Understanding the Functionality of Probiotics on the Edge of Artificial Intelligence (AI) Era
Abstract
1. Introduction
2. Current Knowledge of Probiotics
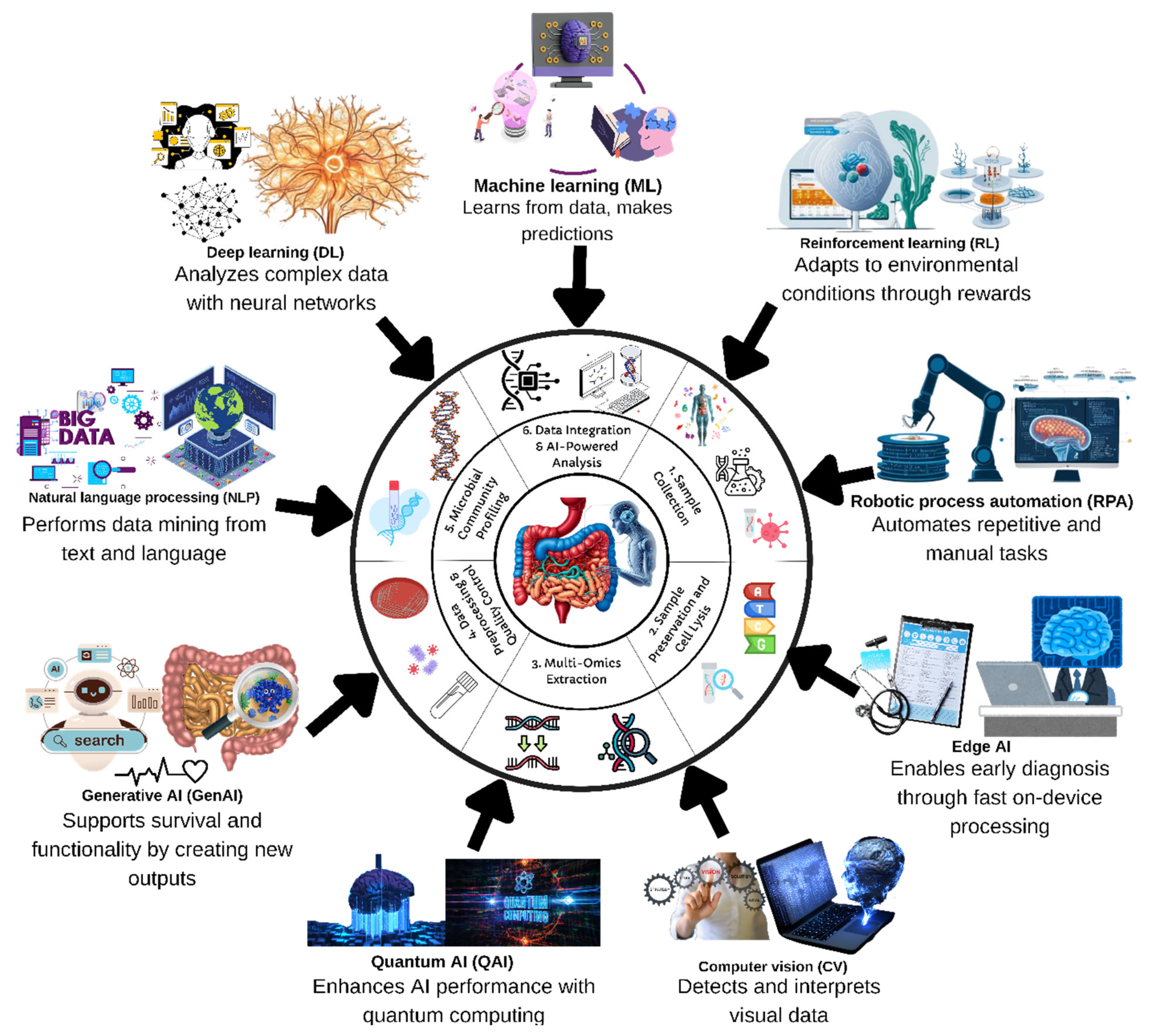
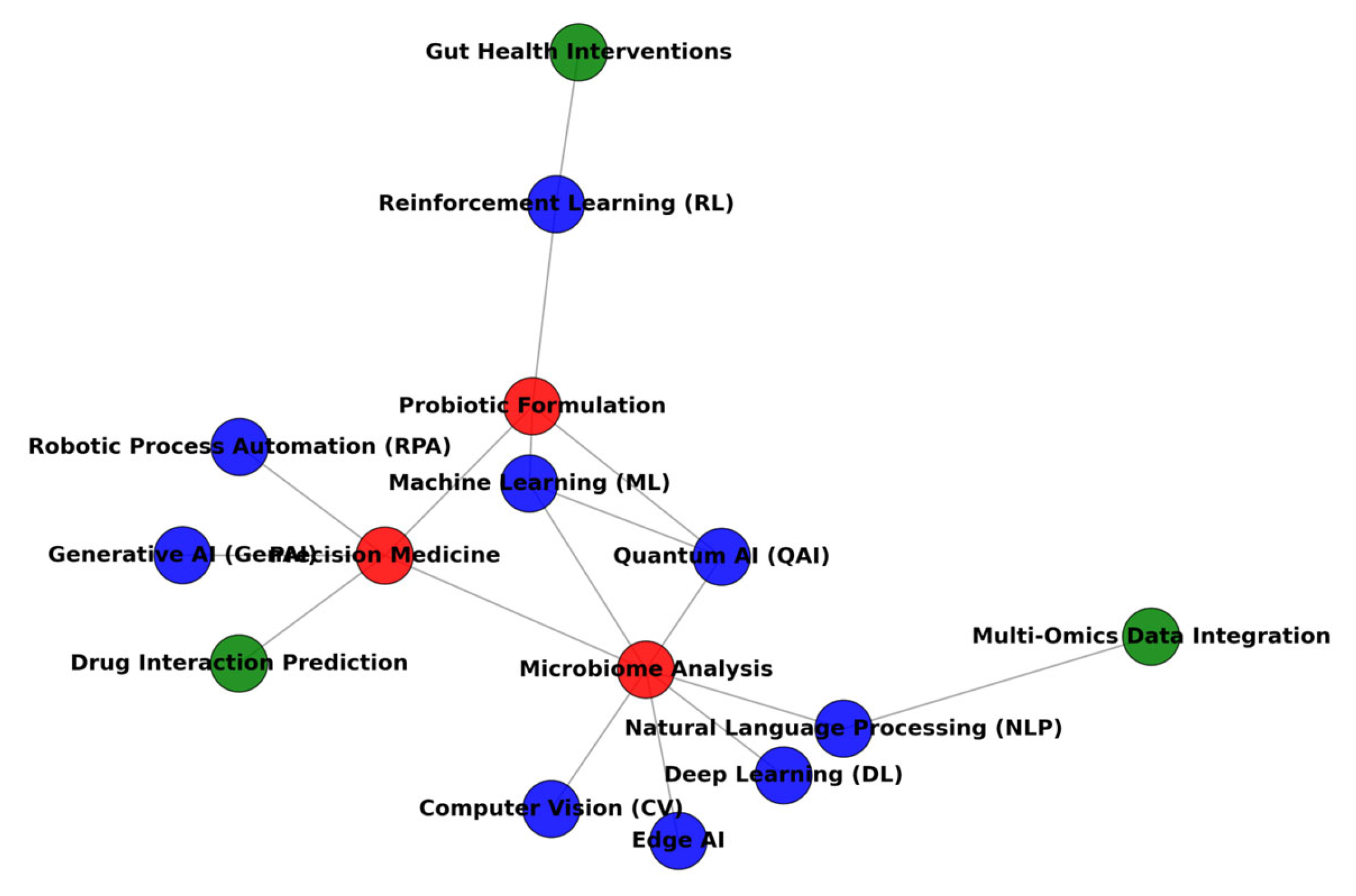
3. Exploring Probiotic Characteristics via AI
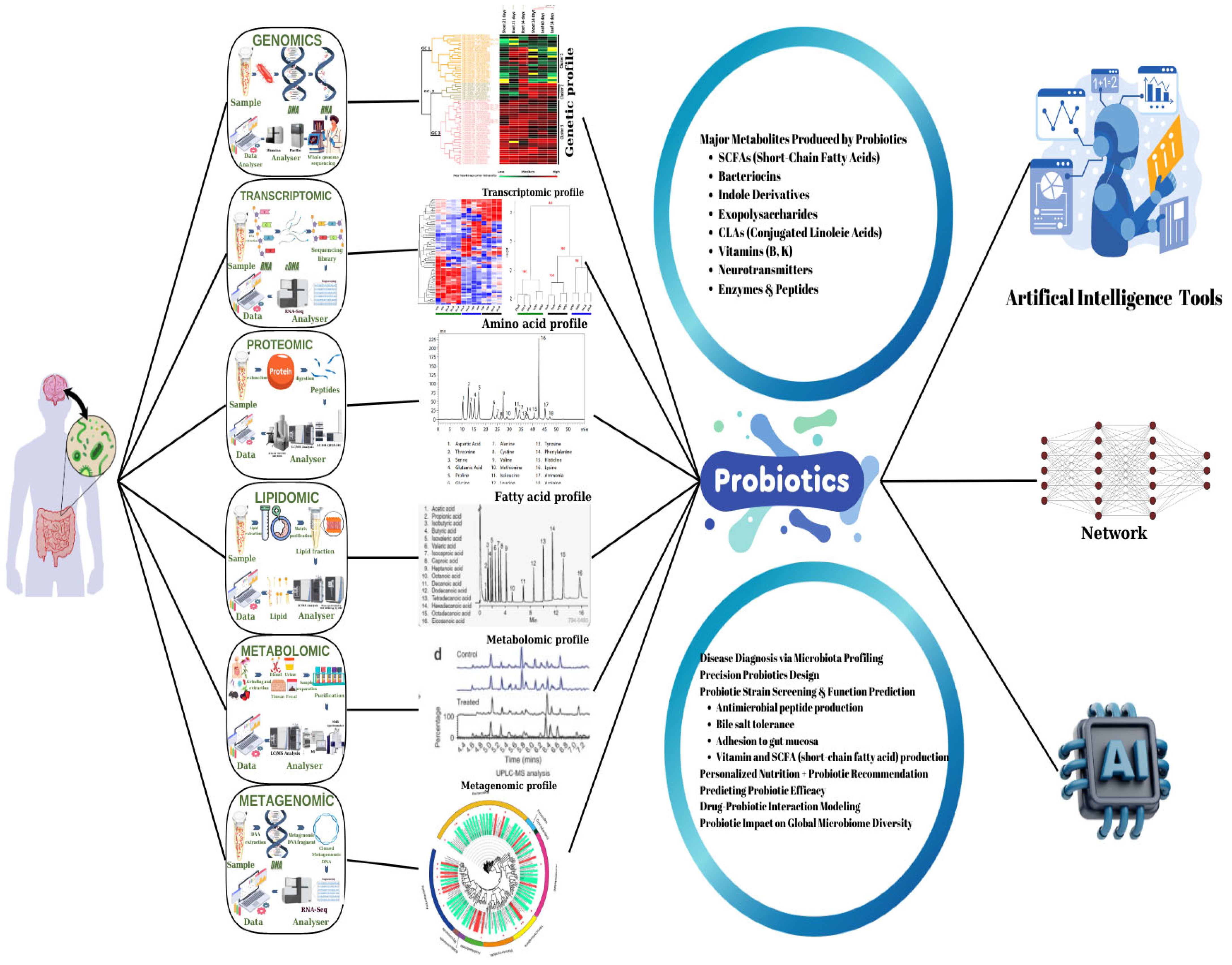
3.1. Omics Technologies
3.2. Mechanism of Action
3.2.1. Role of AI on Microbiota Modulation
3.2.2. Role of AI on Metabolite Production
3.2.3. Role of AI to Understand Immune Modulation
3.2.4. Models and Algorithms in AI-Assisted Probiotic Research
4. Precision of Health Effects
4.1. Intestinal Health
4.2. Anticancerogenic Activities
4.3. Antiaging Roles
4.4. Cardiovascular Health
4.5. Type 2 Diabetes
4.6. Other Roles
5. Technological Perspectives with AI
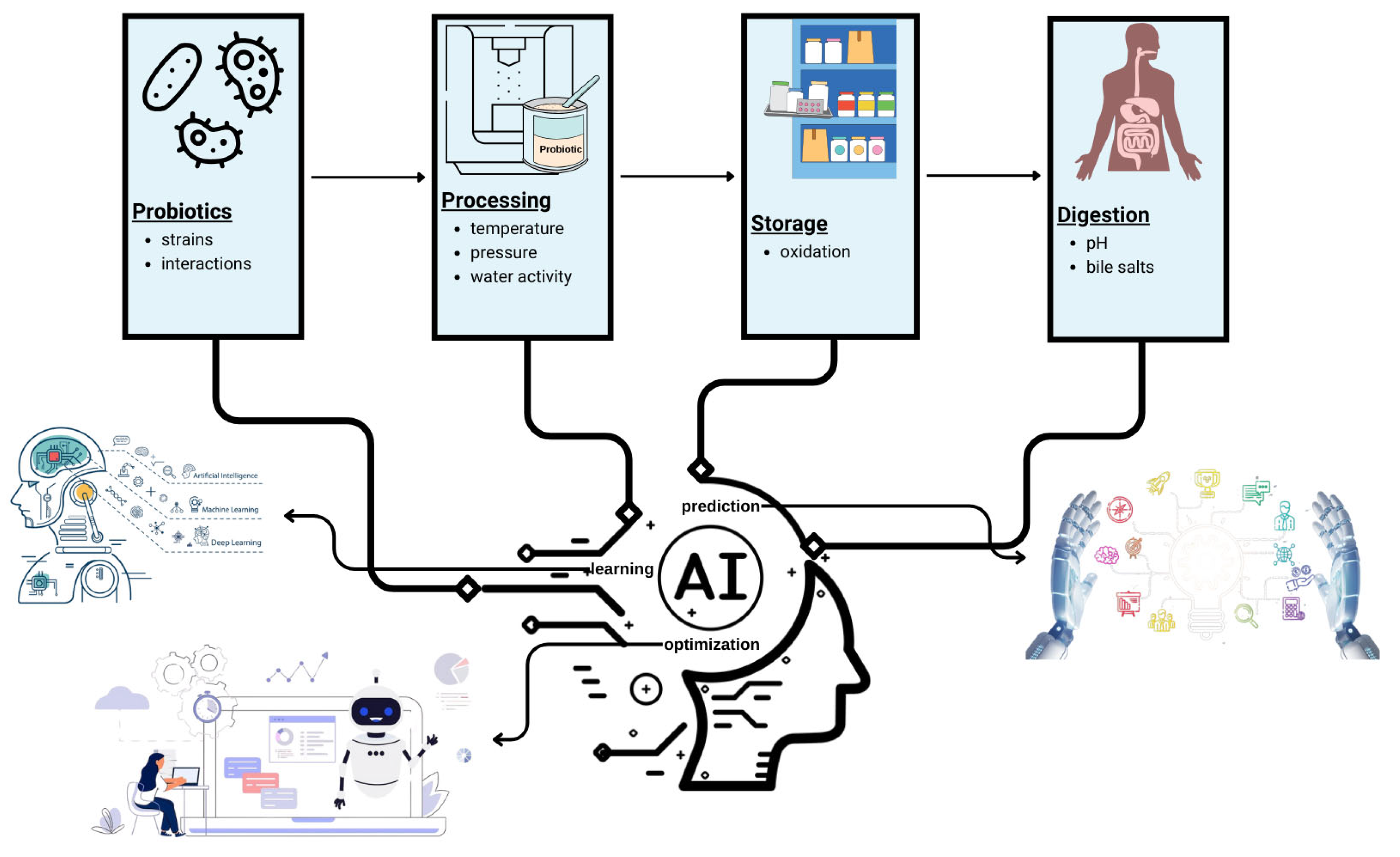
6. Boosting Approach with AI to Increase the Capacity of Probiotics
7. Conclusions and Research Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zendeboodi, F.; Khorshidian, N.; Mortazavian, A.M.; da Cruz, A.G. Probiotic: Conceptualization from a new approach. Curr. Opin. Food Sci. 2020, 32, 103–123. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Díaz, J.; Solís-Urra, P.; Rodríguez-Rodríguez, F.; Olivares-Arancibia, J.; Navarro-Oliveros, M.; Abadía-Molina, F.; Álvarez-Mercado, A.I. The gut barrier, intestinal microbiota, and liver disease: Molecular mechanisms and strategies to manage. Int. J. Mol. Sci. 2020, 21, 8351. [Google Scholar] [CrossRef] [PubMed]
- Al-Fakhrany, O.M.; Elekhnawy, E. Next-generation probiotics: The upcoming biotherapeutics. Mol. Biol. Rep. 2024, 51, 505. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Sharma, V.; Mobeen, F.; Prakash, T. Exploration of survival traits, probiotic determinants, host interactions, and functional evolution of bifidobacterial genomes using comparative genomics. Genes 2018, 9, 477. [Google Scholar] [CrossRef]
- Dianawati, D.; Mishra, V.; Shah, N.P. Survival of microencapsulated probiotic bacteria after processing and during storage: A review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1685–1716. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, H.; Kulyar, M.F.-e.-A.; Pan, H.; Li, K.; Li, A.; Mo, Q.; Wang, Y.; Dong, H.; Bao, Y.; et al. Complete genome analysis of Lactobacillus fermentum YLF016 and its probiotic characteristics. Microb. Pathog. 2022, 162, 105212. [Google Scholar] [CrossRef]
- Josephs-Spaulding, J.; Rajput, A.; Hefner, Y.; Szubin, R.; Balasubramanian, A.; Li, G.; Zielinski Daniel, C.; Jahn, L.; Sommer, M.; Phaneuf, P.; et al. Reconstructing the transcriptional regulatory network of probiotic L. reuteri is enabled by transcriptomics and machine learning. mSystems 2024, 9, e01257-23. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, S.; Chen, L.; Xu, R.; Zhu, J. LC-HRMS-based metabolomics and lipidomics analyses of a novel probiotic Akkermansia muciniphila in response to different nutritional stimulations. J. Microbiol. Methods 2024, 223, 106975. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, Y.; Liu, Z.; Su, M.; Wu, Z.; Zhang, H.; Zhang, C.; Xu, X. Insights into characteristic metabolites and potential bioactive peptides profiles of fresh cheese fermented with three novel probiotics based metabolomics and peptidomics. Food Chem. X 2024, 21, 101147. [Google Scholar] [CrossRef] [PubMed]
- Karakan, T.; Gundogdu, A.; Alagözlü, H.; Ekmen, N.; Ozgul, S.; Tunali, V.; Hora, M.; Beyazgul, D.; Nalbantoglu, O.U. Artificial intelligence-based personalized diet: A pilot clinical study for irritable bowel syndrome. Gut Microbes 2022, 14, 2138672. [Google Scholar] [CrossRef] [PubMed]
- Kwoji, I.D.; Aiyegoro, O.A.; Okpeku, M.; Adeleke, M.A. ‘Multi-omics’ data integration: Applications in probiotics studies. NPJ Sci. Food 2023, 7, 25. [Google Scholar] [CrossRef]
- McCoubrey, L.E.; Seegobin, N.; Elbadawi, M.; Hu, Y.; Orlu, M.; Gaisford, S.; Basit, A.W. Active machine learning for formulation of precision probiotics. Int. J. Pharm. 2022, 616, 121568. [Google Scholar] [CrossRef]
- Dossou, B.F.; Liu, D.; Ji, X.; Jain, M.; van der Sloot, A.M.; Palou, R.; Tyers, M.; Bengio, Y. Graph-based active machine learning method for diverse and novel antimicrobial peptides generation and selection. arXiv 2022, arXiv:2209.13518. [Google Scholar]
- Wu, S.; Feng, J.; Liu, C.; Wu, H.; Qiu, Z.; Ge, J.; Sun, S.; Hong, X.; Li, Y.; Wang, X. Machine learning aided construction of the quorum sensing communication network for human gut microbiota. Nat. Commun. 2022, 13, 3079. [Google Scholar] [CrossRef]
- Pensupa, N.; Treebuppachartsakul, T.; Pechprasarn, S. Machine learning models using data mining for biomass production from Yarrowia lipolytica fermentation. Fermentation 2023, 9, 239. [Google Scholar] [CrossRef]
- Florea, A.; Sipos, A.; Stoisor, M.-C. Applying AI tools for modeling, predicting and managing the white wine fermentation process. Fermentation 2022, 8, 137. [Google Scholar] [CrossRef]
- Bertorello, S.; Cei, F.; Fink, D.; Niccolai, E.; Amedei, A. The future exploring of gut microbiome-immunity interactions: From in vivo/vitro models to in silico innovations. Microorganisms 2024, 12, 1828. [Google Scholar] [CrossRef]
- Sadeghirashed, S.; Kazemi, F.; Taheri, S.; Ebrahimi, M.T.; Arasteh, J. A novel probiotic strain exerts therapeutic effects on mouse model of multiple sclerosis by altering the expression of inflammasome and IDO genes and modulation of T helper cytokine profile. Metab. Brain Dis. 2022, 37, 197–207. [Google Scholar] [CrossRef]
- Han, X.; Liu, Q.; Li, Y.; Zhang, M.; Liu, K.; Kwok, L.-Y.; Zhang, H.; Zhang, W. Synergizing artificial intelligence and probiotics: A comprehensive review of emerging applications in health promotion and industrial innovation. Trends Food Sci. Technol. 2025, 159, 104938. [Google Scholar] [CrossRef]
- Vulpoi, R.A.; Luca, M.; Ciobanu, A.; Olteanu, A.; Bărboi, O.; Iov, D.-E.; Nichita, L.; Ciortescu, I.; Cijevschi Prelipcean, C.; Ștefănescu, G.; et al. The potential use of artificial intelligence in irritable bowel syndrome management. Diagnostics 2023, 13, 3336. [Google Scholar] [CrossRef]
- Karim, M.R.; Morshed, M.N.; Iqbal, S.; Mohammad, S.; Mathiyalagan, R.; Yang, D.C.; Kim, Y.J.; Song, J.H.; Yang, D.U. A network pharmacology and molecular-docking-based approach to identify the probable targets of short-chain fatty-acid-producing microbial metabolites against kidney cancer and inflammation. Biomolecules 2023, 13, 1678. [Google Scholar] [CrossRef]
- Gatineau, G.; Shevroja, E.; Vendrami, C.; Gonzalez-Rodriguez, E.; Leslie, W.D.; Lamy, O.; Hans, D. Development and reporting of artificial intelligence in osteoporosis management. J. Bone Miner. Res. 2024, 39, 1553–1573. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Cao, Z.; Wu, J. Probiotic supplements benefit psoriasis therapy rather than affecting disease risk: Evidence from NHANES machine-learning and meta-analysis study. Probiotics Antimicrob. Proteins 2024, 16, 1–15. [Google Scholar] [CrossRef]
- Rugji, J.; Erol, Z.; Taşçı, F.; Musa, L.; Hamadani, A.; Gündemir, M.G.; Karalliu, E.; Siddiqui, S.A. Utilization of AI—Reshaping the future of food safety, agriculture and food security—A critical review. Crit. Rev. Food Sci. Nutr. 2024, 64, 1–45. [Google Scholar] [CrossRef]
- Bilal, H.; Khan, M.N.; Khan, S.; Shafiq, M.; Fang, W.; Khan, R.U.; Rahman, M.U.; Li, X.; Lv, Q.-L.; Xu, B. The role of artificial intelligence and machine learning in predicting and combating antimicrobial resistance. Comput. Struct. Biotechnol. J. 2025, 27, 423–439. [Google Scholar] [CrossRef]
- Jeong, J.; Kim, S. Application of machine learning for quantitative analysis of industrial fermentation using image processing. Food Sci. Biotechnol. 2025, 34, 373–381. [Google Scholar] [CrossRef]
- Zhang, X.; Elliot, M.A. Unlocking the trove of metabolic treasures: Activating silent biosynthetic gene clusters in bacteria and fungi. Curr. Opin. Microbiol. 2019, 51, 9–15. [Google Scholar] [CrossRef]
- Yue, L.; Song, L.; Zhu, S.; Fu, X.; Li, X.; He, C.; Li, J. Machine learning assisted rational design of antimicrobial peptides based on human endogenous proteins and their applications for cosmetic preservative system optimization. Sci. Rep. 2024, 14, 947. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, W.; Wang, Q.; Cao, Z.; Li, J. Artificially engineered bacteria to treat gastrointestinal disease and cancer. Drug Discov. Today 2023, 28, 103667. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, B.; Liu, Q.; Hou, W.; Cai, J.; Lu, C. Modeling and optimization of sporulation by Bacillus licheniformis BF-002 based on dynamics and recurrent neural networks. Bioresour. Technol. 2024, 398, 130534. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Condò, C.; Tauseef, I.; Idrees, M.; Ghazanfar, S.; Farid, A.; Muzammal, M.; Al Mohaini, M.; Alsalman, A.J.; Al Hawaj, M.A.; et al. Isolation and characterization of a cholesterol-lowering bacteria from Bubalus bubalis raw milk. Fermentation 2022, 8, 163. [Google Scholar] [CrossRef]
- Golnari, M.; Bahrami, N.; Milanian, Z.; Rabbani Khorasgani, M.; Asadollahi, M.A.; Shafiei, R.; Fatemi, S.S.-A. Isolation and characterization of novel Bacillus strains with superior probiotic potential: Comparative analysis and safety evaluation. Sci. Rep. 2024, 14, 1457. [Google Scholar] [CrossRef]
- dos Santos Leandro, E.; Ginani, V.C.; de Alencar, E.R.; Pereira, O.G.; Rose, E.C.P.; do Vale, H.M.M.; Pratesi, R.; Hecht, M.M.; Cavalcanti, M.H.; Tavares, C.S.O. Isolation, identification, and screening of lactic acid bacteria with probiotic potential in silage of different species of forage plants, cocoa beans, and artisanal salami. Probiotics Antimicrob. Proteins 2021, 13, 173–186. [Google Scholar] [CrossRef]
- Amenu, D.; Bacha, K. Probiotic potential and safety analysis of lactic acid bacteria isolated from Ethiopian traditional fermented foods and beverages. Ann. Microbiol. 2023, 73, 37. [Google Scholar] [CrossRef]
- Wu, H.; Shum, T.-F.; Chiou, J. Characterization of the probiotic potential of lactic acid bacteria isolated from kimchi, yogurt, and baby feces in Hong Kong and their performance in soymilk fermentation. Microorganisms 2021, 9, 2544. [Google Scholar] [CrossRef]
- Bihola, A.; Jana, A.H.; Parmar, S.C.; Adil, S. Feasibility study of utilizing Saccharomyces boulardii as an adjunct culture in Mozzarella type cheese and its quality characterization. Discov. Food 2024, 4, 105. [Google Scholar] [CrossRef]
- Vergara Alvarez, S.C.; Pendón, M.D.; Bengoa, A.A.; Leiva Alaniz, M.J.; Maturano, Y.P.; Garrote, G.L. Probiotic potential of yeasts isolated from fermented beverages: Assessment of antagonistic strategies against Salmonella enterica serovar enteritidis. J. Fungi 2024, 10, 878. [Google Scholar] [CrossRef]
- da Silva, M.N.; Tagliapietra, B.L.; do Amaral Flores, V.; dos Santos Richards, N.S.P. In vitro test to evaluate survival in the gastrointestinal tract of commercial probiotics. Curr. Res. Food Sci. 2021, 4, 320–325. [Google Scholar] [CrossRef]
- Abdi, A.; Gatri, E.; Guilbaud, J.; Bouallagui, H.; Fadhlaoui, K.; Garrait, G.; Ayed, L.; Gatri, E.; Guilbaud, J.; Bouallagui, H.; et al. Co-cultivation of potential probiotic strains isolated from water kefir for fermented green tea beverage. Fermentation 2025, 11, 169. [Google Scholar] [CrossRef]
- Shehata, M.G.; Masry, S.H.; Abd El-Aziz, N.M.; Ridouane, F.L.; Mirza, S.B.; El-Sohaimy, S.A. Probiotic potential of lactic acid bacteria isolated from honeybees stomach: Functional and technological insights. Ann. Agric. Sci. 2024, 69, 11–18. [Google Scholar] [CrossRef]
- Sadeghi, M.; Panahi, B.; Mazlumi, A.; Hejazi, M.A.; Komi, D.E.A.; Nami, Y. Screening of potential probiotic lactic acid bacteria with antimicrobial properties and selection of superior bacteria for application as biocontrol using machine learning models. LWT 2022, 162, 113471. [Google Scholar] [CrossRef]
- Xing, T.-l.; Bian, X.; Ma, C.-m.; Yang, Y.; Liu, X.-f.; Wang, Y.; Fan, J.; Zhang, N. In vitro evaluation of probiotic properties of Lactobacillus acidophilus AD125 and antagonism against Escherichia coli O157: H7 adhesion to Caco-2 cell. Food Funct. 2023, 14, 2472–2480. [Google Scholar] [CrossRef]
- Chantanawilas, P.; Pahumunto, N.; Teanpaisan, R. Aggregation and adhesion ability of various probiotic strains and Candida species: An in vitro study. J. Dent. Sci. 2024, 19, 2163–2171. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Q.; Lu, B.; Shen, H.; Liu, S.; Shi, Y.; Leptihn, S.; Li, H.; Wei, J.; Liu, C. Whole-genome analysis of probiotic product isolates reveals the presence of genes related to antimicrobial resistance, virulence factors, and toxic metabolites, posing potential health risks. BMC Genom. 2021, 22, 210. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, S.-I.; Jeong, Y.; Kang, C.-H. Evaluation of safety and probiotic potential of Enterococcus faecalis MG5206 and Enterococcus faecium MG5232 isolated from Kimchi, a Korean fermented cabbage. Microorganisms 2022, 10, 2070. [Google Scholar] [CrossRef]
- Vinayamohan, P.G.; Viju, L.S.; Joseph, D.; Venkitanarayanan, K. Fermented foods as a potential vehicle of antimicrobial-resistant bacteria and genes. Fermentation 2023, 9, 688. [Google Scholar] [CrossRef]
- Gizachew, S.; Van Beeck, W.; Spacova, I.; Dekeukeleire, M.; Alemu, A.; Woldemedhin, W.M.; Mariam, S.H.; Lebeer, S.; Engidawork, E. Antibacterial and Immunostimulatory activity of potential probiotic lactic acid bacteria isolated from Ethiopian fermented dairy products. Fermentation 2023, 9, 258. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Skarveli, A.; Saxami, G.; Mitsou, E.K.; Kotsou, M.; Kyriacou, A. Inhibition of Listeria monocytogenes growth, adherence and invasion in Caco-2 cells by potential probiotic lactic acid bacteria isolated from Fecal samples of healthy neonates. Microorganisms 2023, 11, 363. [Google Scholar] [CrossRef]
- Barzegar, H.; Alizadeh Behbahani, B.; Falah, F. Safety, probiotic properties, antimicrobial activity, and technological performance of Lactobacillus strains isolated from Iranian raw milk cheeses. Food Sci. Nutr. 2021, 9, 4094–4107. [Google Scholar] [CrossRef] [PubMed]
- Ajibade, B.O.; Ajayeoba, T.A.; Sabiu, S.; Moiseenko, K.V.; Mbona, S.V.; Cason, E.D.; Fedorova, T.V.; Ijabadeniyi, O.A. Unveiling the microbial symphony of amasi: A targeted metagenomic 16S rRNA, ITS, and metabolites insights using bovine and caprine milk. Fermentation 2025, 11, 6. [Google Scholar] [CrossRef]
- Chang, H.M.; Foo, H.L.; Loh, T.C.; Lim, E.T.C.; Abdul Mutalib, N.E. Comparative studies of inhibitory and antioxidant activities, and organic acids compositions of postbiotics produced by probiotic Lactiplantibacillus plantarum strains isolated from Malaysian foods. Front. Vet. Sci. 2021, 7, 602280. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.H.; Bamigbade, G.; Tarique, M.; Esposito, G.; Obaid, R.; Abu-Jdayil, B.; Ayyash, M. Physicochemical, rheological, and bioactive properties of exopolysaccharide produced by a potential probiotic Enterococcus faecalis 84B. Int. J. Biol. Macromol. 2023, 240, 124425. [Google Scholar] [CrossRef]
- Cho, H.; Jo, M.; Oh, H.; Lee, Y.; Park, Y. Synergistic antidepressant-like effect of n-3 polyunsaturated fatty acids and probiotics through the brain-gut axis in rats exposed to chronic mild stress. J. Nutr. Biochem. 2023, 116, 109326. [Google Scholar] [CrossRef]
- Noda, M.; Danshiitsoodol, N.; Sakaguchi, T.; Kanno, K.; Sugiyama, M. Exopolysaccharide produced by plant-derived Lactobacillus plantarum SN35N exhibits antiviral activity. Biol. Pharm. Bull. 2021, 44, 1886–1890. [Google Scholar] [CrossRef]
- Nehal, F.; Sahnoun, M.; Smaoui, S.; Jaouadi, B.; Bejar, S.; Mohammed, S. Characterization, high production and antimicrobial activity of exopolysaccharides from Lactococcus lactis F-mou. Microb. Pathog. 2019, 132, 10–19. [Google Scholar] [CrossRef]
- Sugimura, N.; Li, Q.; Chu, E.S.H.; Lau, H.C.H.; Fong, W.; Liu, W.; Liang, C.; Nakatsu, G.; Su, A.C.Y.; Coker, O.O. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut 2022, 71, 2011–2021. [Google Scholar] [CrossRef]
- Afreen, A.; Ahmed, Z.; Khalid, N.; Ferheen, I.; Ahmed, I. Optimization and cholesterol-lowering activity of exopolysaccharide from Lactiplantibacillus paraplantarum NCCP 962. Appl. Microbiol. Biotechnol. 2023, 107, 1189–1204. [Google Scholar] [CrossRef]
- Park, J.M.; Lee, S.C.; Ham, C.; Kim, Y.W. Effect of probiotic supplementation on gastrointestinal motility, inflammation, motor, non-motor symptoms and mental health in Parkinson’s disease: A meta-analysis of randomized controlled trials. Gut Pathog. 2023, 15, 9. [Google Scholar] [CrossRef]
- Juntarachot, N.; Sunpaweravong, S.; Kaewdech, A.; Wongsuwanlert, M.; Ruangsri, P.; Pahumunto, N.; Teanpaisan, R. Characterization of adhesion, anti-adhesion, co-aggregation, and hydrophobicity of Helicobacter pylori and probiotic strains. J. Taibah Univ. Med. Sci. 2023, 18, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bansal, P.; Singh, J.; Dhanda, S. Purification, partial structural characterization and health benefits of exopolysaccharides from potential probiotic Pediococcus acidilactici NCDC 252. Process Biochem. 2020, 99, 79–86. [Google Scholar] [CrossRef]
- Gawande, K.; Kolhekar, M.; Kumari, M.; Kapila, S.; Sharma, P.; Ali, S.A.; Behare, P.V. Lactic acid bacteria based purified exopolysaccharide showed viscofying and hypercholesterolemic capabilites. Food Hydrocoll. Health 2021, 1, 100042. [Google Scholar] [CrossRef]
- Nurkanto, A.; Fanani, A.; Nurcahyanto, D.A.; Mamangkey, J.; Marissa, S.; Pasaribu, K.M.; Afiati, F.; Agustini, N.W.S.; Ahmad, R.Z.; Taufik, M. Anti-hypercholesterolemia properties of exopolysaccharide from Lactiplantibacillus plantarum MI01: Computational and in-vivo approaches. Case Stud. Chem. Environ. Eng. 2025, 11, 101146. [Google Scholar]
- Khalil, M.A.; Sonbol, F.I.; Al-Madboly, L.A.; Aboshady, T.A.; Alqurashi, A.S.; Ali, S.S. Exploring the therapeutic potentials of exopolysaccharides derived from lactic acid bacteria and bifidobacteria: Antioxidant, antitumor, and periodontal regeneration. Front. Microbiol. 2022, 13, 803688. [Google Scholar] [CrossRef]
- Sasikumar, K.; Vaikkath, D.K.; Devendra, L.; Nampoothiri, K.M. An exopolysaccharide (EPS) from a Lactobacillus plantarum BR2 with potential benefits for making functional foods. Bioresour. Technol. 2017, 241, 1152–1156. [Google Scholar] [CrossRef]
- Khromova, N.Y.; Epishkina, J.M.; Karetkin, B.A.; Khabibulina, N.V.; Beloded, A.V.; Shakir, I.V.; Panfilov, V.I. The combination of in vitro assessment of stress tolerance ability, autoaggregation, and vitamin B-producing ability for new probiotic strain introduction. Microorganisms 2022, 10, 470. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Choi, E.-J.; Lee, J.-H.; Yoo, M.-S.; Heo, K.; Shim, J.-J.; Lee, J.-L. Probiotic potential of a novel vitamin B2-overproducing Lactobacillus plantarum strain, HY7715, isolated from Kimchi. Appl. Sci. 2021, 11, 5765. [Google Scholar] [CrossRef]
- Carrizo, S.L.; de LeBlanc, A.d.M.; LeBlanc, J.G.; Rollán, G.C. Quinoa pasta fermented with lactic acid bacteria prevents nutritional deficiencies in mice. Food Res. Int. 2020, 127, 108735. [Google Scholar] [CrossRef]
- Yépez, A.; Russo, P.; Spano, G.; Khomenko, I.; Biasioli, F.; Capozzi, V.; Aznar, R. In situ riboflavin fortification of different kefir-like cereal-based beverages using selected Andean LAB strains. Food Microbiol. 2019, 77, 61–68. [Google Scholar] [CrossRef]
- Thananimit, S.; Pahumunto, N.; Teanpaisan, R. Characterization of short chain fatty acids produced by selected potential probiotic lactobacillus strains. Biomolecules 2022, 12, 1829. [Google Scholar] [CrossRef]
- Alan, Y.; Savcı, A.; Koçpınar, E.F.; Ertaş, M. Postbiotic metabolites, antioxidant and anticancer activities of probiotic Leuconostoc pseudomesenteroides strains in natural pickles. Arch. Microbiol. 2022, 204, 571. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, Y.; Park, S.; Lee, D.; Lee, J.; Hlaing, S.P.; Yoo, J.-W.; Rhee, S.H.; Im, E. Lactobacillus plantarum metabolites elicit anticancer effects by inhibiting autophagy-related responses. Molecules 2023, 28, 1890. [Google Scholar] [CrossRef]
- Abdel-Nasser, A.; Hathout, A.S.; Badr, A.N.; Barakat, O.S.; Fathy, H.M. Extraction and characterization of bioactive secondary metabolites from lactic acid bacteria and evaluating their antifungal and antiaflatoxigenic activity. Biotechnol. Rep. 2023, 38, e00799. [Google Scholar] [CrossRef]
- Sentürk, M.; Ercan, F.; Yalcin, S. The secondary metabolites produced by Lactobacillus plantarum downregulate BCL-2 and BUFFY genes on breast cancer cell line and model organism Drosophila melanogaster: Molecular docking approach. Cancer Chemother. Pharmacol. 2020, 85, 33–45. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Bottacini, F.; Van Sinderen, D.; Gahan, C.G.; Corsetti, A. Comparative genomics of Lactiplantibacillus plantarum: Insights into probiotic markers in strains isolated from the human gastrointestinal tract and fermented foods. Front. Microbiol. 2022, 13, 854266. [Google Scholar] [CrossRef]
- Lau, L.Y.J.; Quek, S.Y. Probiotics: Health benefits, food application, and colonization in the human gastrointestinal tract. Food Bioeng. 2024, 3, 41–64. [Google Scholar] [CrossRef]
- Lalowski, P.; Zielińska, D. The most promising next-generation probiotic candidates—Impact on human health and potential application in food technology. Fermentation 2024, 10, 444. [Google Scholar] [CrossRef]
- Saarela, M.H. Safety aspects of next generation probiotics. Curr. Opin. Food Sci. 2019, 30, 8–13. [Google Scholar] [CrossRef]
- Cai, X.; Wen, J.-s.; Long, H.; Ren, W.; Zhang, X.; Huang, A.-y.; Xie, Z.-y. The probiotic effects, dose, and duration of lactic acid bacteria on disease resistance in Litopenaeus vannamei. Aquac. Rep. 2022, 26, 101299. [Google Scholar] [CrossRef]
- Dai, X.; Shen, L. Advances and trends in omics technology development. Front. Med. 2022, 9, 911861. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M. Multi-omics study for interpretation of genome-wide association study. J. Hum. Genet. 2021, 66, 3–10. [Google Scholar] [CrossRef]
- Vahabi, N.; Michailidis, G. Unsupervised multi-omics data integration methods: A comprehensive review. Front. Genet. 2022, 13, 854752. [Google Scholar] [CrossRef]
- Jan, R.; Hussain, A.; Assad, A.; Khurshid, S.; Macha, M.A. Chapter 10—Challenges with multi-omics data integration. In Multi-Omics Technology in Human Health and Diseases; Macha, M.A., Bhat, A.A., Masoodi, T.A., Eds.; Academic Press: Cambridge, MA, USA, 2025; pp. 223–242. [Google Scholar]
- Gao, F.; Huang, K.; Xing, Y. Artificial intelligence in omics. Genom. Proteom. Bioinform. 2023, 20, 811–813. [Google Scholar] [CrossRef]
- D’Urso, F.; Broccolo, F. Applications of artificial intelligence in microbiome analysis and probiotic interventions—An overview and perspective based on the current state of the art. Appl. Sci. 2024, 14, 8627. [Google Scholar] [CrossRef]
- Xue, W.; Liu, C.; Liu, Y.; Ding, H.; An, C.; Zhang, S.; Ma, S.; Zhang, Q. Probiotic evaluation of Lactiplantibacillus pentosus 68-1, a rutin conversion strain isolated from jiangshui, by genomic analysis and tests in vitro. Fermentation 2024, 10, 87. [Google Scholar] [CrossRef]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef]
- Parimelzaghan, A.; Anbarasu, A.; Ramaiah, S. Gene network analysis of metallo beta lactamase family proteins indicates the role of gene partners in antibiotic resistance and reveals important drug targets. J. Cell. Biochem. 2016, 117, 1330–1339. [Google Scholar] [CrossRef]
- Zhang, G.; He, M.; Xiao, L.; Jiao, Y.; Han, J.; Li, C.; Miller, M.J.; Zhang, L. Milk fat globule membrane protects Bifidobacterium longum ssp. infantis ATCC 15697 against bile stress by modifying global transcriptional responses. J. Dairy Sci. 2024, 107, 91–104. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, R.; Zhang, Z.; Jin, R.; Xie, T.; Liu, X.; Chai, J.; Howe, S.; Zhao, J.; Li, Y. Metagenomic and meta-transcriptomic analysis reveal the colonization and expression profile of probiotic strains in humans and animals. Fermentation 2023, 9, 417. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, J.; Liu, W.; Zhang, H.; Sun, Z. Metagenomic and metatranscriptomic profiling of Lactobacillus casei Zhang in the human gut. NPJ Biofilms Microbiomes 2021, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, Z.; Miyao, S.; Zhang, W. Unraveling the flavor profile and microbial roles during industrial Sichuan radish paocai fermentation by molecular sensory science and metatranscriptomics. Food Biosci. 2022, 48, 101815. [Google Scholar] [CrossRef]
- Bianchi, L.; Laghi, L.; Correani, V.; Schifano, E.; Landi, C.; Uccelletti, D.; Mattei, B. A combined proteomics, metabolomics and in vivo analysis approach for the characterization of probiotics in large-scale production. Biomolecules 2020, 10, 157. [Google Scholar] [CrossRef]
- Siciliano, R.A.; Lippolis, R.; Mazzeo, M.F. Proteomics for the investigation of surface-exposed proteins in probiotics. Front. Nutr. 2019, 6, 52. [Google Scholar] [CrossRef]
- Beck, H.C.; Madsen, S.M.; Glenting, J.; Petersen, J.; Israelsen, H.; Nørrelykke, M.R.; Antonsson, M.; Hansen, A.M. Proteomic analysis of cell surface-associated proteins from probiotic Lactobacillus plantarum. FEMS Microbiol. Lett. 2009, 297, 61–66. [Google Scholar] [CrossRef]
- Klotz, C.; O’Flaherty, S.; Goh, Y.J.; Barrangou, R. Investigating the effect of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front. Microbiol. 2017, 8, 2174. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.; Bridgland, A. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Sun, Z.; Ning, Z.; Figeys, D. The landscape and perspectives of the human gut metaproteomics. Mol. Cell. Proteom. 2024, 23, 100763. [Google Scholar] [CrossRef]
- Kolmeder, C.A.; Salojärvi, J.; Ritari, J.; De Been, M.; Raes, J.; Falony, G.; Vieira-Silva, S.; Kekkonen, R.A.; Corthals, G.L.; Palva, A. Faecal metaproteomic analysis reveals a personalized and stable functional microbiome and limited effects of a probiotic intervention in adults. PLoS ONE 2016, 11, e0153294. [Google Scholar] [CrossRef]
- Hendrickx, D.; An, R.; Boeren, S.; Mutte, S.; Wopereis, H.; Belzer, C.; PRESTO Study Team. Trackability of proteins from probiotic Bifidobacterium spp. in the gut using metaproteomics. Benef. Microbes 2023, 14, 269–280. [Google Scholar] [CrossRef]
- Rueangsri, N.; Roytrakul, S.; Muangnoi, C.; Tongkhao, K.; Sae-Tan, S.; Treesuwan, K.; Sirivarasai, J. Metaproteomic analysis of fermented vegetable formulations with lactic acid bacteria: A comparative study from initial stage to 15 days of production. Foods 2025, 14, 1148. [Google Scholar] [CrossRef] [PubMed]
- Sydor, S.; Dandyk, C.; Schwerdt, J.; Manka, P.; Benndorf, D.; Lehmann, T.; Schallert, K.; Wolf, M.; Reichl, U.; Canbay, A. Discovering biomarkers for non-alcoholic steatohepatitis patients with and without hepatocellular carcinoma using fecal metaproteomics. Int. J. Mol. Sci. 2022, 23, 8841. [Google Scholar] [CrossRef] [PubMed]
- Westfall, S.; Carracci, F.; Estill, M.; Zhao, D.; Wu, Q.L.; Shen, L.; Simon, J.; Pasinetti, G.M. Optimization of probiotic therapeutics using machine learning in an artificial human gastrointestinal tract. Sci. Rep. 2021, 11, 1067. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Suzuki, S.; Murakami, S.; Nishimoto, Y.; Higashi, K.; Watarai, N.; Umetsu, J.; Ishii, C.; Ito, Y.; Mori, Y.; et al. Integrated gut microbiome and metabolome analyses identified fecal biomarkers for bowel movement regulation by Bifidobacterium longum BB536 supplementation: A RCT. Comput. Struct. Biotechnol. J. 2022, 20, 5847–5858. [Google Scholar] [CrossRef]
- Walczak-Skierska, J.; Ludwiczak, A.; Sibińska, E.; Pomastowski, P. Environmental influence on bacterial lipid composition: Insights from pathogenic and probiotic strains. ACS Omega 2024, 9, 37789–37801. [Google Scholar] [CrossRef]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef]
- Lin, W.-J.; Shen, P.-C.; Liu, H.-C.; Cho, Y.-C.; Hsu, M.-K.; Lin, I.-C.; Chen, F.-H.; Yang, J.-C.; Ma, W.-L.; Cheng, W.-C. LipidSig: A web-based tool for lipidomic data analysis. Nucleic Acids Res. 2021, 49, W336–W345. [Google Scholar] [CrossRef]
- Schoug, Å.; Fischer, J.; Heipieper, H.J.; Schnürer, J.; Håkansson, S. Impact of fermentation pH and temperature on freeze-drying survival and membrane lipid composition of Lactobacillus coryniformis Si3. J. Ind. Microbiol. Biotechnol. 2008, 35, 175–181. [Google Scholar] [CrossRef]
- Schifano, E.; Cicalini, I.; Pieragostino, D.; Heipieper, H.J.; Del Boccio, P.; Uccelletti, D. In vitro and in vivo lipidomics as a tool for probiotics evaluation. Appl. Microbiol. Biotechnol. 2020, 104, 8937–8948. [Google Scholar] [CrossRef]
- Yang, F.; Zhu, W.; Edirisuriya, P.; Ai, Q.; Nie, K.; Ji, X.; Zhou, K. Characterization of metabolites and biomarkers for the probiotic effects of Clostridium cochlearium on high-fat diet-induced obese C57BL/6 mice. Eur. J. Nutr. 2022, 61, 2217–2229. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Fitzgerald, G.F.; van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef] [PubMed]
- Ströher, J.A.; Oliveira, W.D.C.; de Freitas, A.S.; Salazar, M.M.; da Silva, L.D.F.F.; Bresciani, L.; Flôres, S.H.; Malheiros, P.D.S. A global review of geographical diversity of kefir microbiome. Fermentation 2025, 11, 150. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef] [PubMed]
- Carasi, P.; Racedo, S.M.; Jacquot, C.; Elie, A.M.; Serradell, M.L.; Urdaci, M.C. Enterococcus durans EP1 a promising anti-inflammatory probiotic able to stimulate siga and to increase Faecalibacterium prausnitzii abundance. Front. Immunol. 2017, 8, 88. [Google Scholar] [CrossRef]
- Rozera, T.; Pasolli, E.; Segata, N.; Ianiro, G. Machine learning and artificial intelligence in the multi-omics approach to gut microbiota. Gastroenterology 2025, 13, 1–15. [Google Scholar] [CrossRef]
- Das, A.; Behera, R.N.; Kapoor, A.; Ambatipudi, K. The potential of meta-proteomics and artificial intelligence to establish the next generation of probiotics for personalized healthcare. J. Agric. Food Chem. 2023, 71, 17528–17542. [Google Scholar] [CrossRef]
- Feng, Y.; Soni, A.; Brightwell, G.; Reis, M.M.; Wang, Z.; Wang, J.; Wu, Q.; Ding, Y. The potential new microbial hazard monitoring tool in food safety: Integration of metabolomics and artificial intelligence. Trends Food Sci. Technol. 2024, 149, 104555. [Google Scholar] [CrossRef]
- Wani, A.K.; Roy, P.; Kumar, V.; Mir, T.u.G. Metagenomics and artificial intelligence in the context of human health. Infect. Genet. Evol. 2022, 100, 105267. [Google Scholar] [CrossRef]
- Shehata, H.R.; Hassane, B.; Newmaster, S.G. Real-time polymerase chain reaction methods for strain specific identification and enumeration of strain Lacticaseibacillus paracasei 8700:2. Front. Microbiol. 2023, 13, 1076631. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef]
- Goodswen, S.J.; Barratt, J.L.N.; Kennedy, P.J.; Kaufer, A.; Calarco, L.; Ellis, J.T. Machine learning and applications in microbiology. FEMS Microbiol. Rev. 2021, 45, fuab015. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K. Applications of machine learning in human microbiome studies: A review on feature selection, biomarker identification, disease prediction and treatment. Front. Microbiol. 2021, 12, 634511. [Google Scholar] [CrossRef] [PubMed]
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Al-Adham, I.S.; Agha, A.S.; Al-Akayleh, F.; Al-Remawi, M.; Jaber, N.; Al Manasur, M.; Collier, P.J. Prebiotics Beyond the Gut: Omics Insights, Artificial Intelligence, and Clinical Trials in Organ-Specific Applications. Probiotics Antimicrob. Proteins 2025, 1–22. [Google Scholar] [CrossRef]
- Krupitzer, C. Generative artificial intelligence in the agri-food value chain-overview, potential, and research challenges. Front. Food Sci. Technol. 2024, 4, 1473357. [Google Scholar] [CrossRef]
- Badidi, E. Edge AI for early detection of chronic diseases and the spread of infectious diseases: Opportunities, challenges, and future directions. Future Internet 2023, 15, 370. [Google Scholar] [CrossRef]
- Banerjee, N.; Chatterjee, K. Quantum AI in healthcare: Revolutionizing diagnosis, treatment and drug discovery. Int. J. Sci. Res. Sci. Technol. 2024, 11, 815–836. [Google Scholar] [CrossRef]
- Abdelwahab, S.I.; Taha, M.M.E.; Jerah, A.A.; Farasani, A.; Abdullah, S.M.; Aljahdali, I.A.; Ibrahim, R.; Oraibi, O.; Oraibi, B.; Alfaifi, H.A.; et al. Artificial intelligence and microbiome research: Evolution of hotspots, research trends, and thematic-based narrative review. Cell Mol. Biol. 2024, 70, 182–192. [Google Scholar] [CrossRef]
- Patil, A.; Singh, N.; Patwekar, M.; Patwekar, F.; Patil, A.; Gupta, J.K.; Elumalai, S.; Priya, N.S.; Sahithi, A. AI-driven insights into the microbiota: Figuring out the mysterious world of the gut. Intell. Pharm. 2025, 3, 46–52. [Google Scholar] [CrossRef]
- McCoubrey, L.E.; Elbadawi, M.; Orlu, M.; Gaisford, S.; Basit, A.W. Harnessing machine learning for development of microbiome therapeutics. Gut Microbes 2021, 13, 1872323. [Google Scholar] [CrossRef]
- Abavisani, M.; Foroushan, S.K.; Ebadpour, N.; Sahebkar, A. Deciphering the gut microbiome: The revolution of artificial intelligence in microbiota analysis and intervention. Curr. Res. Biotechnol. 2024, 7, 100211. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Uchida, T.; Inoue, T.; Iwasaki, Y.; Ito, R.; Saito, K.; Akiyama, H.; Tsuda, K.; Yoshida, K. Analysis of 50 years of health food research trends using natural language processing and generative AI. Procedia Comput. Sci. 2024, 246, 1875–1884. [Google Scholar] [CrossRef]
- Myers, B.K.; Lamichhane, A.; Kvitko, B.H.; Dutta, B. Natural language processing-like deep learning aided in identification and validation of thiosulfinate tolerance clusters in diverse bacteria. bioRxiv 2024. [Google Scholar] [CrossRef]
- Danielsen, J.; Nordenfelt, P. Computer vision-based image analysis of bacteria. In Bacterial Pathogenesis: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 161–172. [Google Scholar]
- Loddo, A.; Di Ruberto, C.; Armano, G.; Manconi, A. Automatic monitoring cheese ripeness using computer vision and artificial intelligence. IEEE Access 2022, 10, 122612–122626. [Google Scholar] [CrossRef]
- Treloar, N.J.; Fedorec, A.J.; Ingalls, B.; Barnes, C.P. Deep reinforcement learning for the control of microbial co-cultures in bioreactors. PLoS Comput. Biol. 2020, 16, e1007783. [Google Scholar] [CrossRef]
- Dhatterwal, J.S.; Kaswan, K.S.; Kumar, N. Robotic process automation in healthcare. In Confluence of Artificial Intelligence and Robotic Process Automation; Springer: Berlin/Heidelberg, Germany, 2023; pp. 157–175. [Google Scholar]
- Das, D.; Roy, A.D.; Gupta, S.D.; Roy, R.D. The use of generative artificial intelligence (AI) in teaching and assessment of postgraduate students in pathology and microbiology. Indian J. Microbiol. Res. 2024, 3, 140–146. [Google Scholar] [CrossRef]
- Sionek, B.; Szydłowska, A.; Zielińska, D.; Neffe-Skocińska, K.; Kołożyn-Krajewska, D. Beneficial bacteria isolated from food in relation to the next generation of probiotics. Microorganisms 2023, 11, 1714. [Google Scholar] [CrossRef]
- Zhai, Y.; Kim, M.; Fan, P.; Rajeev, S.; Kim, S.A.; Driver, J.D.; Galvão, K.N.; Boucher, C.; Jeong, K.C. Machine learning-enhanced assessment of potential probiotics from healthy calves for the treatment of neonatal calf diarrhea. Front. Microbiol. 2024, 15, 1507537. [Google Scholar] [CrossRef]
- Yang, N.J.; Chiu, I.M. Bacterial signaling to the nervous system through toxins and metabolites. J. Mol. Biol. 2017, 429, 587–605. [Google Scholar] [CrossRef]
- Takahashi, M.K.; Tan, X.; Dy, A.J.; Braff, D.; Akana, R.T.; Furuta, Y.; Donghia, N.; Ananthakrishnan, A.; Collins, J.J. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 2018, 9, 3347. [Google Scholar] [CrossRef]
- Marlicz, W.; Skonieczna-Żydecka, K.; Dabos, K.J.; Łoniewski, I.; Koulaouzidis, A. Emerging concepts in non-invasive monitoring of Crohn’s disease. Ther. Adv. Gastroenterol. 2018, 11, 1756284818769076. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Liu, S.; Yang, G.; Hou, X.; Fang, Y. Next-generation probiotics: Innovations in safety assessments. Curr. Opin. Food Sci. 2024, 61, 101238. [Google Scholar] [CrossRef]
- Wei, X.; Xi, P.; Chen, M.; Wen, Y.; Wu, H.; Wang, L.; Zhu, Y.; Ren, Y.; Gu, Z. Capsule robots for the monitoring, diagnosis, and treatment of intestinal diseases. Mater. Today Bio 2024, 29, 101294. [Google Scholar] [CrossRef] [PubMed]
- Chelliah, R.; Wei, S.; Daliri, E.B.-M.; Rubab, M.; Elahi, F.; Yeon, S.-J.; Jo, K.h.; Yan, P.; Liu, S.; Oh, D.H. Development of nanosensors based intelligent packaging systems: Food quality and medicine. Nanomaterials 2021, 11, 1515. [Google Scholar] [CrossRef]
- McCusker, R.H.; Kelley, K.W. Immune–neural connections: How the immune system’s response to infectious agents influences behavior. J. Exp. Biol. 2013, 216, 84–98. [Google Scholar] [CrossRef]
- Curry, K.; Nute, M.; Treangen, T.J. It takes guts to learn: Machine learning techniques for disease detection from the gut microbiome. Emerg. Top. Life Sci. 2021, 5, 815–827. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Zheng, L.; Li, J.; Hong, Y.; Liang, P.; Kwok, L.-Y.; Zuo, Y.; Zhang, W.; Zhang, H. iProbiotics: A machine learning platform for rapid identification of probiotic properties from whole-genome primary sequences. Brief. Bioinform. 2022, 23, bbab477. [Google Scholar] [CrossRef]
- Bergamini, C.M.; Bianchi, N.; Giaccone, V.; Catellani, P.; Alberghini, L.; Stella, A.; Biffani, S.; Yaddehige, S.K.; Bobbo, T.; Taccioli, C. Machine learning algorithms highlight trna information content and chargaff’s second parity rule score as important features in discriminating probiotics from non-probiotics. Biology 2022, 11, 1024. [Google Scholar] [CrossRef]
- Maftei, N.M.; Raileanu, C.R.; Balta, A.A.; Ambrose, L.; Boev, M.; Marin, D.B.; Lisa, E.L. The potential impact of probiotics on human health: An update on their health-promoting properties. Microorganisms 2024, 12, 234. [Google Scholar] [CrossRef]
- Mazziotta, C.; Tognon, M.; Martini, F.; Torreggiani, E.; Rotondo, J.C. Probiotics mechanism of action on immune cells and beneficial effects on human health. Cells 2023, 12, 184. [Google Scholar] [CrossRef]
- Berson, E.; Chung, P.; Espinosa, C.; Montine, T.J.; Aghaeepour, N. Unlocking human immune system complexity through AI. Nat. Methods 2024, 21, 1400–1402. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Rajeshwari, S.; Shukla, P. Artificial intelligence and synthetic biology approaches for human gut microbiome. Crit. Rev. Food Sci. Nutr. 2022, 62, 2103–2121. [Google Scholar] [CrossRef] [PubMed]
- Ozcelik, F.; Dundar, M.S.; Yildirim, A.B.; Henehan, G.; Vicente, O.; Sánchez-Alcázar, J.A.; Gokce, N.; Yildirim, D.T.; Bingol, N.N.; Karanfilska, D.P.; et al. The impact and future of artificial intelligence in medical genetics and molecular medicine: An ongoing revolution. Funct. Integr. Genom. 2024, 24, 138. [Google Scholar] [CrossRef]
- Wu, S.; Feng, T.; Tang, W.; Qi, C.; Gao, J.; He, X.; Wang, J.; Zhou, H.; Fang, Z. metaProbiotics: A tool for mining probiotic from metagenomic binning data based on a language model. Brief. Bioinform. 2024, 25, bbae085. [Google Scholar] [CrossRef]
- Orkkatteri Krishnan, A.; Mudgal, L.N.; Soni, V.; Prakash, T. ProbML: A machine learning-based genome classifier for identifying probiotic organisms. Mol. Nutr. Food Res. 2025, e70025. [Google Scholar] [CrossRef]
- Kang, M.; Tian, J. Machine Learning: Data Pre-processing. In Prognostics and Health Management of Electronics; John Wiley and Sons: Hoboken, NJ, USA, 2018; pp. 111–130. [Google Scholar]
- Varoquaux, G.; Colliot, O. Evaluating machine learning models and their diagnostic value. In Machine Learning for Brain Disorders; Humana: New York, NY, USA, 2023; pp. 601–630. [Google Scholar]
- Li, P.; Luo, H.; Ji, B.; Nielsen, J. Machine learning for data integration in human gut microbiome. Microb. Cell Factories 2022, 21, 241. [Google Scholar] [CrossRef]
- Sulaimany, S.; Farahmandi, K.; Mafakheri, A. Computational prediction of new therapeutic effects of probiotics. Sci. Rep. 2024, 14, 11932. [Google Scholar] [CrossRef]
- Erion, G.; Janizek, J.D.; Hudelson, C.; Utarnachitt, R.B.; McCoy, A.M.; Sayre, M.R.; White, N.J.; Lee, S.-I. A cost-aware framework for the development of AI models for healthcare applications. Nat. Biomed. Eng. 2022, 6, 1384–1398. [Google Scholar] [CrossRef]
- Choudhary, R.; Mahadevan, R. Toward a systematic design of smart probiotics. Curr. Opin. Biotechnol. 2020, 64, 199–209. [Google Scholar] [CrossRef]
- Pasolli, E.; Truong, D.T.; Malik, F.; Waldron, L.; Segata, N. Machine learning meta-analysis of large metagenomic datasets: Tools and biological insights. PLoS Comput. Biol. 2016, 12, e1004977. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, B.; Liu, X.; Gu, W.; Xu, F.; Wang, J.; Liu, Q.; Wang, R.; Hu, Y.; Liu, J. An intelligent intestine-on-a-chip for rapid screening of probiotics with relief-enteritis function. Adv. Mater. 2024, 36, 2408485. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-B.; Kim, H.; Kim, S.; Kim, J.; Park, S.-K.; Lee, C.-W.; Kim, K.O.; Seo, G.-S.; Kim, M.S.; Cha, J.M.; et al. Potential oral microbial markers for differential diagnosis of Crohn’s disease and ulcerative colitis using machine learning models. Microorganisms 2023, 11, 1665. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: Looking into the future. Cancer Cell Int. 2021, 21, 270. [Google Scholar] [CrossRef]
- Dlamini, Z.; Francies, F.Z.; Hull, R.; Marima, R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct. Biotechnol. J. 2020, 18, 2300–2311. [Google Scholar] [CrossRef]
- Freitas, P.; Silva, F.; Sousa, J.V.; Ferreira, R.M.; Figueiredo, C.; Pereira, T.; Oliveira, H.P. Machine learning-based approaches for cancer prediction using microbiome data. Sci. Rep. 2023, 13, 11821. [Google Scholar] [CrossRef]
- Huang, A.; Huo, Y.; Zhong, Y.; Yang, W. AI Technology for Anti-Aging: An Overview. In Proceedings of the 2023 International Conference on Intelligent Supercomputing and BioPharma (ISBP), Zhuhai, China, 6–8 January 2023; pp. 54–62. [Google Scholar]
- Wu, Z.; Feng, C.; Hu, Y.; Zhou, Y.; Li, S.; Zhang, S.; Hu, Y.; Chen, Y.; Chao, H.; Ni, Q. HALD, a human aging and longevity knowledge graph for precision gerontology and geroscience analyses. Sci. Data 2023, 10, 851. [Google Scholar] [CrossRef]
- Oniszczuk, A.; Oniszczuk, T.; Gancarz, M.; Szymańska, J. Role of gut microbiota, probiotics and prebiotics in the cardiovascular diseases. Molecules 2021, 26, 1172. [Google Scholar] [CrossRef]
- Tao, Y.-W.; Gu, Y.-L.; Mao, X.-Q.; Zhang, L.; Pei, Y.-F. Effects of probiotics on type II diabetes mellitus: A meta-analysis. J. Transl. Med. 2020, 18, 30. [Google Scholar] [CrossRef]
- Oudat, Q.; Okour, A. The role of probiotics in modulating gut microbiota and metabolic health for weight management: A mini review. Acta Microbiol. Hell. 2025, 70, 5. [Google Scholar] [CrossRef]
- Wan, Y.; Zuo, T.; Xu, Z.; Zhang, F.; Zhan, H.; Chan, D.; Leung, T.-F.; Yeoh, Y.K.; Chan, F.K.; Chan, R. Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut 2022, 71, 910–918. [Google Scholar] [CrossRef]
- Wang, S.; Naumovski, N.; Ajlouni, S.; Ayyash, M.; Silva, R.; Balthazar, C.F.; Esmerino, E.A.; Freitas, M.Q.; da Silva, M.C.; Sant’Ana, A.S. Nonbovine milk and its products as sources of probiotics delivery: An overview of its viability, functionality and product quality characteristics. Int. J. Dairy Technol. 2023, 76, 482–511. [Google Scholar] [CrossRef]
- Ge, S.; Han, J.; Sun, Q.; Zhou, Q.; Ye, Z.; Li, P.; Gu, Q. Research progress on improving the freeze-drying resistance of probiotics: A review. Trends Food Sci. Technol. 2024, 147, 104425. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Kim, J.-S.; Kwon, H.-J.; Kang, C.-H. The effect of a glutathione (gsh)-containing cryo-protectant on the viability of probiotic cells using a freeze-drying process. Fermentation 2023, 9, 187. [Google Scholar] [CrossRef]
- Chen, H.; Tian, M.; Chen, L.; Cui, X.; Meng, J.; Shu, G. Optimization of composite cryoprotectant for freeze-drying Bifidobacterium bifidum BB01 by response surface methodology. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Kavak, A.E.; Zent, İ.; Özdemir, A.; Dertli, E. Optimization of cryoprotectant formulation to enhance the viability of Lactiplantibacillus plantarum NBC99 isolated from human origin. Prep. Biochem. Biotechnol. 2024, 54, 958–966. [Google Scholar] [CrossRef]
- Yusuff, A.S.; Ishola, N.B.; Gbadamosi, A.O. Artificial intelligence techniques and response surface methodology for the optimization of methyl ester sulfonate synthesis from used cooking oil by sulfonation. ACS Omega 2023, 8, 19287–19301. [Google Scholar] [CrossRef]
- Bozkurt Keser, S.; Buruk Sahin, Y. Response surface methodology to tune artificial neural network hyper-parameters. Expert Syst. 2021, 38, e12792. [Google Scholar] [CrossRef]
- Gul, L.B.; Gul, O.; Yilmaz, M.T.; Dertli, E.; Con, A.H. Optimization of cryoprotectant formulation to enhance the viability of Lactobacillus brevis ED25: Determination of storage stability and acidification kinetics in sourdough. J. Food Process. Preserv. 2020, 44, e14400. [Google Scholar] [CrossRef]
- Bobba, S.; Harguindeguy, M.; Colucci, D.; Fissore, D. Diffuse interface model of the freeze-drying process of individually frozen products. Dry. Technol. 2020, 38, 758–774. [Google Scholar] [CrossRef]
- Van Chuyen, H.; Le Minh, T.; Phu, Q.P.; Giurgiulescu, L.; Tan, D. Mathematical model study to optimize the freeze drying process for production of dried yogurt. Carpathian J. Food Sci. Technol. 2024, 16, 151–163. [Google Scholar]
- Zhu, H.; Jin, D.; Fu, N.; Chen, X.D.; Xiao, J. Neural network modeling of the dynamic inactivation of probiotics during single droplet drying for improved cell viability. Powder Technol. 2023, 413, 118042. [Google Scholar] [CrossRef]
- Chutrtong, J.; Kularbphettong, K.; Sirichokworrakit, S. Investigation into the impact of varying inulin levels on the survival of probiotic microorganisms. In Proceedings of the 2023 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 13–15 December 2023; pp. 577–581. [Google Scholar]
- Reker, D. Practical considerations for active machine learning in drug discovery. Drug Discov. Today Technol. 2019, 32, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, R.; Abedin, M.M.; Chiring Phukon, L.; Sahoo, D.; Singh, S.P.; Rai, A.K. Biotechnological approaches for the production of designer cheese with improved functionality. Compr. Rev. Food Sci. Food Saf. 2021, 20, 960–979. [Google Scholar] [CrossRef]
- Yang, S.; Bai, M.; Kwok, L.-Y.; Zhong, Z.; Sun, Z. The intricate symbiotic relationship between lactic acid bacterial starters in the milk fermentation ecosystem. Crit. Rev. Food Sci. Nutr. 2025, 65, 728–745. [Google Scholar] [CrossRef]
- Yang, S.; Bai, M.; Liu, W.; Li, W.; Zhong, Z.; Kwok, L.-Y.; Dong, G.; Sun, Z. Predicting Lactobacillus delbrueckii subsp. bulgaricus-Streptococcus thermophilus interactions based on a highly accurate semi-supervised learning method. Sci. China Life Sci. 2025, 68, 558–574. [Google Scholar] [CrossRef]
- Hernández Medina, R.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine learning and deep learning applications in microbiome research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
- Oh, M.; Zhang, L. DeepMicro: Deep representation learning for disease prediction based on microbiome data. Sci. Rep. 2020, 10, 6026. [Google Scholar] [CrossRef]
- Yang, J.; Shin, T.-S.; Kim, J.S.; Jee, Y.-K.; Kim, Y.-K. A new horizon of precision medicine: Combination of the microbiome and extracellular vesicles. Exp. Mol. Med. 2022, 54, 466–482. [Google Scholar] [CrossRef]
- Hao, X.; Xia, Y.; Wang, Y.; Zhang, X.; Liu, L. The addition of probiotic promotes the release of ACE-I peptide of Cheddar cheese: Peptide profile and molecular docking. Int. Dairy J. 2023, 137, 105507. [Google Scholar] [CrossRef]
- Ul Ain, N.; Naveed, M.; Aziz, T.; Shabbir, M.A.; Al Asmari, F.; Abdi, G.; Sameeh, M.Y.; Alhhazmi, A.A. Mix-match synthesis of nanosynbiotics from probiotics and prebiotics to counter gut dysbiosis via AI integrated formulation profiling. Sci. Rep. 2024, 14, 18397. [Google Scholar] [CrossRef]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D. Shaping the future of probiotics and prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Shen, J.; Manickam, S.; Li, S.; Tao, Y.; Li, D.; Liu, D.; Han, Y. Investigation of blueberry juice fermentation by mixed probiotic strains: Regression modeling, machine learning optimization and comparison with fermentation by single strain in the phenolic and volatile profiles. Food Chem. 2023, 405, 134982. [Google Scholar] [CrossRef] [PubMed]
- Martinović, A.; Martinović, I.; Mora, D.; Arioli, S. Uncovering the Probiotic supplement landscape: Market offerings, sales patterns, and future forecasts using machine learning approach—A case study of montenegro. Probiotics Antimicrob. Proteins 2024, 1–24. [Google Scholar]
| Metabolite | Type | Microorganism | Biological Activity/Key Outcomes | Reference |
|---|---|---|---|---|
| Exopolysaccharides | α-glucan | Pediococcus acidilactici NCDC 252 | Anticancer (human colon cancer cell line) | [62] |
| Heteropolysaccharide | Lactiplantibacillus paraplantarum NCCP 962 | Cholesterol-lowering | [59] | |
| Heteropolysaccharide | Limosilactobacillus fermentum NCDC400 | Cholesterol-lowering | [63] | |
| - | Lactiplantibacillus plantarum MI01 | Anticholesterol | [64] | |
| - | Lactobacillus delbrueckii ssp. bulgaricus DSM 20081 | Antioxidant, antitumor, periodontal regeneration | [65] | |
| Negatively charged acidic | Lactiplantibacillus plantarum SN35N | Antiviral | [56] | |
| Galactoglucan and levan | Lactococcus lactis F-mou | Antimicrobial | [57] | |
| Glucomannan | Lactiplantibacillus plantarum BR2 | Antidiabetic, cholesterol-lowering, and antioxidant | [66] | |
| Vitamin | Vitamin B | Bifidobacteria spp. | Secretion pyridoxine (B6): 0.988–26.060 mg/L | [67] |
| Lactic acid bacteria spp. | Secretion pyridoxine (B6): 1.100–11.400 mg/L Secretion pantothenic acid (B3): 3.966–138.600 mg/L Secretion thiamine (B1): 14.720–19.540 mg/L | |||
| Lactiplantibacillus plantarum HY7715 | Secretion riboflavin (B2): 34.5 ± 2.41 mg/L | [68] | ||
| Vitamin B2 and B9 | Leuconostoc mesenteroides subsp. mesenteroides, Lactiplantibacillus plantarum, Lacticaseibacillus rhamnosus | About 1.7–32-fold increase in quinoa sourdough | [69] | |
| Riboflavin (B2) | Lactiplantibacillus plantarum M5MA1-B2 | About 2.5-fold increase in oat kefir | [70] | |
| Short-chain fatty acids | Butyrate | Lacticaseibacillus paracasei SD1 and Lacticaseibacillus rhamnosus SD11 | Anticancer and anti-inflammation | [71] |
| Clostridium butyricum | ||||
| Postbiotic metabolites | Organic acids, acetoin, 2,3- butanediol | Leuconostoc pseudomesenteroides Y6 | Anticancer | [72] |
| - | Lactobacillus plantarum | Anticancer | [73] | |
| Organic acids | Lactiplantibacillus plantarum, Lacticaseibacillus rhamnosus, Lactobacillus gasseri | Antifungal | [74] | |
| Organic acids, volatile organic compounds, polyphenols | Lacticaseibacillus rhamnosus | Antiaflatoxigenic | [74] | |
| 3-phenyl-1,2,4-benzotriazine | Lactiplantibacillus plantarum | Anticancer | [75] |
| Omics Approach | Data | Evaluation Output | Application Area | AI Tools | Reference |
|---|---|---|---|---|---|
| Genomics | Whole sequence, DNA sequences, annotated genes | Identification of functional genes, prediction of probiotic traits | Strain selection, probiotic characterization | Machine learning, deep learning, natural language processing | [13,116] |
| Transcriptomics | Raw RNA reads, single-cell RNA-seq data, mRNA expression levels, etc. | Gene expression profiling under stress, prediction of stress responses | Strain selection, probiotic characterization | Machine learning and clustering algorithms (such as hierarchical clustering or K-means) | [9] |
| Metatranscriptomics | Total RNA expression profiles in microbiota, RNA-seq data | Functional activities of probiotics within microbiota, host interaction | Strain selection, probiotic characterization, personalized diet, formulation | Machine learning, deep learning | [116] |
| Proteomics | Proteins, peptide sequences | Protein structure-function prediction, adaptation analysis | Viability, strain selection, functional food design, disease diagnostics, drug development | Machine learning, deep learning, natural language processing | [13,116] |
| Metaproteomics | Collective protein profiles, functional markers | Functional protein markers linked to probiotic activity | Health biomarker identification, survival | Machine learning, deep learning | [117] |
| Lipidomics | Lipid composition, lipid profiles | Membrane lipid profiling, adaptation indicators, host interaction | Strain selection, probiotic characterization | Machine learning, deep learning, natural language processing | [13,116] |
| Metabolomics | Metabolite concentrations, metabolic fingerprints | Metabolite profiles (e.g., SCFAs, vitamins), prediction of health effects | Strain selection, functional food design, personalized diet | Machine learning, deep learning | [13,118] |
| Metagenomics | Microbial abundance tables (OTUs, taxa profiles) | Microbial abundance shifts | Strain selection, microbiota composition optimization, functional food design, functional food design | Machine learning, deep learning (such as Meta-Signer, DeepMicro, mAML, PaPrBaG, MicrobiomeAnalystR, mothur, QIIME, BiomMiner, Scikit-learn, and MIPMLP) | [119] |
| Key Findings | AI Technology | Significance | Reference |
|---|---|---|---|
| PCA used to visualize excipients’ chemical structure impact. The model started with 6 excipients, predicted effects on 111 ones. After 3 rounds of AML, achieved 67.7% accuracy, identifying 3/4 tested excipients. | Active ML to predict the effects of pharmaceutical excipients on Lacticaseibacillus paracasei. | Demonstrates ML’s power in pharmaceutical formulation even with small datasets. | [14] |
| 144 LAB strains were evaluated for low pH, bile salt resistance, and antimicrobial activity. Best strains identified: Lacticaseibacillus paracasei S23, Lactiplantibacillus plantarum S57 & S70, Lacticaseibacillus casei S81. Decision Tree algorithm validated biofilm production’s role in gut colonization. | Applied ML algorithms: Information Gain Ratio, Information Gain, PCA, Gini Index, Chi-square, Deviance, Rule-Based Learning, Uncertainty, Correlation, and Relaxation. | ML methods successfully identified top LAB strains with >99% accuracy. | [43] |
| AI used to identify beneficial microbial species, link them to diseases, and develop personalized probiotic formulations. NGPs could be key to personalized medicine. | AI-driven data analytics for large-scale gut microbiota studies across geographic regions. | AI-microbiome integration could revolutionize clinical applications and personalized probiotics. | [140] |
| 91 calf microbiome samples (74 healthy, 17 diarrheic used to train random forest model. Limosilactobacillus reuteri administration restored gut microbiota in diarrheic calves. Model identified health-associated bacteria. | Whole-genome sequencing of 22 Limosilactobacillus reuteri strains; random forest model to analyze gut microbiome profiles. | Probiotic treatment confirmed effective in restoring gut health; ML model validated findings. | [141] |
| Key Findings | AI Technology | Database Used | Applications | Reference |
|---|---|---|---|---|
| Applied ABIOME model to the optimization of probiotic formulation; used MARS algorithm; reported synergistic interactions in metabolite production. | ML algorithm for probiotic synergism | ABIOME bioreactor, MARS algorithm, probiotic-metabolite interactions | Development of next-generation probiotics, personalized formulations | [104] |
| Developed machine learning model (SVM) to predict probiotic potential based on genomic k-mer analysis; achieved 97.77% accuracy. | Support vector machine (SVM) algorithm model | Genomic k-mer analysis, SVM model, incremental feature selection | Probiotic genome classification, food and supplement industry applications | [150] |
| Analyzed 89 bacterial genomes; applied multiple ML models; neural networks achieved 95.1% accuracy in probiotic classification. | Multiple ML models (GLM, RF, SVM, NN) are used to predict the ability to classify bacteria | NCBI GenBank data | Probiotic identification for food and health industries | [151] |
| Used ICA to analyze Limosilactobacillus reuteri transcriptional regulation; identified 35 iModulons; discovered bistable regulatory mechanisms. | ML model | RNA-seq datasets, independent component analysis (ICA) | Optimization of probiotic properties, microbial food production strategies | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asar, R.; Erenler, S.; Devecioglu, D.; Ispirli, H.; Karbancioglu-Guler, F.; Ozturk, H.I.; Dertli, E. Understanding the Functionality of Probiotics on the Edge of Artificial Intelligence (AI) Era. Fermentation 2025, 11, 259. https://doi.org/10.3390/fermentation11050259
Asar R, Erenler S, Devecioglu D, Ispirli H, Karbancioglu-Guler F, Ozturk HI, Dertli E. Understanding the Functionality of Probiotics on the Edge of Artificial Intelligence (AI) Era. Fermentation. 2025; 11(5):259. https://doi.org/10.3390/fermentation11050259
Chicago/Turabian StyleAsar, Remziye, Sinem Erenler, Dilara Devecioglu, Humeyra Ispirli, Funda Karbancioglu-Guler, Hale Inci Ozturk, and Enes Dertli. 2025. "Understanding the Functionality of Probiotics on the Edge of Artificial Intelligence (AI) Era" Fermentation 11, no. 5: 259. https://doi.org/10.3390/fermentation11050259
APA StyleAsar, R., Erenler, S., Devecioglu, D., Ispirli, H., Karbancioglu-Guler, F., Ozturk, H. I., & Dertli, E. (2025). Understanding the Functionality of Probiotics on the Edge of Artificial Intelligence (AI) Era. Fermentation, 11(5), 259. https://doi.org/10.3390/fermentation11050259






