaction (qPCR) Validation
2.10. Characterization of DEGs and Heterologous Expression of an XOS Inducible Hydrolase
2.11. Enzymatic Activity Assays
2.12. Statistical Analysis
3. Results and Discussion
3.1. XOS Show Dose- and Fraction-Dependent Growth-Promoting Effects in Lb. plantarum B20
3.2. XOS Induced a Large Number of DEGs in Lb. plantarum B20
3.3. Genomic Insight into XOS Catabolism in Lb. plantarum
3.4. An XOS-Inductive ABC Transporter May Mediate Its Uptake
3.5. An Unknown Hydrolase RS06170 Can Degrade XOS
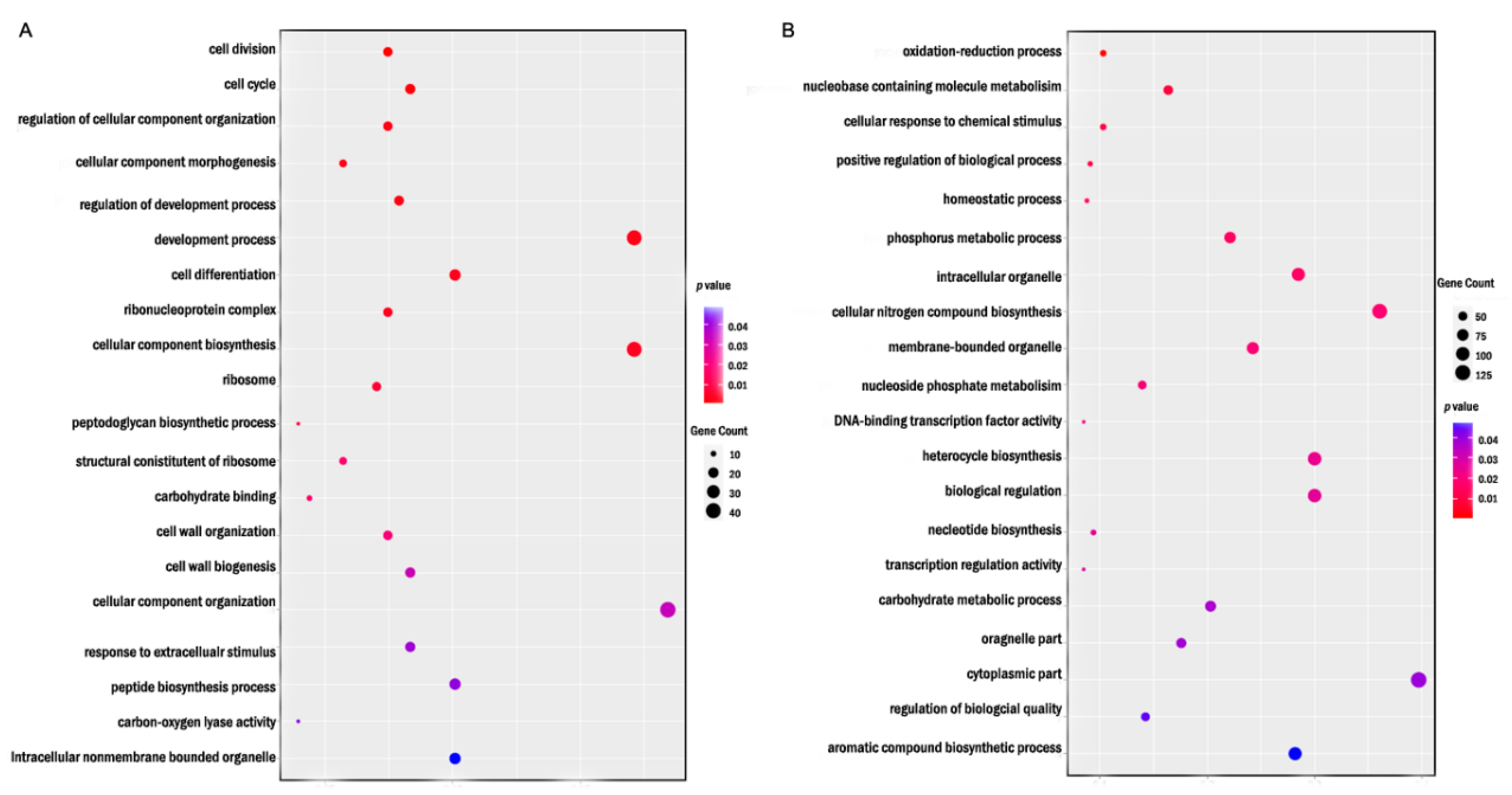
3.6. DEGs Were Enriched in Different GO Terms Under Different Levels of XOS
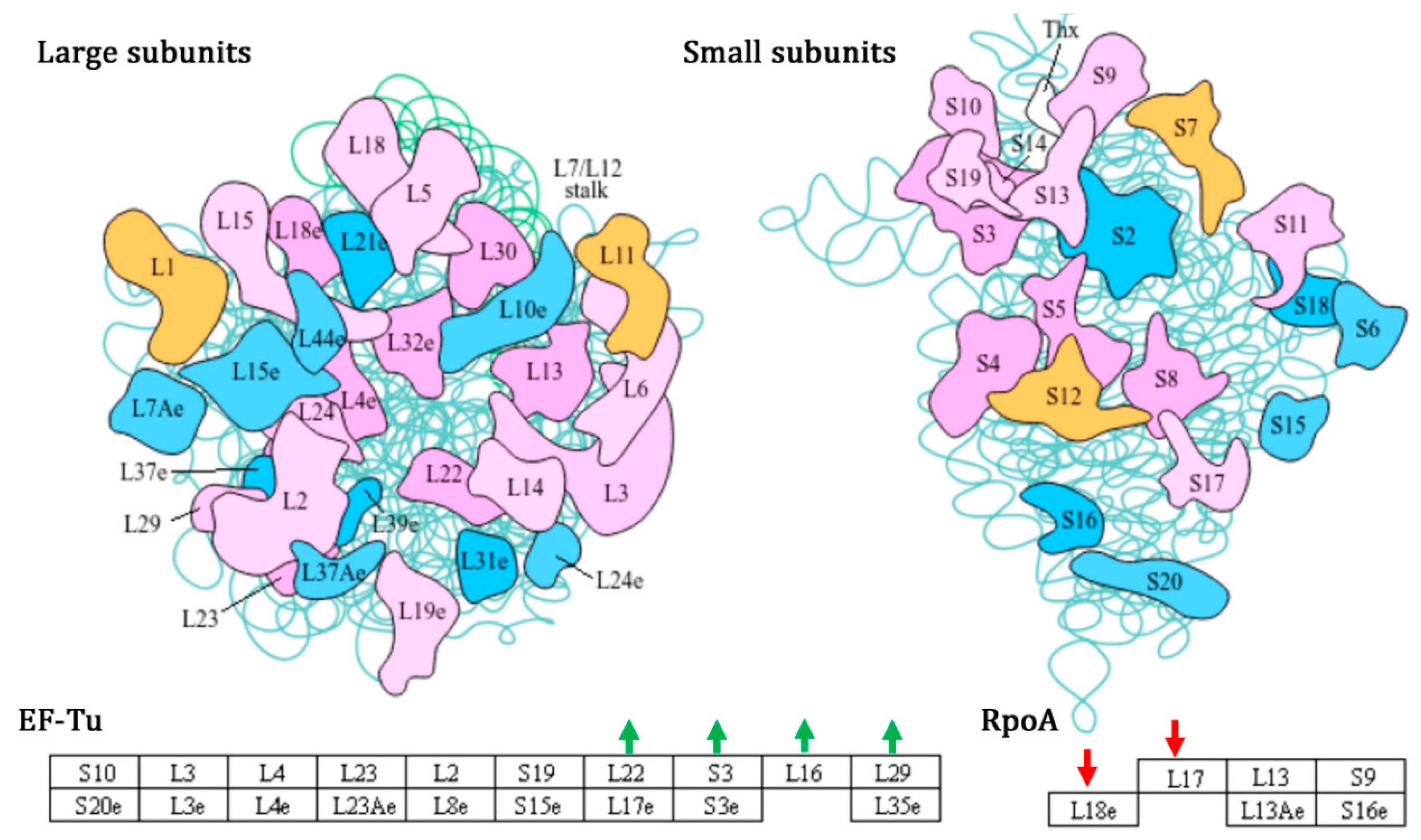
3.7. XOS-Promoted Growth of Lb. plantarum B20 by Impacting Ribosome and Carbohydrate Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, S.; Yang, Z.; Yan, F.; Xue, X.; Lu, J. Preparation of Xylooligosaccharides from Rice Husks and Their Structural Characterization, Antioxidant Activity, and Probiotic Properties. Int. J. Biol. Macromol. 2024, 271, 132575. [Google Scholar] [CrossRef]
- Sun, Z.; Yue, Z.; Liu, E.; Li, X.; Li, C. Assessment of the Bifidogenic and Antibacterial Activities of Xylooligosaccharide. Front. Nutr. 2022, 9, 858949. [Google Scholar] [CrossRef]
- Iliev, I.; Vasileva, T.; Bivolarski, V.; Momchilova, A.; Ivanova, I. Metabolic Profiling of Xylooligosaccharides by Lactobacilli. Polymers 2020, 12, 2387. [Google Scholar] [CrossRef]
- Puja, B.K.; Mallick, S.; Dey, T.; Chanda, S.; Ghosh, S. Xylooligosaccharide Recovery from Sugarcane Bagasse Using β-Xylosidase-Less Xylanase, BsXln1, Produced by Bacillus stercoris DWS1: Characterization, Antioxidant Potential and Influence on Probiotics Growth under Anaerobic Conditions. Int. J. Biol. Macromol. 2025, 285, 138307. [Google Scholar] [CrossRef]
- Li, Z.; Summanen, P.H.; Komoriya, T.; Finegold, S.M. In vitro study of the prebiotic xylooligosaccharide (XOS) on the growth of Bifidobacterium spp and Lactobacillus spp. Int. J. Food Sci. Nutr. 2015, 66, 919–922. [Google Scholar] [CrossRef]
- Ejby, M.; Fredslund, F.; Vujicic-Zagar, A.; Svensson, B.; Slotboom, D.J.; Abou Hachem, M. Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium animalis subsp. lactis Bl-04. J. Mol. Microbiol. Biotechnol. 2013, 90, 1100–1112. [Google Scholar] [CrossRef]
- Mukherjee, S.; Lodha, T.D.; Madhuprakash, J. Comprehensive Genome Analysis of Cellulose and Xylan-Active CAZymes from the Genus Paenibacillus: Special Emphasis on the Novel Xylanolytic Paenibacillus sp. LS1. Microbiol. Spectrum. 2023, 11, e05028-22. [Google Scholar] [CrossRef]
- Gilad, O.; Jacobsen, S.; Stuer-Lauridsen, B.; Pedersen, M.B.; Garrigues, C.; Svensson, B. Combined Transcriptome and Proteome Analysis of Bifidobacterium animalis Subsp. lactis BB-12 Grown on Xylo-Oligosaccharides and a Model of Their Utilization. Appl. Environ. Microbiol. 2010, 76, 7285–7291. [Google Scholar] [CrossRef]
- Zúñiga, M.; Yebra, M.J.; Monedero, V. Complex Oligosaccharide Utilization Pathways in Lactobacillus. Curr. Issues Mol. Biol. 2021, 40, 49–80. [Google Scholar] [CrossRef] [PubMed]
- Michlmayr, H.; Hell, J.; Lorenz, C.; Böhmdorfer, S.; Rosenau, T.; Kneifel, W. Arabinoxylan Oligosaccharide Hydrolysis by Family 43 and 51 Glycosidases from Lactobacillus brevis DSM 20054. Appl. Environ. Microbiol. 2013, 79, 6747–6754. [Google Scholar] [CrossRef] [PubMed]
- Lokman, B.C.; Santen, P.V.; Verdoes, J.C.; Kruse, J.; Leer, R.J.; Posno, M.; Pouwels, P.H. Organization and Characterization of Three Genes Involved in D-Xylose Catabolism in Lactobacillus pentosus. Molec. Gen. Genet. 1991, 230, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wang, M.; Zheng, Y.; Miao, K.; Qu, X. The Carbohydrate Metabolism of Lactiplantibacillus plantarum. Int. J. Mol. Sci. 2021, 22, 13452. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Powell, H.R.; Islam, S.A.; David, A.; Sternberg, M.J.E. Phyre2.2: A Community Resource for Template-Based Protein Structure Prediction. J. Mol. Biol. 2025, 168960. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sun, Z.; Ni, Z.; Qi, Y.; Sun, Q.; Hu, Y.; Li, C. Molecular Identification and Engineering a Salt-Tolerant GH11 Xylanase for Efficient Xylooligosaccharides Production. Biomolecules. 2024, 14, 1188. [Google Scholar] [CrossRef]
- Das, R.; Mishra, P.; Mishra, B.; Jha, R. Effects of in ovo feeding of xylobiose and xylotriose on growth performance, carcass traits, ileal histomorphometry, and immune-related gene expression in broiler chickens. Anim. Feed Sci. Technol. 2024, 313, 115998. [Google Scholar] [CrossRef]
- Pavankumar, T.L. RNase R vs. PNPase: Selecting the best-suited exoribonuclease for environmental adaptation. Extremophiles 2024, 28, 35. [Google Scholar] [CrossRef]
- Dimitrova-Paternoga, L.; Kasvandik, S.; Beckert, B.; Granneman, S.; Tenson, T.; Wilson, D.N.; Paternoga, H. Structural basis of ribosomal 30S subunit degradation by RNase R. Nature 2024, 626, 1133–1140. [Google Scholar] [CrossRef]
- Suman; Chaudhary, M.; Nain, V. In Silico Identification and Evaluation of Bacillus Subtilis Cold Shock Protein B (cspB)-Like Plant RNA Chaperones. J. Biomol. Struct. Dyn. 2021, 39, 841–850. [Google Scholar] [CrossRef]
- Bilsing, F.L.; Anlauf, M.T.; Hachani, E.; Khosa, S.; Schmitt, L. ABC Transporters in Bacterial Nanomachineries. Int. J. Mol. Sci. 2023, 24, 6227. [Google Scholar] [CrossRef]
- Pulianmackal, L.T.; Vecchiarelli, A.G. Positioning of Cellular Components by the ParA/MinD Family of ATPases. Curr. Opin. Microbiol. 2024, 79, 102485. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.J.; Klaenhammer, T.R. Genetic Mechanisms of Prebiotic Oligosaccharide Metabolism in Probiotic Microbes. Annu. Rev. Food Sci. Technol. 2015, 6, 137–156. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Erni, B. Transporters of Glucose and Other Carbohydrates in Bacteria. Pflugers Arch-Eur. J. Physiol. 2020, 472, 1129–1153. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.M.; Barrangou, R.; Hachem, M.A.; Lahtinen, S.J.; Goh, Y.J.; Svensson, B.; Klaenhammer, T.R. Transcriptional Analysis of Oligosaccharide Utilization by Bifidobacterium lactis Bl-04. BMC Genomics 2013, 14, 312. [Google Scholar] [CrossRef]
- Watanabe, A.; Hiraga, K.; Suda, M.; Yukawa, H.; Inui, M. Functional Characterization of Corynebacterium alkanolyticum β-Xylosidase and Xyloside ABC Transporter in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2015, 81, 4173–4183. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, J.; Li, X.; Li, X.; Qiu, S.; Xu, Z.; Yang, J.; Zhu, Y.; Zhang, X.; Yan, L. Phenotypic and Genomic Characterization of Levilactobacillus brevis YT108: A Potential Probiotic Strain Capable of Metabolizing Xylo-Oligosaccharides. Lett. Appl. Microbiol. 2025, 78, ovaf014. [Google Scholar] [CrossRef] [PubMed]
- Shulami, S.; Zaide, G.; Zolotnitsky, G.; Langut, Y.; Feld, G.; Sonenshein, A.L.; Shoham, Y. A Two-Component System Regulates the Expression of an ABC Transporter for Xylo-Oligosaccharides in Geobacillus searothermophilus. Appl. Environ. Microbiol. 2007, 73, 874–884. [Google Scholar] [CrossRef]
- Masulis, I.S.; Sukharycheva, N.A.; Kiselev, S.S.; Andreeva, Z.S.; Ozoline, O.N. Between Computational Predictions and High-Throughput Transcriptional Profiling: In Depth Expression Analysis of the OppB Trans-Membrane Subunit of Escherichia coli OppABCDF Oligopeptide Transporter. Res. Microbiol. 2020, 171, 55–63. [Google Scholar] [CrossRef]
- Yang, X.; Hu, T.; Liang, J.; Xiong, Z.; Lin, Z.; Zhao, Y.; Zhou, X.; Gao, Y.; Sun, S.; Yang, X.; et al. An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality. Nat. Struct. Mol. Biol. 2024, 31, 1072–1082. [Google Scholar] [CrossRef]
- Nanavati, D.M.; Thirangoon, K.; Noll, K.M. Several Archaeal Homologs of Putative Oligopeptide-Binding Proteins Encoded by Thermotoga maritima Bind Sugars. Appl. Environ. Microbiol. 2006, 72, 1336–1345. [Google Scholar] [CrossRef]
- Chandravanshi, M.; Sharma, A.; Dasgupta, P.; Mandal, S.K.; Kanaujia, S.P. Identification and characterization of ABC transporters for carbohydrate uptake in Thermus thermophilus HB8. Gene 2019, 696, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Li, L.; Du, G.; Zhang, Y.; Wang, X.; Li, S.; Xiang, W. A previously unidentified sugar transporter for engineering of high-yield Streptomyces. Appl. Microbiol. Biotechnol. 2024, 108, 72. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.; Aronsson, A.; Karlsson, E.N.; Adlercreutz, P. Xylo- and arabinoxylooligosaccharides from wheat bran by endoxylanases, utilisation by probiotic bacteria, and structural studies of the enzymes. Appl. Microbiol Biotechnol. 2018, 102, 3105–3120. [Google Scholar] [CrossRef]
- Khangwal, I.; Skariyachan, S.; Uttarkar, A.; Muddebihalkar, A.G.; Niranjan, V.; Shukla, P. Understanding the Xylooligosaccharides Utilization Mechanism of Lactobacillus brevis and Bifidobacterium adolescentis: Proteins Involved and Their Conformational Stabilities for Effectual Binding. Mol. Biotechnol. 2022, 64, 75–89. [Google Scholar] [CrossRef]
- Saberi, M.; Dekkers, R.; Passerini, L.; Huber, M.; Overhand, M.; Ubbink, M. Terminal spin labeling of xylotriose strongly affects interactions in the active site of xylanase BcX. J. Biomol. NMR, 2025; advance online publication. [Google Scholar] [CrossRef]
- Ferenczy, G.G.; Kellermayer, M. Contribution of hydrophobic interactions to protein mechanical stability. Comput. Struct. Biotechnol. J. 2022, 20, 1946–1956. [Google Scholar] [CrossRef]
- Orihara, K.; Yahagi, K.; Saito, Y.; Watanabe, Y.; Sasai, T.; Hara, T.; Tsukuda, N.; Oki, K.; Fujimoto, J.; Matsuki, T. Characterization of Bifidobacterium kashiwanohense that utilizes both milk- and plant-derived oligosaccharides. Gut Microbes 2023, 15, 2207455. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, Y.; Ishikawa, S.; Sakane, I.; Yoshida, K.I.; Osawa, R. Identification of genes encoding a novel ABC transporter in Lactobacillus delbrueckii for inulin polymers uptake. Sci. Rep. 2021, 11, 16007. [Google Scholar] [CrossRef]
- Yan, F.; Dong, S.; Liu, Y.J.; Yao, X.; Chen, C.; Xiao, Y.; Bayer, E.A.; Shoham, Y.; You, C.; Cui, Q.; et al. Deciphering Cellodextrin and Glucose Uptake in Clostridium thermocellum. mBio 2022, 13, e0147622. [Google Scholar] [CrossRef]
- Skariyachan, S.; Khangwal, I.; Niranjan, V.; Kango, N.; Shukla, P. Deciphering effectual binding potential of xylo-substrates towards xylose isomerase and xylokinase through molecular docking and molecular dynamic simulation. J. Biomol. Struct. Dyn. 2021, 39, 3948–3957. [Google Scholar] [CrossRef]
- Singh, R.P.; Bhaiyya, R.; Thakur, R.; Niharika, J.; Singh, C.; Latousakis, D.; Saalbach, G.; Nepogodiev, S.A.; Singh, P.; Sharma, S.C.; et al. Biochemical Basis of Xylooligosaccharide Utilisation by Gut Bacteria. Int. J. Mol. Sci. 2022, 23, 2992. [Google Scholar] [CrossRef] [PubMed]
- Rohman, A.; Dijkstra, B.W.; Puspaningsih, N.N.T. β-Xylosidases: Structural Diversity, Catalytic Mechanism, and Inhibition by Monosaccharides. Int. J. Mol. Sci. 2019, 20, 5524. [Google Scholar] [CrossRef] [PubMed]
- Lasrado, L.D.; Gudipati, M. Purification and Characterization of β-D-Xylosidase from Lactobacillus brevis Grown on Xylo-Oligosaccharides. Carbohydr. Polym. 2013, 92, 1978–1983. [Google Scholar] [CrossRef]
- Pontonio, E.; Mahony, J.; Di Cagno, R.; O’Connell Motherway, M.; Lugli, G.A.; O’Callaghan, A.; De Angelis, M.; Ventura, M.; Gobbetti, M.; van Sinderen, D. Cloning, Expression and Characterization of a β-D-Xylosidase from Lactobacillus rossiae DSM 15814T. Microb. Cell Fact. 2016, 15, 72. [Google Scholar] [CrossRef]
- Mahone, C.R.; Goley, E.D. Bacterial Cell Division at a Glance. J. Cell Sci. 2020, 133, jcs237057. [Google Scholar] [CrossRef]
- Cameron, T.A.; Margolin, W. Insights into the Assembly and Regulation of the Bacterial Divisome. Nat. Rev. Microbiol. 2024, 22, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Villanelo, F.; Ordenes, A.; Brunet, J.; Lagos, R.; Monasterio, O. A Model for the Escherichia coli FtsB/FtsL/FtsQ Cell Division Complex. BMC Struct. Biol. 2011, 11, 28. [Google Scholar] [CrossRef]
- Ren, L.; Ye, H.; Fang, J.; Cao, Q.; Zhang, C.; Dong, Z.; Feng, D.; Zuo, J.; Wang, W. Xylooligosaccharide Interferes with the Cell Cycle and Reduces the Antibiotic Tolerance of Avian Pathogenic Escherichia coli by Associating with Its Potential Antimetabolic Actions. Poult. Sci. 2024, 103, 104405. [Google Scholar] [CrossRef]
- Fasnacht, M.; Polacek, N. Oxidative Stress in Bacteria and the Central Dogma of Molecular Biology. Front. Mol. Biosci. 2021, 8, 671037. [Google Scholar] [CrossRef]
- Nasseri, S.A.; Lazarski, A.C.; Lemmer, I.L.; Zhang, C.Y.; Brencher, E.; Chen, H.-M.; Sim, L.; Panwar, D.; Betschart, L.; Worrall, L.J.; et al. An Alternative Broad-Specificity Pathway for Glycan Breakdown in Bacteria. Nature 2024, 631, 199–206. [Google Scholar] [CrossRef]
- Jüttner, M.; Ferreira-Cerca, S. Ribosome Biogenesis: Methods and Protocols, 1st ed.; Humana Press: New York, NY, USA, 2022; pp. 3–22. ISBN 978-01-4939-2685-7. [Google Scholar]
- Levin, B.R.; McCall, I.C.; Perrot, V.; Weiss, H.; Ovesepian, A.; Baquero, F. A Numbers Game: Ribosome Densities, Bacterial Growth, and Antibiotic-Mediated Stasis and Death. mBio 2017, 8, e02253-16. [Google Scholar] [CrossRef] [PubMed]
- Serbanescu, D.; Ojkic, N.; Banerjee, S. Nutrient-Dependent Trade-Offs between Ribosomes and Division Protein Synthesis Control Bacterial Cell Size and Growth. Cell Rep. 2020, 32, 108183. [Google Scholar] [CrossRef] [PubMed]
- Øvrebø, J.I.; Ma, Y.; Edgar, B.A. Cell growth and the cell cycle: New insights about persistent questions. BioEssays News Rev. Mol. Cell. Dev. Biol. 2022, 44, e2200150. [Google Scholar] [CrossRef] [PubMed]
- Durica-Mitic, S.; Göpel, Y.; Görke, B. Carbohydrate Utilization in Bacteria: Making the Most Out of Sugars with the Help of Small Regulatory RNAs. Microbiol. Spectr. 2018, 6, 229–248. [Google Scholar] [CrossRef]
- Cylke, A.; Serbanescu, D.; Banerjee, S. Energy allocation theory for bacterial growth control in and out of steady state. bioRxiv 2024. [CrossRef]
- Pony, P.; Rapisarda, C.; Terradot, L.; Marza, E.; Fronzes, R. Filamentation of the Bacterial Bi-Functional Alcohol/Aldehyde Dehydrogenase AdhE is Essential for Substrate Channeling and Enzymatic Regulation. Nat. Commun. 2020, 11, 1426. [Google Scholar] [CrossRef]
- Yu, S.; Meng, S.; Xiang, M.; Ma, H. Phosphoenolpyruvate Carboxykinase in Cell Metabolism: Roles and Mechanisms beyond Gluconeogenesis. Mol. Metab. 2021, 53, 101257. [Google Scholar] [CrossRef]
- Latorre-Muro, P.; Baeza, J.; Hurtado-Guerrero, R.; Hicks, T.; Delso, I.; Hernández-Ruiz, C.; Velázquez-Campoy, A.; Lawton, A.J.; Angulo, J.; Denu, J.M.; et al. Self-Acetylation at the Active Site of Phosphoenolpyruvate Carboxykinase (PCK1) Controls Enzyme Activity. J. Biol. Chem. 2021, 296, 100205. [Google Scholar] [CrossRef]


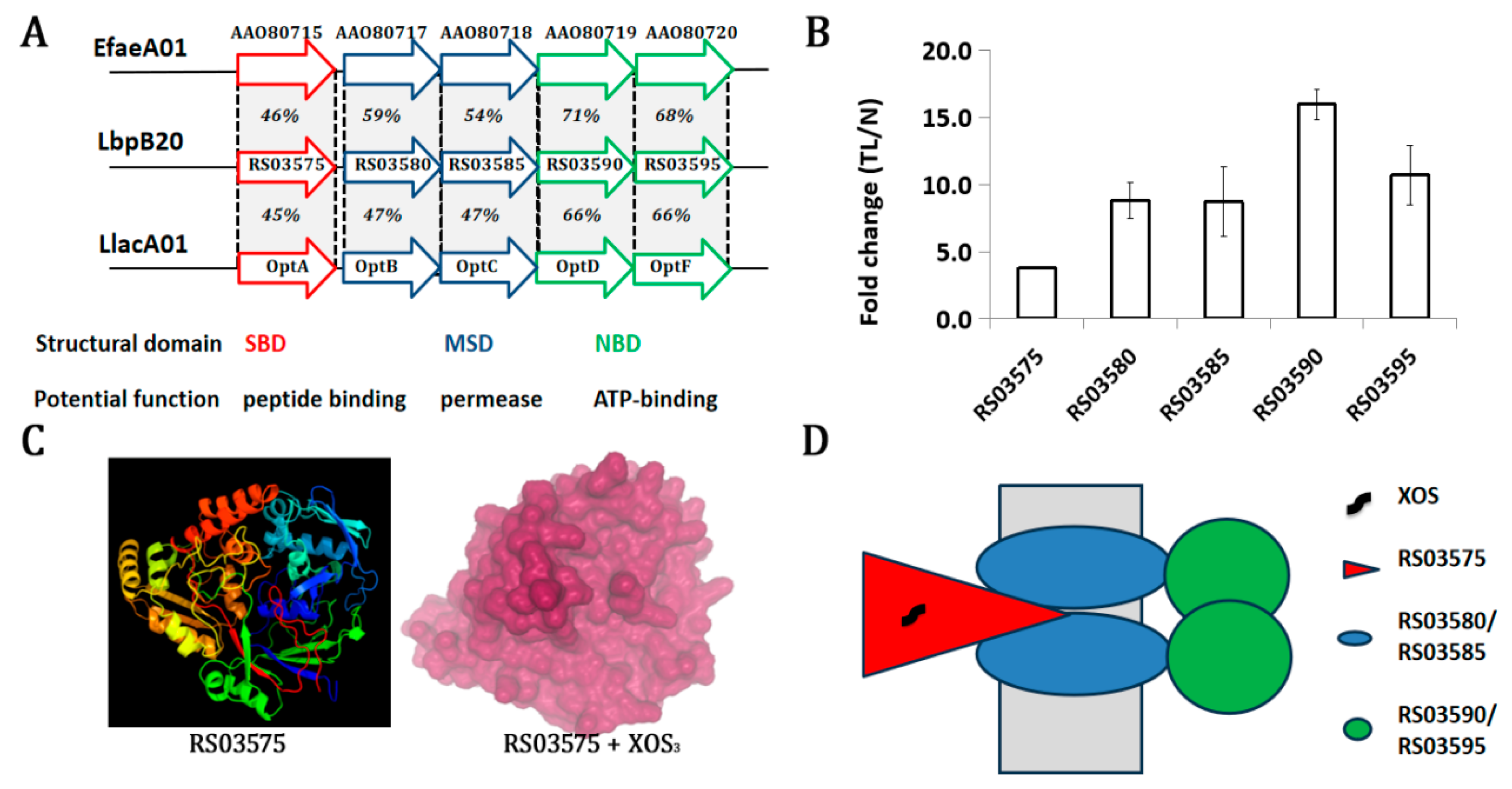
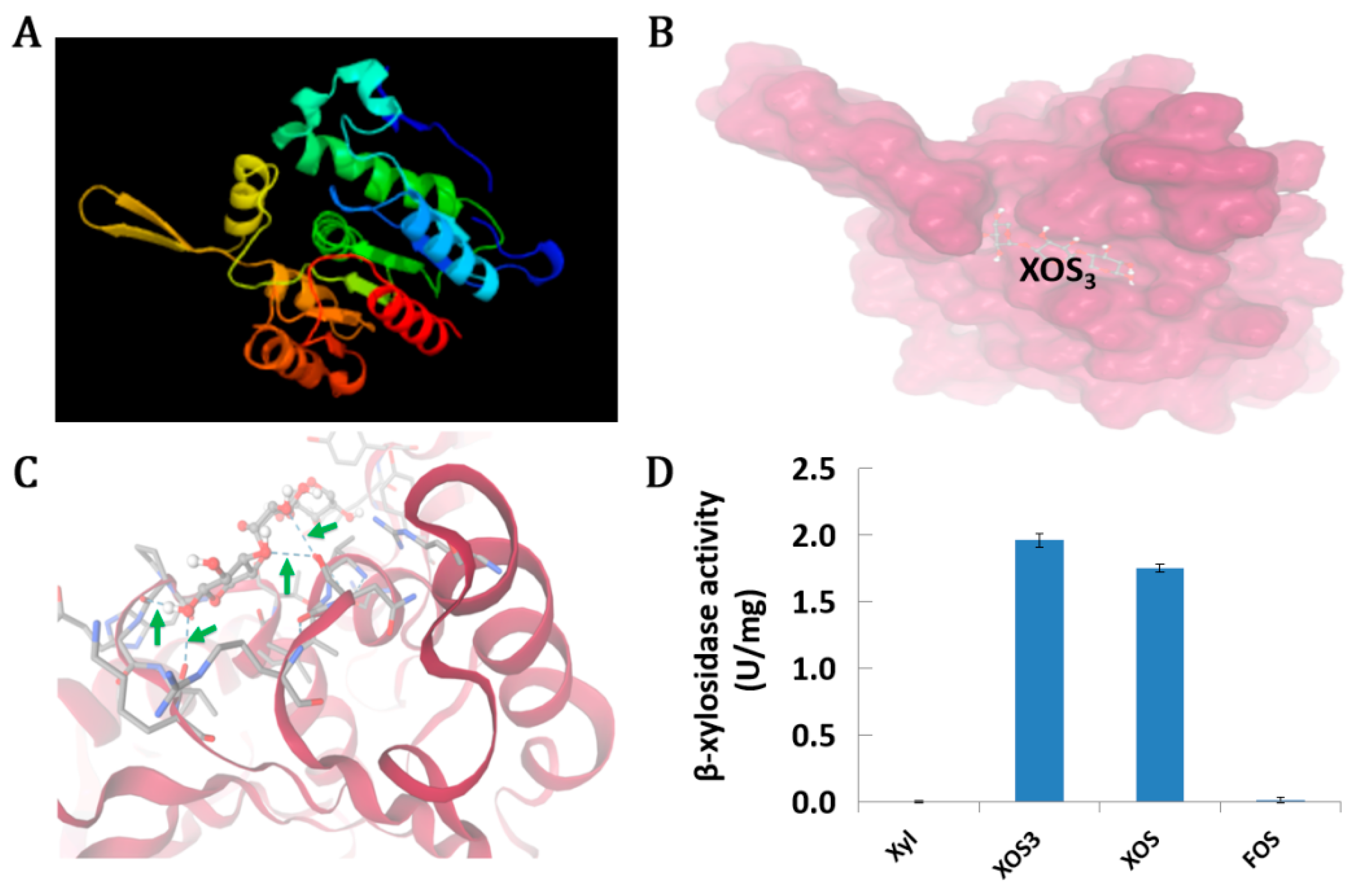
| Seq. id | log2FC(TH/N) | p-Value | log2FC(TL/N) | p-Value | Function |
|---|---|---|---|---|---|
| RS06170 | 7.7 | 0.000 | 8.1 | 0.000 | alpha/beta fold hydrolase |
| RS01605 | 6.9 | 0.000 | 6.8 | 0.000 | ribonuclease R |
| RS11975 | 5.8 | 0.000 | 7.2 | 0.000 | ParA family protein |
| RS01790 | 6.9 | 0.000 | 3.7 | 0.002 | hypothetical protein HMPREF0531_11053 |
| RS12305 | 3.7 | 0.000 | 5.0 | 0.000 | AI-2E family transport protein |
| RS13070 | 3.3 | 0.000 | 5.1 | 0.000 | UDP-glucose 4-epimerase GalE |
| RS05470 | 4.2 | 0.000 | 4.1 | 0.000 | ACP S-malonyltransferase |
| RS07205 | 5.0 | 0.000 | 3.1 | 0.000 | Chaperone protein DnaK |
| RS03035 | 5.2 | 0.000 | 2.9 | 0.016 | trans-hexaprenyltranstransferase, component II |
| RS13065 | 3.7 | 0.000 | 3.8 | 0.000 | UDP-glucose-hexose-1-phosphate uridylyltransferase |
| RS05485 | 3.1 | 0.005 | 4.3 | 0.000 | acetyl-CoA carboxylase, biotin carboxyl carrier protein |
| RS15065 | 3.8 | 0.000 | 3.4 | 0.008 | DUF2089 family protein |
| RS03590 | 2.5 | 0.000 | 3.9 | 0.000 | ABC transporter ATP-binding protein |
| RS05080 | 2.2 | 0.001 | 4.0 | 0.000 | hypothetical protein |
| RS04790 | 3.0 | 0.022 | 2.7 | 0.006 | 50S ribosomal protein L35 |
| RS03995 | 2.7 | 0.000 | 2.5 | 0.000 | hypothetical protein HMPREF0531_12577 |
| RS02650 | 2.4 | 0.000 | 2.5 | 0.004 | 50S ribosomal protein L16 |
| RS01150 | 2.3 | 0.006 | 2.4 | 0.029 | recombination protein RecR |
| RS07280 | 2.1 | 0.045 | 2.6 | 0.022 | YlxR family protein |
| RS11970 | 2.0 | 0.000 | 2.5 | 0.000 | ParB/RepB/Spo0J family partition protein, DNA-binding protein |
| Function | Query | Description | Top Hit | Identity (%) | Reference |
|---|---|---|---|---|---|
| Uptake | BIF_00212 | Sugar-binding protein | WP_063847148.1 | 23.23 | [8] |
| BIF_00257 BIF_00258 | ABC transporter permease | POD88560.1 WP_072539333.1 | 28.38 34.78 | ||
| BALAC_0514 | Sugar-binding protein | WP_063847148.1 | 23.23 | [6] | |
| BALAC_0515 BALAC_0516 | ABC transporter permease | POD88560.1 WP_072539333.1 | 28.38 34.78 | [24] | |
| xylE (AJY53615.1) | Solute-binding protein | WP_063847148.1 | 25.63 | [25] | |
| xylF (AJY53616.1) xylG (AJY53617.1) | ABC transporter permease | POD88560.1 WP_072539333.1 | 27.27 32.91 | ||
| Degradation | BIF_00928 BIF_00633 | Endo-1,4-β-xylanase | WP_097558172.1 WP_063486650.1 | 37.01 22.44 | [8] |
| BIF_00405 BIF_00092 | β-xylosidase | BBM21911.1 WP_063486650.1 | 27.49 30.11 | ||
| xylD (AJY53618.1) | β-xylosidase | WP_114648652.1 | 30.03 | [25] | |
| xynB1 (LVIS_0375) | β-xylosidase | BBM21911.1 | 27.96 | [10] | |
| xynB2 (LVIS_2285) | BBM21911.1 | 29.17 | |||
| Conversion | BIF_00501 | Xylose isomerase | WP_021337395.1 | 70.14 | [8] |
| BIF_00829 | Xylulose kinase | WP_021337394.1 | 26.28 | ||
| BALAC_0521 | Xylulose kinase | WP_021337394.1 | 26.28 | [24] | |
| xylA (P21938.1) | Xylose isomerase | WP_021337395.1 | 99.11 | [11] | |
| xylB (P21939.1) | Xylulose kinase | WP_021337394.1 | 98.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Wang, H.; Sun, Z.; Ni, Z.; Li, C. Catabolism Mechanism and Growth-Promoting Effect of Xylooligosaccharides in Lactiplantibacillus plantarum Strain B20. Fermentation 2025, 11, 280. https://doi.org/10.3390/fermentation11050280
Shi Y, Wang H, Sun Z, Ni Z, Li C. Catabolism Mechanism and Growth-Promoting Effect of Xylooligosaccharides in Lactiplantibacillus plantarum Strain B20. Fermentation. 2025; 11(5):280. https://doi.org/10.3390/fermentation11050280
Chicago/Turabian StyleShi, Yini, Huan Wang, Zhongke Sun, Zifu Ni, and Chengwei Li. 2025. "Catabolism Mechanism and Growth-Promoting Effect of Xylooligosaccharides in Lactiplantibacillus plantarum Strain B20" Fermentation 11, no. 5: 280. https://doi.org/10.3390/fermentation11050280
APA StyleShi, Y., Wang, H., Sun, Z., Ni, Z., & Li, C. (2025). Catabolism Mechanism and Growth-Promoting Effect of Xylooligosaccharides in Lactiplantibacillus plantarum Strain B20. Fermentation, 11(5), 280. https://doi.org/10.3390/fermentation11050280





