Microbial Community, Fatty Acid Composition, and Health Potential of Horse Oil Fermented with Barley Nuruk
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Fatty Acid Analysis of Horse Oil Hexane Extracts and Fermentation Samples
2.3. Preparation of Horse Oil Ferments and Extracts
2.4. Amplification and Sequencing
2.5. Antidiabetic Capacity
2.6. Molecular Docking Simulations Analysis
2.7. Human Skin Irritation Test
2.8. Data Analysis
3. Results and Discussion
3.1. Fatty Acid Composition of Horse Oil and Related Extracts
3.2. Microbial Community Dynamics During Fermentation of Horse Oil Using Barley Nuruks
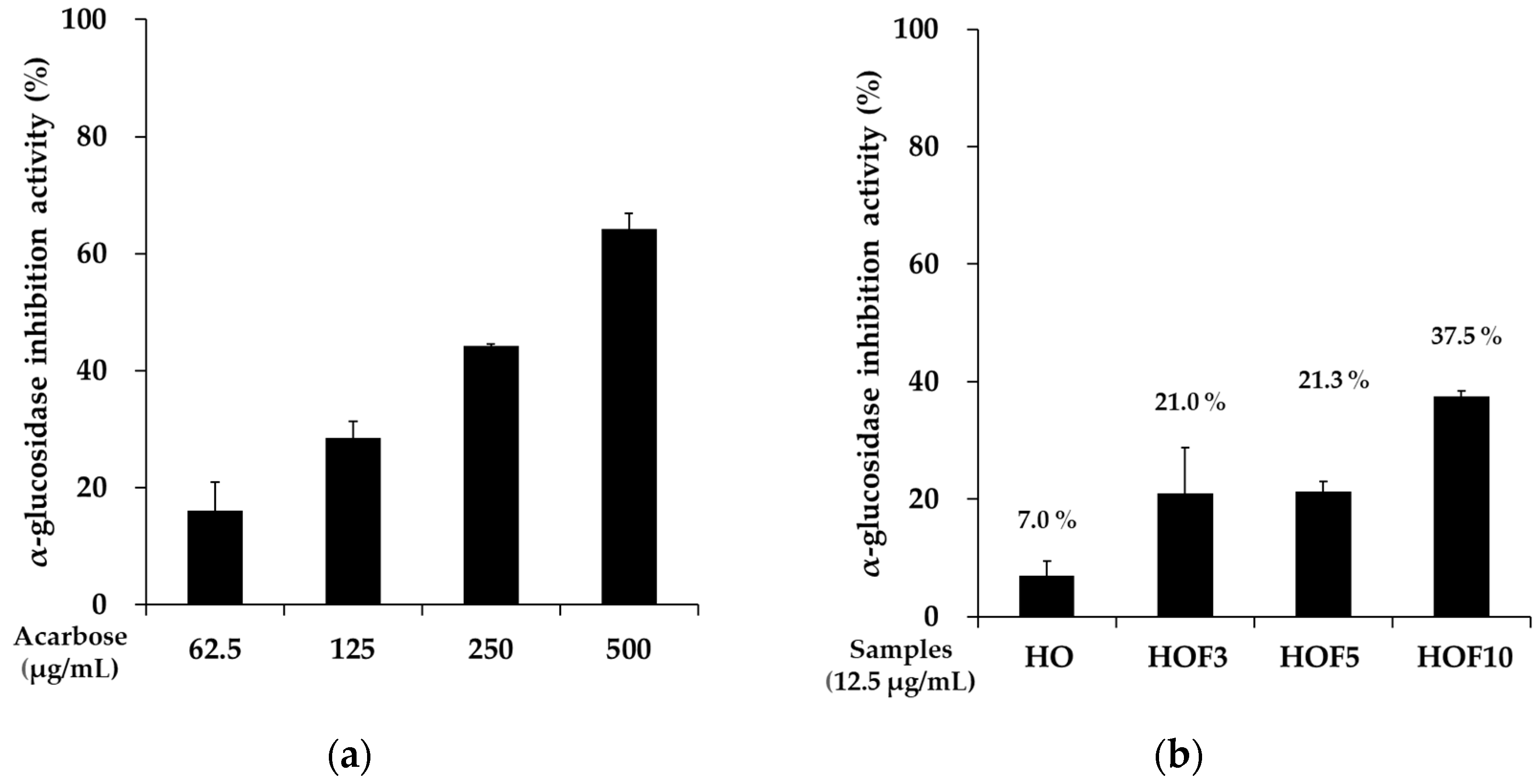
3.3. Evaluation of α-Glucosidase Inhibitory Activity in Fermented Extracts
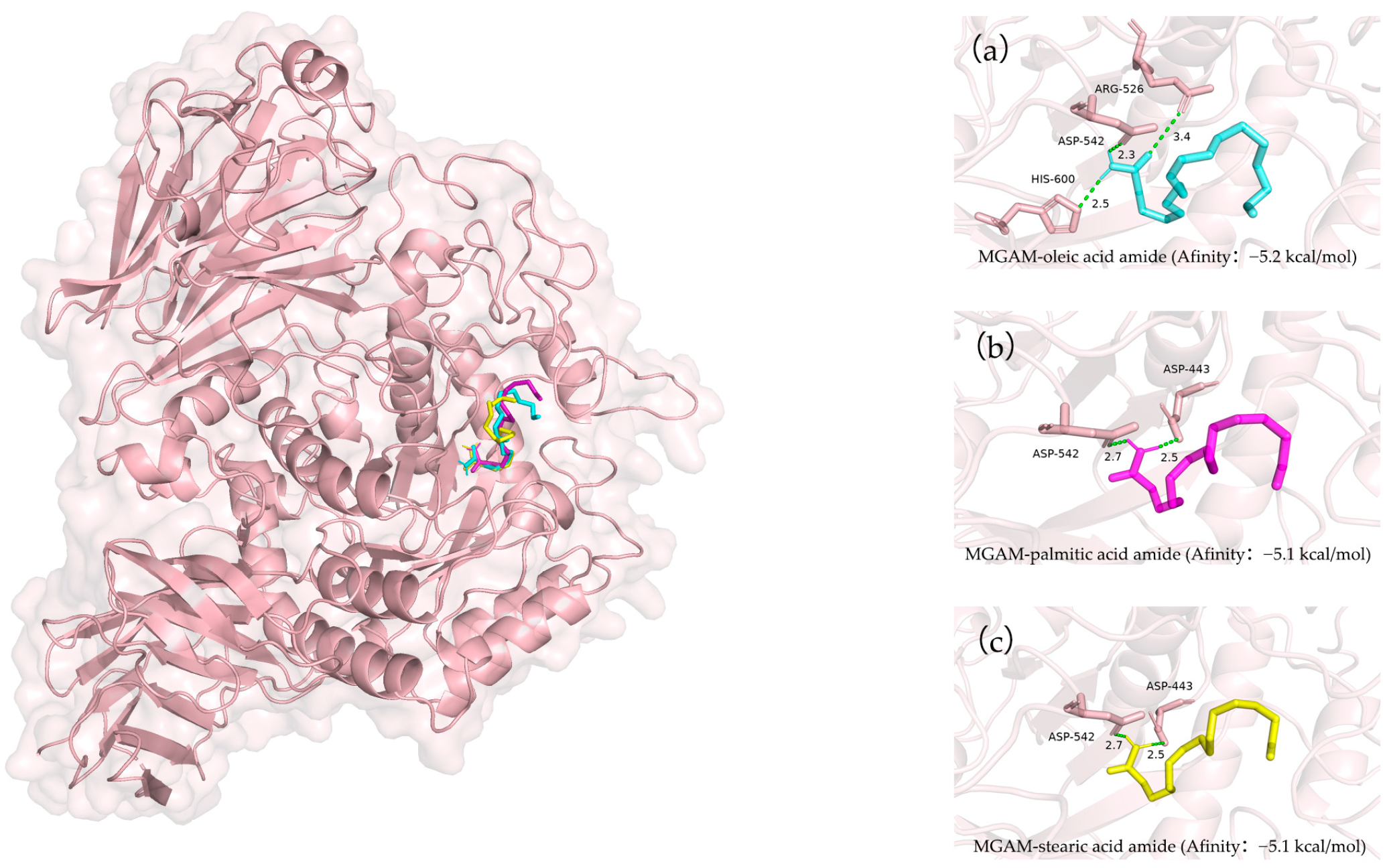
3.4. Molecular Docking Simulations
3.5. Human Skin Primary Irritation Test for Safety Evaluation of Functional Ingredients
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, X.; Dong, M.; Liu, J.; Guo, N.; Li, J.; Shi, Y.; Yang, Y. Fermentation: Improvement of pharmacological effects and applications of botanical drugs. Front. Pharmacol. 2024, 15, 1430238. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mazumder, R.; Rani, A.; Pandey, P.; Khurana, N. Novel Approaches for the Management of Type 2 Diabetes Mellitus: An Update. Curr. Diabetes Rev. 2024, 20, e051023221768. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Sharma, M.; Singh, S.; Goyal, A. Recent Advances of α-Glucosidase Inhibitors: A Comprehensive Review. Curr. Top. Med. Chem. 2022, 22, 2069–2086. [Google Scholar] [CrossRef]
- Kumar, S.; Narwal, S.; Kumar, V.; Prakash, O. α-glucosidase inhibitors from plants: A natural approach to treat diabetes. Pharmacogn. Rev. 2011, 5, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.C.; Ko, J.H.; Kang, H.-K.; Kim, S.; Kang, C.I.; Lee, J.N.; Park, S.M.; Hyun, C.G. Antimelanogenic Effects of Polygonum tinctorium Flower Extract from Traditional Jeju Fermentation via Upregulation of Extracellular Signal-Regulated Kinase and Protein Kinase B Activation. Int. J. Mol. Sci. 2018, 19, 2895. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hyun, S.B.; Yun, S.H.; Chung, Y.C.; Hyun, C.G. Anti-oxidant and Anti-inflammatory Effects of the Fermented Rhododendron weyrichii Flower Extracts in Shindari, a Traditional Jeju Fermented Drink. Microbiol. Biotechnol. Lett. 2020, 48, 471–479. [Google Scholar] [CrossRef]
- Lee, J.C.; Park, G.R.; Choi, B.S.; Lee, Y.; Han, C.H. Restoration of the inflammatory gene expression by horse oil in DNCB-treated mice skin. J. Vet. Sci. 2020, 21, e15. [Google Scholar] [CrossRef]
- Kim, I.W.; Jeong, H.S.; Yun, H.Y.; Baek, K.J.; Kwon, N.S.; Kim, D.S. Efficacy of horse oil on lipopolysaccharide-induced inflammation in human keratinocyte. J. Tradit. Chin. Med. 2021, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kim, S.; Jang, M.; Kim, H.; Lee, S.; Kim, Y.; Eom, Y.A.; Kang, G.; Chiang, L.; Baek, J.H.; et al. Two-phase delivery using a horse oil and adenosine-loaded dissolving microneedle patch for skin barrier restoration, moisturization, and wrinkle improvement. J. Cosmet. Dermatol. 2019, 18, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Maeng, Y.; Kim, K.T.; Zhou, X.; Jin, L.; Kim, K.S.; Kim, Y.H.; Lee, S.; Park, J.H.; Chen, X.; Kong, M.; et al. A novel microbial technique for producing high-quality sophorolipids from horse oil suitable for cosmetic applications. Microb. Biotechnol. 2018, 11, 917–929. [Google Scholar] [CrossRef]
- Park, Y.H.; Kim, H.J. Formulation and stability of horse oil-in-water emulsion by HLB system. Food Sci. Biotechnol. 2021, 16, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.A.; Liang, X.; Xu, Y. Analysis of the Setomimycin Biosynthetic Gene Cluster from Streptomyces nojiriensis JCM3382 and Evaluation of Its α-Glucosidase Inhibitory Activity Using Molecular Docking and Molecular Dynamics Simulations. Int. J. Mol. Sci. 2024, 25, 10758. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Dvořáková, M.; Svobodová, L.; Rucki, M.; Ševčík, V.; Hošíková, B.; Chrz, J.; Bendová, H.; Kejlová, K.; Očadlíková, D.; Malý, M.; et al. The Safety Assessment of Cosmetic Perfumes by Using in Chemico and In Vitro Methods in Combination with GC-MS/MS Analysis. Altern. Lab. Anim. 2023, 51, 224–248. [Google Scholar] [CrossRef]
- Ali, S.; Ekbbal, R.; Salar, S.; Yasheshwar, A.S.A.; Jaiswal, A.K.; Singh, M.; Yadav, D.K.; Kumar, S. Gaurav Quality Standards and Pharmacological Interventions of Natural Oils: Current Scenario and Future Perspectives. ACS Omega 2023, 8, 39945–39963. [Google Scholar] [CrossRef] [PubMed]
- Duchateau, C.; Canfyn, M.; Desmedt, B.; Kauffmann, J.M.; Stévigny, C.; De Braekeleer, K.; Deconinck, E. CBD oils on the Belgian market: A validated MRM GC-MS/MS method for routine quality control using QuEChERS sample clean up. J. Pharm. Biomed. Anal. 2021, 205, 114344. [Google Scholar] [CrossRef] [PubMed]
- Czarnowski, P.; Mikula, M.; Ostrowski, J.; Żeber-Lubecka, N. Gas Chromatography-Mass Spectrometry-Based Analyses of Fecal Short-Chain Fatty Acids (SCFAs): A Summary Review and Own Experience. Biomedicines 2024, 12, 1904. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.H.; Kuo, C.H. Gas chromatography-mass spectrometry-based analytical strategies for fatty acid analysis in biological samples. J. Food Drug Anal. 2020, 28, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, P.; Chittiboyina, A.G.; Chen, D.; Zhao, J.; Avula, B.; Wang, Y.H.; Khan, I.A. Characterization, Quantification and Quality Assessment of Avocado (Persea americana Mill.) Oils. Molecules 2020, 25, 1453. [Google Scholar] [CrossRef] [PubMed]
- Franceschetti, L.; Lodetti, G.; Blandino, A.; Amadasi, A.; Bugelli, V. Exploring the role of the human microbiome in forensic identification: Opportunities and challenges. Int. J. Leg. Med. 2024, 138, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Hoisington, A.J.; Stamper, C.E.; Ellis, J.C.; Lowry, C.A.; Brenner, L.A. Quantifying variation across 16S rRNA gene sequencing runs in human microbiome studies. Appl. Microbiol. Biotechnol. 2024, 108, 367. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Lu, H.; Liu, X.; He, J.; Li, B.; Wang, Z.; Zhao, Y.; Zhang, X.; Yu, X. Unravelling the enigma of the human microbiome: Evolution and selection of sequencing technologies. Microb. Biotechnol. 2024, 17, e14364. [Google Scholar] [CrossRef] [PubMed]
- Bertolo, A.; Valido, E.; Stoyanov, J. Optimized bacterial community characterization through full-length 16S rRNA gene sequencing utilizing MinION nanopore technology. BMC Microbiol. 2024, 24, 58. [Google Scholar] [CrossRef] [PubMed]
- Curry, K.D.; Soriano, S.; Nute, M.G.; Villapol, S.; Dilthey, A.; Treangen, T.J. Microbial Community Profiling Protocol with Full-length 16S rRNA Sequences and Emu. Curr. Protoc. 2024, 4, e978. [Google Scholar] [CrossRef]
- Buetas, E.; Jordán-López, M.; López-Roldán, A.; D’Auria, G.; Martínez-Priego, L.; De Marco, G.; Carda-Diéguez, M.; Mira, A. Full-length 16S rRNA gene sequencing by PacBio improves taxonomic resolution in human microbiome samples. BMC Genom. 2024, 25, 310. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Song, J.; Liu, C.; Lin, R.; Liang, D.; Aweya, J.J.; Weng, W.; Zhu, L.; Shang, J.; Yang, S. Synthetic microbial communities: Novel strategies to enhance the quality of traditional fermented foods. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13388. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.E.; Bae, S.; Park, S.J.; Lee, P.; Hyun, C.G. Microbial Consortium of Jeju Traditional Fermented Foods and Their Cosmetic Ingredient Potential. Fermentation 2024, 10, 345. [Google Scholar] [CrossRef]
- Valentino, V.; Magliulo, R.; Farsi, D.; Cotter, P.D.; O’Sullivan, O.; Ercolini, D.; De Filippis, F. Fermented foods, their microbiome and its potential in boosting human health. Microb. Biotechnol. 2024, 17, e14428. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Nawazish, H.; Farid, M.S.; Abdul Qadoos, K.; Habiba, U.E.; Muzamil, M.; Tanveer, M.; Sienkiewicz, M.; Lichota, A.; Łopusiewicz, Ł. Health-Promoting Effects of Lactobacillus acidophilus and Its Technological Applications in Fermented Food Products and Beverages. Fermentation 2024, 10, 380. [Google Scholar] [CrossRef]
- Gao, H.; Li, X.; Chen, X.; Hai, D.; Wei, C.; Zhang, L.; Li, P. The Functional Roles of Lactobacillus acidophilus in Different Physiological and Pathological Processes. J. Microbiol. Biotechnol. 2022, 32, 1226–1233. [Google Scholar] [CrossRef]
- Hossain, U.; Das, A.K.; Ghosh, S.; Sil, P.C. An overview on the role of bioactive α-glucosidase inhibitors in ameliorating diabetic complications. Food Chem. Toxicol. 2020, 145, 111738. [Google Scholar] [CrossRef] [PubMed]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2022, 21, 1049–1079. [Google Scholar] [CrossRef]
- Zhang, K.Q.; Lin, L.L.; Xu, H.J. Research on antioxidant performance of diglucosyl gallic acid and its application in emulsion cosmetics. Int. J. Cosmet. Sci. 2022, 44, 177–188. [Google Scholar] [CrossRef]
- Ćorković, I.; Gašo-Sokač, D.; Pichler, A.; Šimunović, J.; Kopjar, M. Dietary Polyphenols as Natural Inhibitors of α-Amylase and α-Glucosidase. Life 2022, 12, 1692. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.C.; Hyun, C.G. Inhibitory Effects of Pinostilbene on Adipogenesis in 3T3-L1 Adipocytes: A Study of Possible Mechanisms. Int. J. Mol. Sci. 2021, 22, 13446. [Google Scholar] [CrossRef]
- Sugiyama, M.; Akita, M.; Alépée, N.; Fujishiro, M.; Hagino, S.; Handa, Y.; Ikeda, H.; Imai, N.; Jitsukawa, S.; Katoh, M.; et al. Comparative assessment of 24-hr primary skin irritation test and human patch test data with in vitro skin irritation tests according to OECD Test Guideline 439 (for quasi-drugs in Japan). J. Toxicol. Sci. 2018, 43, 751–768. [Google Scholar] [CrossRef] [PubMed]
- Na, M.; Ritacco, G.; O’Brien, D.; Lavelle, M.; Api, A.M.; Basketter, D. Fragrance Skin Sensitization Evaluation and Human Testing: 30-Year Experience. Dermatitis 2021, 32, 339–352. [Google Scholar] [CrossRef] [PubMed]
- DeKoven, J.G.; Warshaw, E.M.; Reeder, M.J.; Atwater, A.R.; Silverberg, J.I.; Belsito, D.V.; Sasseville, D.; Zug, K.A.; Taylor, J.S.; Pratt, M.D.; et al. North American Contact Dermatitis Group Patch Test Results: 2019-2020. Dermatitis 2023, 34, 90–104. [Google Scholar] [CrossRef]

| Ingredient | Area (%) |
|---|---|
| 5-Methylundecane | 5.3 |
| 4,6-Dimethyldodecane | 5.7 |
| Heneicosane | 6.0 |
| Docosane | 5.7 |
| Myristic acid | 1.4 |
| Stearyl aldehyde | 2.1 |
| Docosane | 3.5 |
| Palmitic acid | 22.5 |
| Stearyl aldehyde | 3.0 |
| Docosane | 3.2 |
| Oleic acid | 19.5 |
| Stearic acid | 1.8 |
| Palmitoyl chloride | 2.6 |
| (Z)-octadec-13-enal | 0.9 |
| Palmitic acid diglycerin ester | 5.7 |
| Dotriacontane | 2.4 |
| 1-Monolinolein (1-linoleoyl glycerol) | 0.8 |
| 3-Hydroxypropyl oleate | 2.1 |
| Linoleic acid | 1.6 |
| cis-9-Hexadecenal | 4.5 |
| Ingredient | Hexane Extracts 1 | |||
|---|---|---|---|---|
| HO | HOF3 | HOF5 | HOF10 | |
| Palmitic amide | 10.0 | 14.6 | 15.1 | 24.0 |
| Oleic acid amide | 43.8 | 72.3 | 84.9 | 73.9 |
| Stearic acid amide | 2.1 | |||
| Palmitic acid (16:0) | 20.4 | 5.2 | ||
| Oleic acid (18:1) | 11.9 | |||
| Squalene | 6.5 | 2.3 | ||
| Cholesterol | 3.5 | 0.9 | ||
| Myristic aldehyde | 1.1 | |||
| 3-Hydroxypropyl oleate | 1.2 | |||
| di-(9-octadecenoyl)-glycerol | 1.5 | |||
| Ethyl palmitate | 1.4 | |||
| Ethyl linoleate | 1.2 | |||
| Vinyl palmitate | 1.1 | |||
| 1,2-Dipalmitoyl-sn-glycerol | 1.2 | |||
| Scientific Names | HOF3 | HOF5 | HOF10 |
|---|---|---|---|
| Bifidobacterium animalis | 0 | 0 | 0.5 |
| Bacillus subtilis | 0 | 2.4 | 0.7 |
| Staphylococcus warneri | 1.5 | 1.4 | 0 |
| Enterococcus faecalis | 32.9 | 29.1 | 0.3 |
| Enterococcus faecium | 4.8 | 9.3 | 0 |
| Enterococcus gallinarum | 0.7 | 0 | 0 |
| Enterococcus hirae | 1.2 | 2.2 | 0 |
| Enterococcus innesii | 0 | 0.7 | 0 |
| Enterococcus mundtii | 1.5 | 1.3 | 0 |
| Lactobacillus acidophilus | 0 | 0 | 95.2 |
| Leuconostoc mesenteroides | 0.4 | 0.4 | 0 |
| Pediococcus pentosaceus | 0.4 | 0.3 | 0 |
| Weissella paramesenteroides | 1.4 | 1.8 | 0 |
| Lactococcus lactis | 0.2 | 0.3 | 0 |
| Lactococcus taiwanensis | 0.2 | 0.2 | 0 |
| Clostridium beijerinckii | 0 | 0.3 | 0 |
| Clostridium butyricum | 0 | <0.1 | <0.1 |
| Clostridium diolis | 0 | 4.4 | 2.6 |
| Crassifilum sonorensis | <0.1 | 0.2 | 0 |
| Acetobacter persici | 0 | 0 | 0.5 |
| Gluconobacter japonicus | <0.1 | 0 | 0 |
| Atlantibacter hermannii | 6.6 | 9.3 | 0 |
| Enterobacter kobei | <0.1 | 0 | 0 |
| Enterobacter quasihormaechei | 24 | 20.2 | 1.40 |
| Phytobacter palmae | 1.80% | 0 | 0 |
| Salmonella enterica | 19.4 | 14.8 | 0 |
| Other | 0 | 1.5 | 0 |
| No | Samples | Responders | 1st Assessment | 2nd Assessment | Reaction Grade (R) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| +1 | +2 | +3 | +4 | +1 | +2 | +3 | +4 | ||||
| 1 | HO (12.5 μg/mL) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | HOF3 (12.5 μg/mL) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | HOF5 (12.5 μg/mL) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | HOF10 (12.5 μg/mL) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-H.; Bae, S.-E.; Kang, H.-M.; Ha, Y.-J.; Hyun, C.-G. Microbial Community, Fatty Acid Composition, and Health Potential of Horse Oil Fermented with Barley Nuruk. Fermentation 2025, 11, 1. https://doi.org/10.3390/fermentation11010001
Lee J-H, Bae S-E, Kang H-M, Ha Y-J, Hyun C-G. Microbial Community, Fatty Acid Composition, and Health Potential of Horse Oil Fermented with Barley Nuruk. Fermentation. 2025; 11(1):1. https://doi.org/10.3390/fermentation11010001
Chicago/Turabian StyleLee, Jeong-Ha, Sung-Eun Bae, Ho-Min Kang, Yu-Jin Ha, and Chang-Gu Hyun. 2025. "Microbial Community, Fatty Acid Composition, and Health Potential of Horse Oil Fermented with Barley Nuruk" Fermentation 11, no. 1: 1. https://doi.org/10.3390/fermentation11010001
APA StyleLee, J.-H., Bae, S.-E., Kang, H.-M., Ha, Y.-J., & Hyun, C.-G. (2025). Microbial Community, Fatty Acid Composition, and Health Potential of Horse Oil Fermented with Barley Nuruk. Fermentation, 11(1), 1. https://doi.org/10.3390/fermentation11010001






