Retinal Production by Precision Fermentation of Saccharomyces cerevisiae
Abstract
1. Introduction
2. Material and Methods
2.1. Strains and Plasmids
2.2. Media and Culture Conditions
2.3. Construction of Plasmids
2.4. Construction of Recombinant Strains
2.5. Fermentation Condition
2.6. Extraction and Quantitative Analysis Methods
3. Results and Discussion
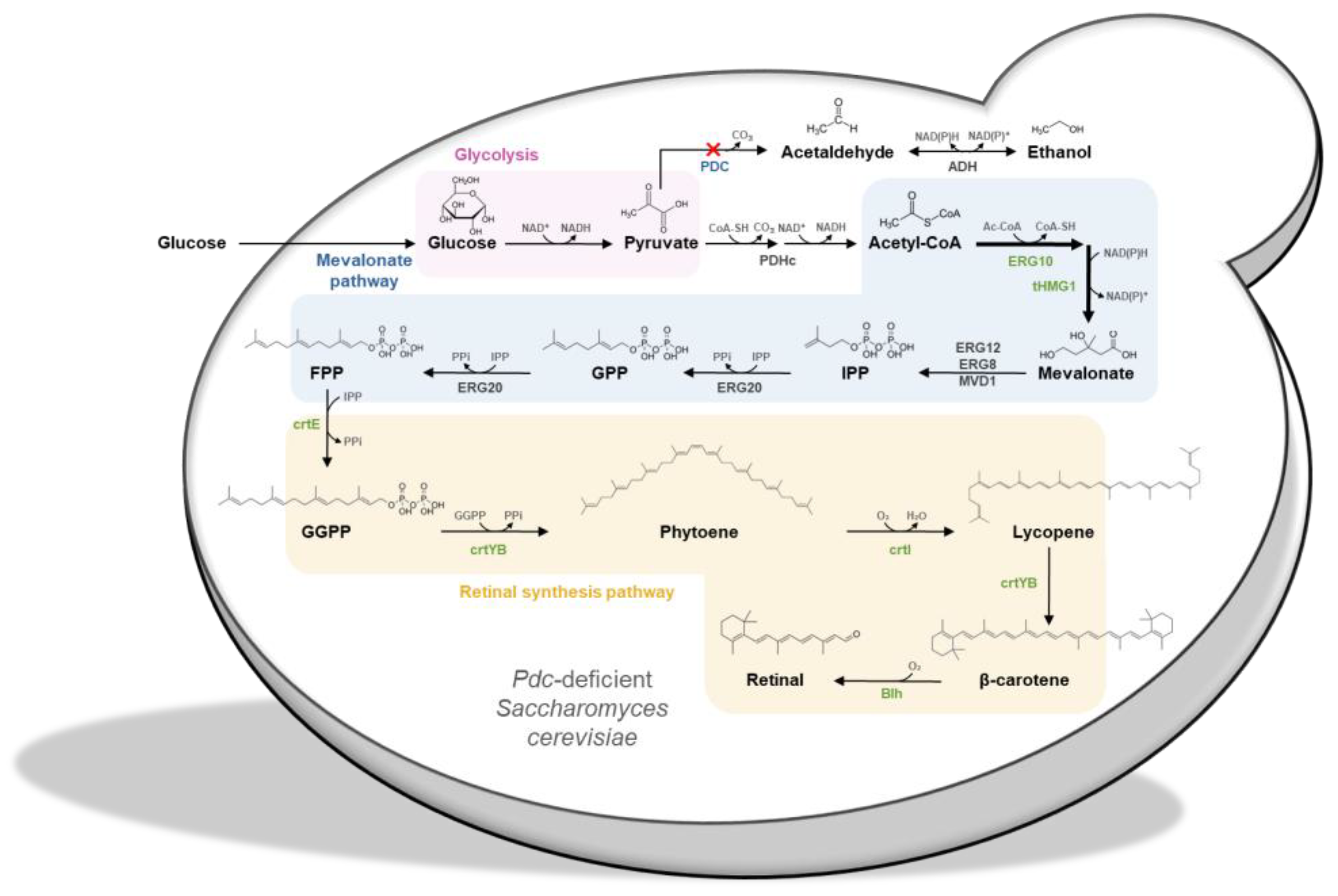
3.1. Introduction of Retinal Biosynthetic Pathway into S. cerevisiae
3.2. Enhancement of Mevalonate Pathway for Increasing Retinal Production
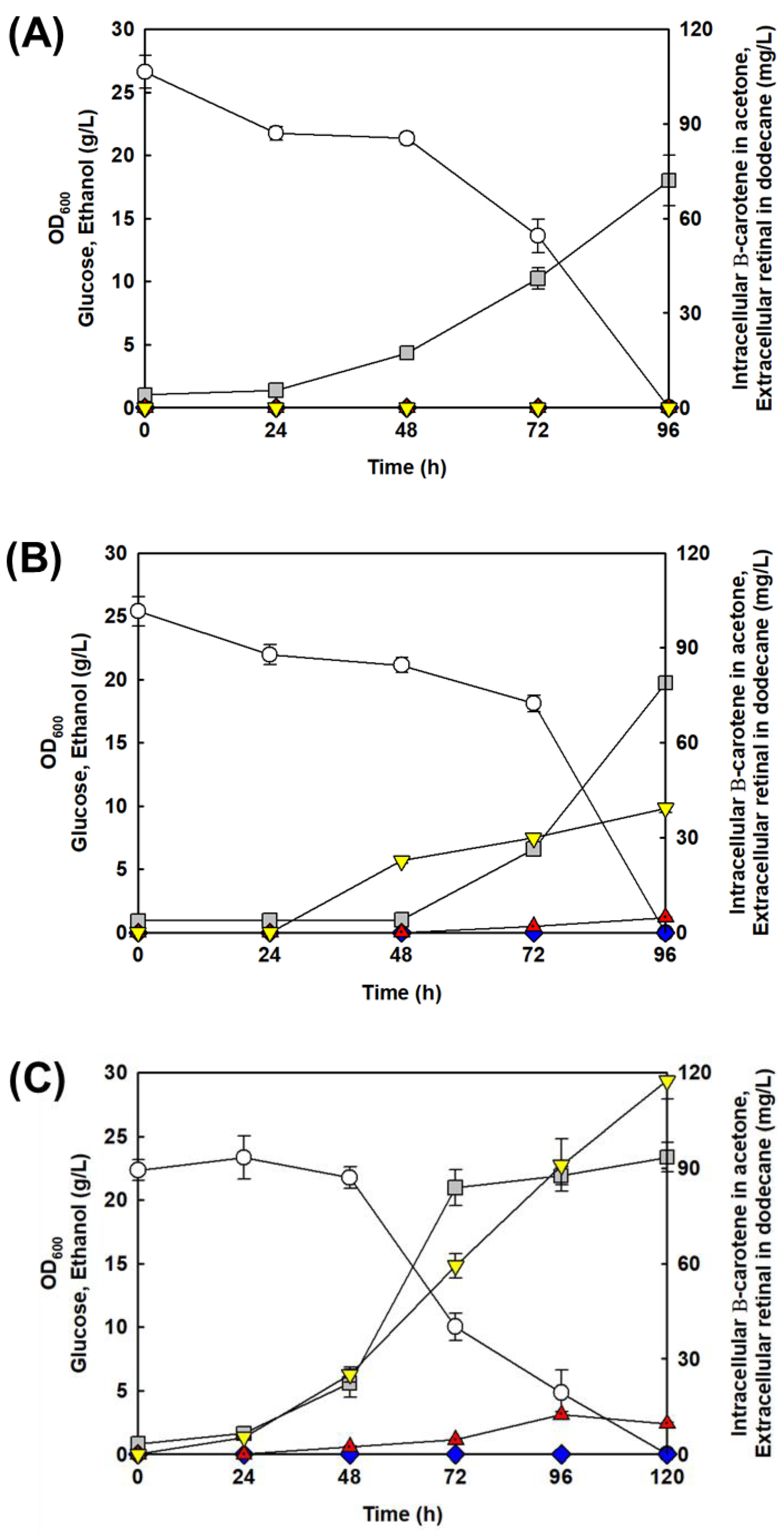
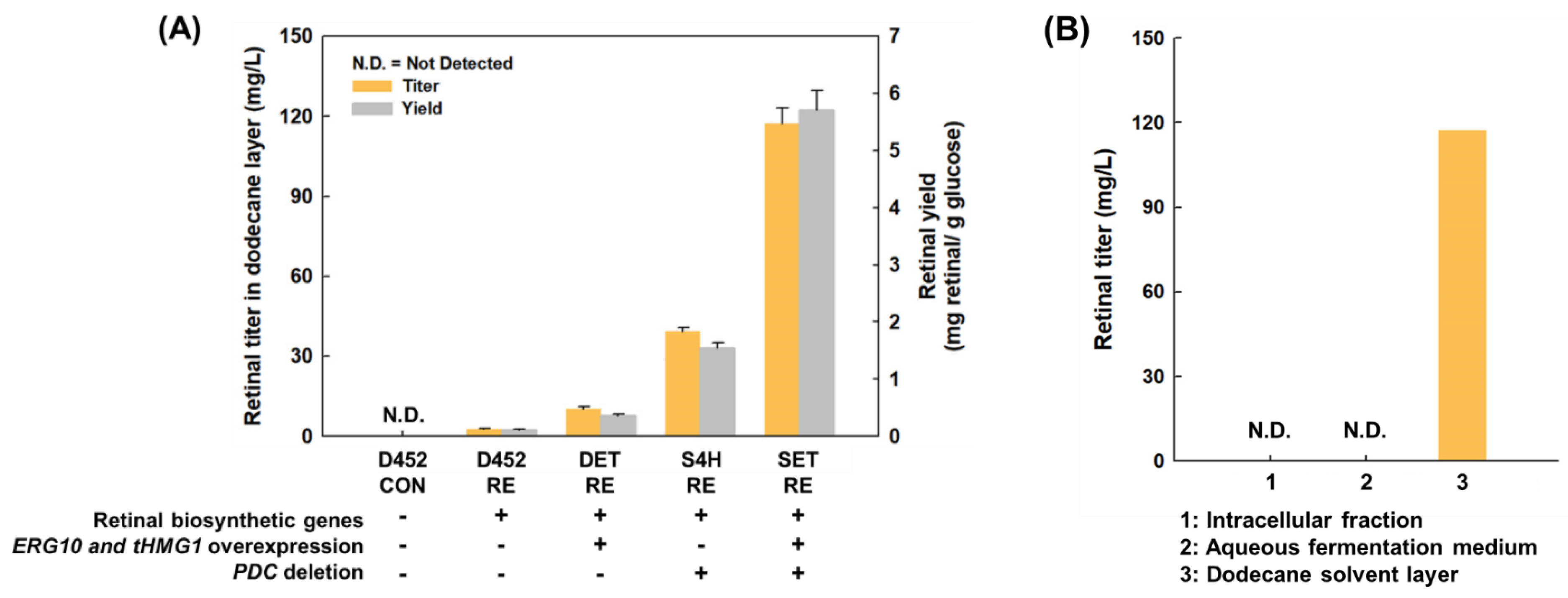
3.3. Increased Retinal Production in S. cerevisiae by Elimination of Ethanol Pathway
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, L.; Kwak, S.; Jin, Y.-S. Vitamin A Production by Engineered Saccharomyces cerevisiae from Xylose via Two-Phase in Situ Extraction. ACS Synth. Biol. 2019, 8, 2131–2140. [Google Scholar] [CrossRef]
- Sorg, O.; Antille, C.; Kaya, G.; Saurat, J.H. Retinoids in cosmeceuticals. Dermatol. Ther. 2006, 19, 289–296. [Google Scholar] [CrossRef]
- Serri, R.; Iorizzo, M. Cosmeceuticals: Focus on topical retinoids in photoaging. Clin. Dermatol. 2008, 26, 633–635. [Google Scholar] [CrossRef]
- Kim, B.-H. Safety Evaluation and Anti-wrinkle Effects of Retinoids on Skin. Toxicol. Res. 2010, 26, 61–66. [Google Scholar] [CrossRef]
- Parker, G.L.; Smith, L.K.; Baxendale, I.R. Development of the industrial synthesis of vitamin A. Tetrahedron 2016, 72, 1645–1652. [Google Scholar] [CrossRef]
- Mercier, C.; Chabardes, P. Organometallic chemistry in industrial vitamin A and vitamin E synthesis. Pure Appl. Chem. 1994, 66, 1509–1518. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Laudert, D.; Letinois, U.; McClymont, T.; Medlock, J.; Netscher, T.; Bonrath, W. One hundred years of vitamins—A success story of the natural sciences. Angew. Chem. Int. Ed. 2012, 51, 12960–12990. [Google Scholar] [CrossRef]
- Lee, Y.G.; Kim, C.; Sun, L.; Lee, T.H.; Jin, Y.S. Selective production of retinol by engineered Saccharomyces cerevisiae through the expression of retinol dehydrogenase. Biotechnol. Bioeng. 2022, 119, 399–410. [Google Scholar] [CrossRef]
- Mo, Q.; Song, W.; Xue, Z.; Yuan, J. Multi-level engineering of Saccharomyces cerevisiae for the synthesis and accumulation of retinal. Green Chem. 2022, 24, 8259–8263. [Google Scholar] [CrossRef]
- Seo, S.O.; Jin, Y.S. Next-Generation Genetic and Fermentation Technologies for Safe and Sustainable Production of Food Ingredients: Colors and Flavorings. Annu. Rev. Food Sci. Technol. 2022, 13, 463–488. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Ye, L. From β-Carotene to Retinoids: A Review of Microbial Production of Vitamin A. J. Agric. Food Chem. 2024, 72, 20752–20762. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D.; Ajikumar, P.K.; Stephanopoulos, G. Engineering microbial cell factories for biosynthesis of isoprenoid molecules: Beyond lycopene. TRENDS Biotechnol. 2007, 25, 417–424. [Google Scholar] [CrossRef]
- Jang, H.-J.; Yoon, S.-H.; Ryu, H.-K.; Kim, J.-H.; Wang, C.-L.; Kim, J.-Y.; Oh, D.-K.; Kim, S.-W. Retinoid production using metabolically engineered Escherichia coli with a two-phase culture system. Microb. Cell Factories 2011, 10, 59. [Google Scholar] [CrossRef]
- Hong, K.-K.; Nielsen, J. Metabolic engineering of Saccharomyces cerevisiae: A key cell factory platform for future biorefineries. Cell. Mol. Life Sci. 2012, 69, 2671–2690. [Google Scholar] [CrossRef]
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56. [Google Scholar] [CrossRef]
- Kwak, S.; Kim, S.R.; Xu, H.; Zhang, G.C.; Lane, S.; Kim, H.; Jin, Y.S. Enhanced isoprenoid production f rom xylose by engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 2017, 114, 2581–2591. [Google Scholar] [CrossRef]
- Özaydın, B.; Burd, H.; Lee, T.S.; Keasling, J.D. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab. Eng. 2013, 15, 174–183. [Google Scholar] [CrossRef]
- Li, T.; Liu, G.-S.; Zhou, W.; Jiang, M.; Ren, Y.-H.; Tao, X.-Y.; Liu, M.; Zhao, M.; Wang, F.-Q.; Gao, B. Metabolic engineering of Saccharomyces cerevisiae to overproduce squalene. J. Agric. Food Chem. 2020, 68, 2132–2138. [Google Scholar] [CrossRef]
- Kim, S.-J.; Seo, S.-O.; Jin, Y.-S.; Seo, J.-H. Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresour. Technol. 2013, 146, 274–281. [Google Scholar] [CrossRef]
- Hosaka, K.; Nikawa, J.-i.; Kodaki, T.; Yamashita, S. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J. Biochem. 1992, 111, 352–358. [Google Scholar] [CrossRef]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Kang, M.-J.; Hwang, H.-S.; Baek, K.-R.; Seo, S.-O. Reduction of Ethyl Carbamate in an Alcoholic Beverage by CRISPR/Cas9-Based Genome Editing of the Wild Yeast. Foods 2023, 12, 102. [Google Scholar] [CrossRef]
- Kwak, S. Yeast Engineering for Pharmaceutical and Nutraceutical Purposes; University of Illinois at Urbana-Champaign: Champaign, IL, USA, 2017. [Google Scholar]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Boulet, L.; Alex, B.; Clavey, N.; Martinez, J.; Ducros, V. Simultaneous analysis of retinol, six carotenoids, two tocopherols, and coenzyme Q10 from human plasma by HPLC. J. Chromatogr. B 2020, 1151, 122158. [Google Scholar] [CrossRef]
- Hu, Q.; Yu, H.; Ye, L. Production of retinoic acid by engineered Saccharomyces cerevisiae using an endogenous aldehyde dehydrogenase. Biotechnol. Bioeng. 2022, 119, 3241–3251. [Google Scholar] [CrossRef]
- Huang, G.; Li, J.; Lin, J.; Duan, C.; Yan, G. Multi-modular metabolic engineering and efflux engineering for enhanced lycopene production in recombinant Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2024, 51, kuae015. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, T.; Yu, H.; Ye, L. Selective biosynthesis of retinol in S. cerevisiae. Bioresour. Bioprocess. 2022, 9, 22. [Google Scholar] [CrossRef]
- Polakowski, T.; Stahl, U.; Lang, C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 1998, 49, 66–71. [Google Scholar] [CrossRef]
- Kim, T.Y.; Park, H.; Kim, S.-K.; Kim, S.-J.; Park, Y.-C. Production of (−)-α-bisabolol in metabolically engineered Saccharomyces cerevisiae. J. Biotechnol. 2021, 340, 13–21. [Google Scholar] [CrossRef]
- Wegner, S.A.; Chen, J.-M.; Ip, S.S.; Zhang, Y.; Dugar, D.; Avalos, J.L. Engineering acetyl-CoA supply and ERG9 repression to enhance mevalonate production in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab050. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Bu, X.; Lan, Y.-B.; Duan, C.-Q.; Yan, G.-L. Combined metabolic engineering and lipid droplets degradation to increase vitamin A production in Saccharomyces cerevisiae. Microb. Cell Factories 2024, 23, 317. [Google Scholar] [CrossRef]
- Chen, Y.; Partow, S.; Scalcinati, G.; Siewers, V.; Nielsen, J. Enhancing the copy number of episomal plasmids in Saccharomyces cerevisiae for improved protein production. FEMS Yeast Res. 2012, 12, 598–607. [Google Scholar] [CrossRef]
- He, S.; Zhang, Z.; Lu, W. Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2023, 50, kuac029. [Google Scholar] [CrossRef]
- Sasaki, Y.; Mitsui, R.; Yamada, R.; Ogino, H. Secretory overexpression of the endoglucanase by Saccharomyces cerevisiae via CRISPR-δ-integration and multiple promoter shuffling. Enzym. Microb. Technol. 2019, 121, 17–22. [Google Scholar] [CrossRef]


| Name | Description | Reference |
|---|---|---|
| Strains | ||
| Escherichia coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen (Carlsbad, CA, USA) |
| Saccharomyces cerevisiae D452-2 | MATα, his3, leu2, ura3, can1 | [20] |
| Saccharomyces cerevisiae SOS4 | S. cerevisiae D452-2 Δpdc1, Δpdc5 | [19] |
| Saccharomyces cerevisiae S4H | S. cerevisiae SOS4, evolved for cell growth | In this study |
| Saccharomyces cerevisiae DET | S. cerevisiae D452-2 CS5::PGKp-tHMG1-TEFt, CS6::TEFp-ERG10-CYCt | In this study |
| Saccharomyces cerevisiae SET | S. cerevisiae SOS4 CS5::PGKp-tHMG1-TEFt, CS6::TEFp-ERG10-CYCt | In this study |
| D452-CON | S. cerevisiae D452-2 (pRS423GPD, pRS425GPD, pRS426GPD, pRS42KGPD) | In this study |
| D452-RE | S. cerevisiae D452-2 (pRS423GPD-crtYB, pRS425GPD-crtE, pRS426GPD-crtI, pRS42KGPD-Blh) | In this study |
| DET-RE | S. cerevisiae DET (pRS423GPD-crtYB, pRS425GPD-crtE, pRS426GPD-crtI, pRS42KGPD-Blh) | In this study |
| S4H-CON | S. cerevisiae S4H (pRS423GPD, pRS425GPD, pRS426GPD, pRS42KGPD) | In this study |
| S4H-RE | S. cerevisiae S4H (pRS423GPD-crtYB, pRS425GPD-crtE, pRS426GPD-crtI, pRS42KGPD-Blh) | In this study |
| SET-RE | S. cerevisiae SET (pRS423GPD-crtYB, pRS425GPD-crtE, pRS426GPD-crtI, pRS42KGPD-Blh) | In this study |
| Plasmids | ||
| pRS423GPD | GPDp, CYCt, 2u origin, HIS3, Ampr | [21] |
| pRS425GPD | GPDp, CYCt, 2u origin, LEU2, Ampr | [21] |
| pRS426GPD | GPDp, CYCt, 2u origin, URA3, Ampr | [21] |
| pRS42KGPD | GPDp, CYCt, 2u origin, KanMX, Ampr | In this study |
| pRS423GPD-crtYB | pRS423GPD harboring the crtYB gene | In this study |
| pRS425GPD-crtE | pRS425GPD harboring the crtE gene | In this study |
| pRS426GPD-crtI | pRS426GPD harboring the crtI gene | In this study |
| pRS42KGPD-Blh | pRS42HGPD harboring the Blh gene | In this study |
| pRS426PGKp-tHMG1-TEFt | PGKp, TEFt, URA3, Ampr, harboring the tHMG1 gene | In this study |
| pRS426TEFp-ERG10-CYCt | TEFp, CYCt, URA3, Ampr, harboring the ERG10 gene | In this study |
| pCas-Hyg | Cas9 expression plasmid, Hygromycin B marker | [22] |
| pRS42K-CS5 | pRS42K, gRNA expression cassette targeting the intergenic site on Chr XV | In this study |
| pRS42K-CS6 | pRS42K, gRNA expression cassette targeting the intergenic site on Chr VII | In this study |
| Strain | Retinal Biosynthetic Genes | Overexpression of ERG10 and tHMG1 | Pdc Deletion | Retinal (mg/L) | Ethanol (g/L) | Yield of Retinal (mg Retinal/g Glucose) |
|---|---|---|---|---|---|---|
| D452-CON | − | − | − | N.D. | 12.6 | N.D. |
| D452-RE | + | − | − | 2.7 | 12.7 | 0.1 |
| DET-RE | + | + | − | 10.2 | 13.9 | 0.3 |
| S4H-CON | − | − | + | N.D. | N.D. | N.D. |
| S4H-RE | + | − | + | 39.2 | N.D. | 1.4 |
| SET-RE | + | + | + | 117.4 | N.D. | 5.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, H.-S.; Baek, K.-R.; Seo, S.-O. Retinal Production by Precision Fermentation of Saccharomyces cerevisiae. Fermentation 2025, 11, 214. https://doi.org/10.3390/fermentation11040214
Hwang H-S, Baek K-R, Seo S-O. Retinal Production by Precision Fermentation of Saccharomyces cerevisiae. Fermentation. 2025; 11(4):214. https://doi.org/10.3390/fermentation11040214
Chicago/Turabian StyleHwang, Hye-Seon, Kwang-Rim Baek, and Seung-Oh Seo. 2025. "Retinal Production by Precision Fermentation of Saccharomyces cerevisiae" Fermentation 11, no. 4: 214. https://doi.org/10.3390/fermentation11040214
APA StyleHwang, H.-S., Baek, K.-R., & Seo, S.-O. (2025). Retinal Production by Precision Fermentation of Saccharomyces cerevisiae. Fermentation, 11(4), 214. https://doi.org/10.3390/fermentation11040214







