Characterization of the Gamma-Aminobutyric Acid (GABA) Biosynthetic Gene Cluster in High GABA-Producing Enterococcus avium G-15
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. DNA Manipulations and Sequencing
2.3. Overexpression and Purification of GAD Proteins
2.4. Kinetic Study of the GAD Protein
2.5. Settings and Locations Electrophoretic Mobility Shift Assay (EMSA)
2.6. Construction of Plasmids Carrying a Modified gad Cluster
2.7. Transformation of LAB
3. Results
3.1. Kinetic Parameters of Each GAD
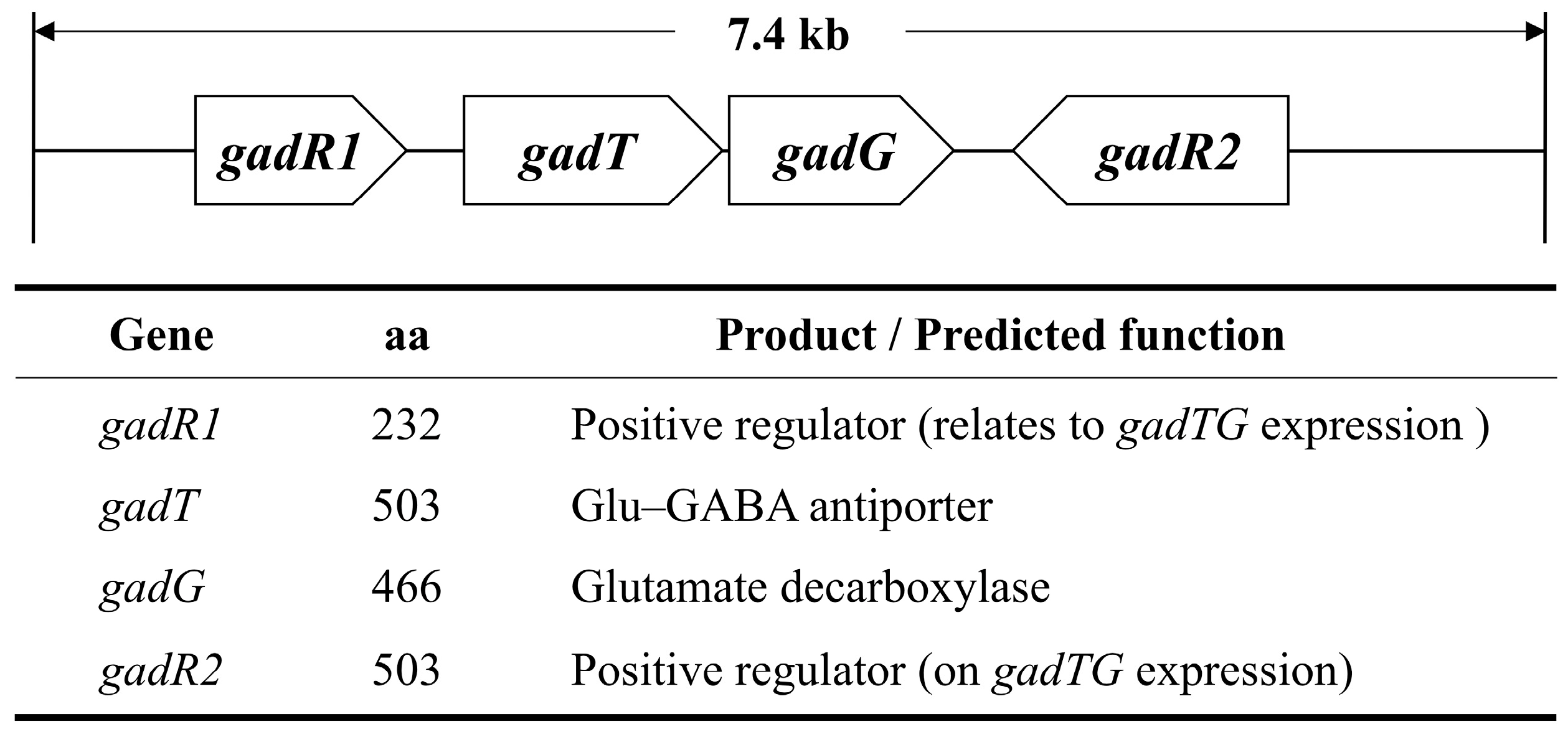
3.2. DNA-Binding Assay of Putative Transcriptional Regulator, GadR1
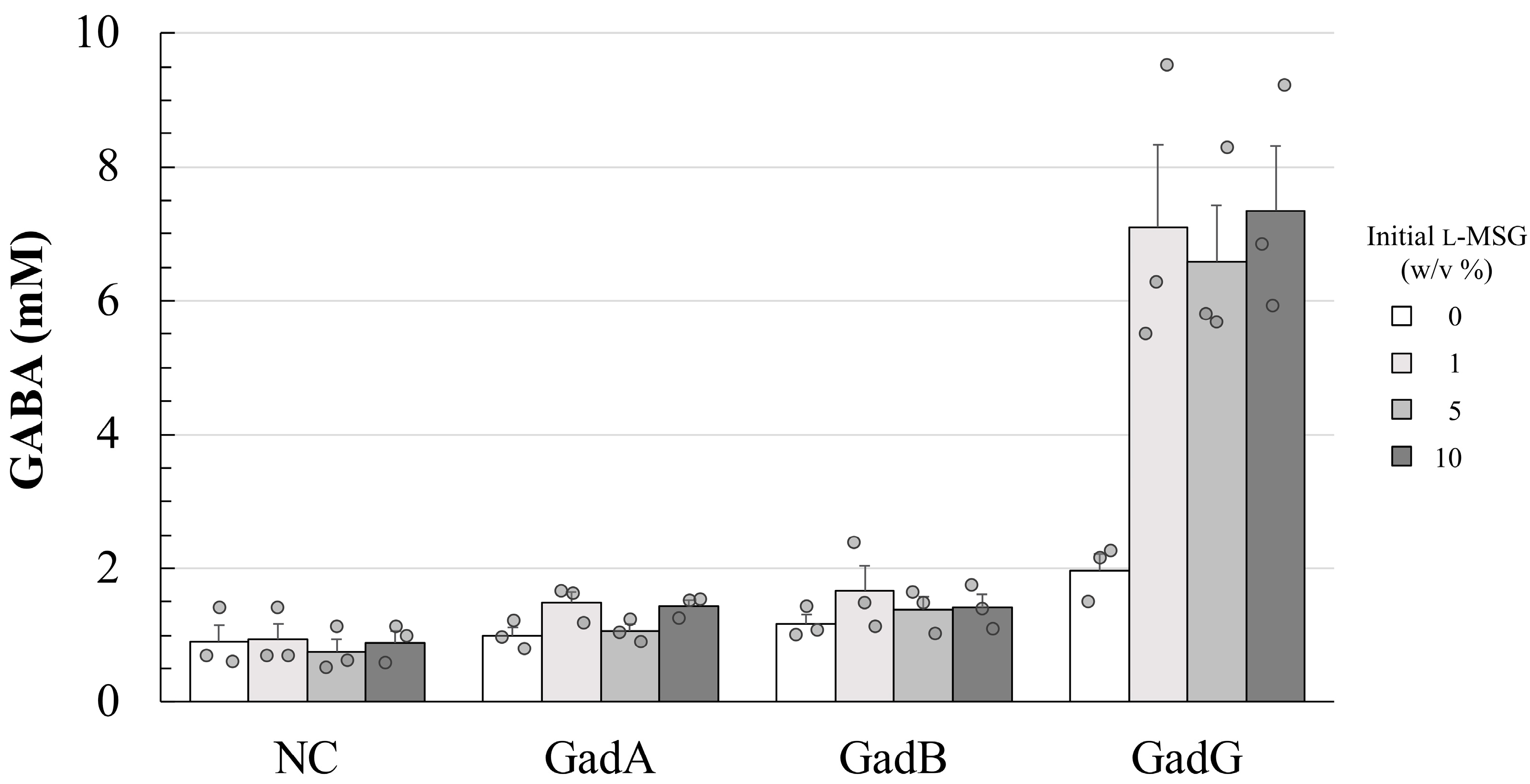
3.3. Effect of Promoter Differences on GABA Productivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdoua, A.M.; Higashiguchi, S.; Horie, K.; Kima, M.; Hatta, H.; Yokogoshi, H. Relaxation and immunity enhancement effects of gamma-aminobutyric acid (GABA) administration in humans. Biofactors 2006, 26, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Diana, M.; Quílez, J.; Rafecas, M. Gamma-aminobutyric acid as a bioactive compound in foods: A review. J. Funct. Foods 2014, 10, 407–420. [Google Scholar] [CrossRef]
- Boonstra, E.; de Kleijn, R.; Colzato, L.S.; Alkemade, A.; Forstmann, B.U.; Nieuwenhuis, S. Neurotransmitters as food supplements: The effects of GABA on brain and behavior. Front. Psychol. 2015, 6, 1520. [Google Scholar] [CrossRef] [PubMed]
- Hepsomali, P.; Groeger, J.A.; Nishihira, J.; Scholey, A. Effects of oral gamma-aminobutyric acid (GABA) administration on stress and sleep in humans: A systematic review. Front. Neurosci. 2020, 14, 923. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Kimura, M.; Kasaha, K.; Matsumoto, K.; Sansawa, H.; Yamori, Y. Effect of a γ-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Br. J. Nutr. 2004, 92, 411–417. [Google Scholar] [CrossRef]
- Kawakami, K.; Yamada, K.; Yamada, T.; Nabika, T.; Nomura, M. Antihypertensive effect of γ-aminobutyric acid-enriched brown rice on spontaneously hypertensive rats. J. Nutr. Sci. Vitaminol. 2018, 64, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Heli, Z.; Hongyu, C.; Dapeng, B.; Yee Shin, T.; Yejun, Z.; Xi, Z.; Yingying, W. Recent advances of γ-aminobutyric acid: Physiological and immunity function, enrichment, and metabolic pathway. Front. Nutr. 2022, 9, 1076223. [Google Scholar] [CrossRef]
- Dhakal, R.; Bajpai, V.K.; Baek, K.H. Production of gaba (γ-aminobutyric acid) by microorganisms: A review. Braz. J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef]
- Li, H.; Cao, Y. Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 2010, 39, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Capitani, G.; De Biase, D.; Aurizi, C.; Gut, H.; Bossa, F.; Grütter, M.G. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 2003, 22, 4027–4037. [Google Scholar] [CrossRef]
- Ma, D.; Lu, P.; Yan, C.; Fan, C.; Yin, P.; Wang, J.; Shi, Y. Structure and mechanism of a glutamate-GABA antiporter. Nature 2012, 483, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Noda, M.; Ozaki, M.; Maruyama, M.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Establishment of an efficient fermentation system of gamma-aminobutyric acid by a lactic acid bacterium, Enterococcus avium G-15, isolated from carrot leaves. Biol. Pharm. Bull. 2010, 33, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Noda, M.; Danshiitsoodol, N.; Inoue, Y.; Okamoto, T.; Sultana, N.; Sugiyama, M. Antibiotic susceptibility of plant-derived lactic acid bacteria conferring health benefits to human. J. Antibiot. 2019, 72, 834–842. [Google Scholar] [CrossRef] [PubMed]
- McIver, K.S.; Myles, R.L. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 2002, 43, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Pabo, C.O.; Sauer, R.T. Protein-DNA recognition. Annu. Rev. Biochem. 1984, 53, 293–321. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.W.; Leenhouts, K.; Burghoorn, J.; Brands, J.R.; Venema, G.; Kok, J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 1998, 27, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Gut, H.; Pennacchietti, E.; John, R.A.; Bossa, F.; Capitani, G.; De Biase, D.; Grütter, M.G. Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. EMBO J. 2006, 25, 2643–2651. [Google Scholar] [CrossRef]
- Noda, M.; Miyauchi, R.; Danshiitsoodol, N.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Expression of genes involved in bacteriocin production and self-resistance in Lactobacillus brevis 174A is mediated by two regulatory proteins. Appl. Environ. Microbiol. 2018, 84, e02707-17. [Google Scholar] [CrossRef]
- Bachmann, B.J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 1972, 36, 525–557. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Jeon, H.J.; Noda, M.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Crystal structure and mutagenic analysis of a bacteriocin immunity protein, Mun-im. Biochem. Biophys. Res. Commun. 2009, 378, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, W.B. Enzymes of γ-aminobutyrate metabolism (bacterial). Methods Enzymol. 1962, 5, 765–778. [Google Scholar]
- Wilkinson, G.N. Statistical estimations in enzyme kinetics. Biochem. J. 1961, 80, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Duggleby, R.G. A nonlinear regression program for small computers. Anal. Biochem. 1981, 110, 9–18. [Google Scholar] [CrossRef]
- Dunny, G.M.; Lee, L.N.; LeBlanc, D.J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 1991, 57, 1194–1201. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.; Collins, M.A. Improvement in electroporation efficiency for Lactobacillus plantarum by the inclusion of high concentrations of glycine in the growth medium. J. Microbiol. Methods 1996, 26, 73–79. [Google Scholar] [CrossRef]
- Ma, Z.; Richard, H.; Tucker, D.L.; Conway, T.; Foster, J.W. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 2002, 184, 7001–7012. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, H.; Lin, Z.; Xu, P. Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol. Adv. 2015, 33, 1484–1492. [Google Scholar] [CrossRef]
- Shabayek, S.; Spellerberg, B. Acid stress response mechanisms of group B Streptococci. Front. Cell. Infect. Microbiol. 2017, 7, 395. [Google Scholar] [CrossRef]
- He, A.; Penix, S.R.; Basting, P.J.; Griffith, J.M.; Creamer, K.E.; Camperchioli, D.; Clark, M.W.; Gonzales, A.S.; Chávez Erazo, J.S.; George, N.S.; et al. Acid evolution of Escherichia coli K-12 eliminates amino acid decarboxylases and reregulates catabolism. Appl. Environ. Microbiol. 2017, 83, e00442-17. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Ladero, V.; Alvarez, M.A.; Lucas, P.M. Putrescine production via the ornithine decarboxylation pathway improves the acid stress survival of Lactobacillus brevis and is part of a horizontally transferred acid resistance locus. Int. J. Food Microbiol. 2014, 175, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Ren, C.; Xu, Y. Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb. Cell Fact. 2019, 18, 108. [Google Scholar] [CrossRef] [PubMed]
- van de Guchte, M.; Serror, P.; Chervaux, C.; Smokvina, T.; Ehrlich, S.D.; Maguin, E. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 2002, 82, 187–216. [Google Scholar] [CrossRef]
- Wang, S.; Meng, S.; Yang, X.; Zhang, B.; Zhang, J.H.; Bai, Z.K.; Song, M.X. The mechanism of acid resistance by ornithine decarboxylase in Trichinella spiralis. Exp. Parasitol. 2023, 249, 108503. [Google Scholar] [CrossRef]
- Richard, H.; Foster, J.W. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 2004, 186, 6032–6041. [Google Scholar] [CrossRef]
- Small, P.L. Shigella and Escherichia coli strategies for survival at low pH. Jpn. J. Med. Sci. Biol. 1998, 51, S81–S89. [Google Scholar] [CrossRef]
- Tan, Q.; Gou, L.; Fan, T.P.; Cai, Y. Enzymatic properties of ornithine decarboxylase from Clostridium aceticum DSM1496. Biotechnol. Appl. Biochem. 2024, 71, 525–535. [Google Scholar] [CrossRef]
- Bao, X.; Wang, F.; Yang, R.; Zhang, Y.; Fu, L.; Wang, Y. Ornithine decarboxylation system of Shewanella baltica regulates putrescine production and acid resistance. J. Food Prot. 2021, 84, 303–309. [Google Scholar] [CrossRef]
- Condon, S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 1987, 46, 269–280. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Fogg, G.C.; Gibson, C.M.; Caparon, M.G. The identification of rofA, a positive-acting regulatory component of prtF expression: Use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 1994, 11, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; McAuliffe, O.; O’Byrne, C.P. A novel RofA-family transcriptional regulator, GadR, controls the development of acid resistance in Listeria monocytogenes. mBio 2023, 14, e0171623. [Google Scholar] [CrossRef] [PubMed]
- Dramsi, S.; Dubrac, S.; Konto-Ghiorghi, Y.; Da Cunha, V.; Couvé, E.; Glaser, P.; Caliot, E.; Débarbouillé, M.; Bellais, S.; Trieu-Cuot, P.; et al. Rga, a RofA-like regulator, is the major transcriptional activator of the PI-2a pilus in Streptococcus agalactiae. Microb. Drug Resist. 2012, 18, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhao, J.; Zhang, R.; Yu, R.; Guo, T.; Kong, J. Molecular analysis of glutamate decarboxylases in Enterococcus avium. Front. Microbiol. 2021, 12, 691968. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Galloway-Peña, J.R.; Sillanpaa, J.; Roh, J.H.; Nallapareddy, S.R.; Chowdhury, S.; Bourgogne, A.; Choudhury, T.; Muzny, D.M.; Buhay, C.J.; et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.W.; Venema, G.; Kok, J.; Leenhouts, K. Identification of a sodium chloride-regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 1998, 257, 681–685. [Google Scholar] [CrossRef][Green Version]
- Laroute, V.; Aubry, N.; Audonnet, M.; Mercier-Bonin, M.; Daveran-Mingot, M.L.; Cocaign-Bousquet, M. Natural diversity of lactococci in γ-aminobutyric acid (GABA) production and genetic and phenotypic determinants. Microb. Cell Fact. 2023, 22, 178. [Google Scholar] [CrossRef]
- Zhang, B.; Ran, L.; Wu, M.; Li, Z.; Jiang, J.; Wang, Z.; Cheng, S.; Fu, J.; Liu, X. Shigella flexneri regulator SlyA controls bacterial acid resistance by directly activating the glutamate decarboxylation system. Front. Microbiol. 2018, 9, 2071. [Google Scholar]
- Castanie-Cornet, M.P.; Foster, J.W. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 2001, 147, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Tun, H.M.; Law, Y.S.; Khafipour, E.; Shah, N.P. Common distribution of gad operon in Lactobacillus brevis and its GadA contributes to efficient GABA synthesis toward cytosolic near-neutral pH. Front. Microbiol. 2017, 8, 206. [Google Scholar] [CrossRef] [PubMed]



| Bacterial Strain/Plasmids | Description | Resource/References | |
|---|---|---|---|
| E. coli | K12 W3110 | Used as a template for PCR to amplify the GAD. | [19] |
| DH5α | Used in construction of plasmid. | [20] | |
| BL21(DE3) pLysS | Used as protein over-expressing strain. | Promega | |
| HST04 (dam− and dcm−) | Used in preparation of methylation free plasmid. | Takara Bio Inc. | |
| E. avium | G-15 | GABA hyper-producing strain. | [12] |
| E. hirae | IFO 3128 | Used as a recipient strain for LAB transformation. | NBRC |
| E. mundtii | 15-1A | Used as a template for PCR to amplify the Pmun. | [21] |
| Plasmids | pUC19 | E. coli cloning vector. | [22] |
| pGEM-T | E. coli TA-cloning vector. | Promega | |
| pET-21a(+) | E. coli over-expression vector. | Merck | |
| pLES003-b | E. coli–LAB shuttle vector. | [18] | |
| Purpose | Name | Sequence (5′→3′) | Restriction Site | Target |
|---|---|---|---|---|
| Protein expression | Ex-gadA-F | CATATGGACCAGAAGCTGTTAACGGAT | NdeI | gadA (E. coli K12 W3110) |
| Ex-gadA-R | CTCGAGGGTGTGTTTAAAGCTGTTCTGC | XhoI | gadA (E. coli K12 W3110) | |
| Ex-gadB-F | CATATGGATAAGAAGCAAGTAACGGATT | NdeI | gadB (E. coli K12 W3110) | |
| Ex-gadB-R | CTCGAGGGTATGTTTAAAGCTGTTCTGT | XhoI | gadB (E. coli K12 W3110) | |
| Ex-gadG-F | CATATGTTATATGGAAAAGAAAATCGTGA | NdeI | gadG (E. avium G-15) | |
| Ex-gadG-R | CTCGAGATGCGTAAATCCGTAAGTTTTG | XhoI | gadG (E. avium G-15) | |
| EMSA | EMSA-gadT-F | Biotin-GTAAAACAAAAAGGTTCTAGTTTT CTTCTG | upstream of gadTG | |
| EMSA-gadT-R | AGATACGCTTCTTTCCTTTTCTTTAAT | upstream of gadTG | ||
| Modified gad cluster construction | N-gadTG-F | GCCCCCGGGCTACATTTTTTTTATTTTTCT | SmaI | gadTG (E. avium G-15) |
| N-gadTG-R | CGTAAGAGCTCTCATCGTGTGTGAC | SacI | gadTG (E. avium G-15) | |
| N-gadR1-F | GCTCTAGACAACAAGCGGAGGATCAA | XbaI | gadR1 (E. avium G-15) | |
| N-gadR1-R | TCCCCCGGGGCACCTCTTTCCTTACAA | SmaI | gadR1 (E. avium G-15) | |
| gadTG-gadR2-F | GCAGGTCGACTCTAGACTTCTGATTCAGA AGGAGCTA | XbaI | gadTG and gadR2 (E. avium G-15) | |
| gadTG-gadR2-R | CGGCCAGTGAATTCGAGCTCAAGTTTCTC CCTTCAAACTCT | SacI | gadTG and gadR2 (E. avium G-15) | |
| P-gadR1-F | CGTCTAGATAGTTCTTATGTCTTCTTGC | XbaI | Pmun (E. mundtii 15-1A) | |
| P-gadR1-R | CGGGATCCAACCTTCTCTCATTTTTTGAACA | BamHI | Pmun (E. mundtii 15-1A) | |
| gadR1-F | CGCGGATCCATGAAAAAAGTAATCATTTT GAC | BamHI | gadR1 (E. avium G-15) | |
| gadTG-F | CGCGGTACCATGAATCAGAAAAAATTATC | KpnI | gadTG (E. avium G-15) | |
| P-gadTG-F | AATCCCGGGTAGTTCTTATGTCTTCTTGC | SmaI | Pmun (E. mundtii 15-1A) | |
| P-gadTG-R | ACTGGTACCAACCTTCTCTCATTTTTTGAAC | KpnI | Pmun (E. mundtii 15-1A) |
| by Michaelis–Menten Equation | by Hanes–Woolf Linearization | |||||
|---|---|---|---|---|---|---|
| Km (mM) | kcat (min−1) | kcat/Km | Km (mM) | kcat (min−1) | kcat/Km | |
| GadA | 5.1 | 623 | 122 | 5.1 | 637 | 125 |
| GadB | 3.2 | 530 | 168 | 3.1 | 527 | 171 |
| GadG | 7.8 | 847 | 109 | 6.6 | 839 | 127 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noda, M.; Ozaki, M.; Ogura, S.; Danshiitsoodol, N.; Nakashima, E.; Sugiyama, M. Characterization of the Gamma-Aminobutyric Acid (GABA) Biosynthetic Gene Cluster in High GABA-Producing Enterococcus avium G-15. Fermentation 2024, 10, 379. https://doi.org/10.3390/fermentation10080379
Noda M, Ozaki M, Ogura S, Danshiitsoodol N, Nakashima E, Sugiyama M. Characterization of the Gamma-Aminobutyric Acid (GABA) Biosynthetic Gene Cluster in High GABA-Producing Enterococcus avium G-15. Fermentation. 2024; 10(8):379. https://doi.org/10.3390/fermentation10080379
Chicago/Turabian StyleNoda, Masafumi, Moeko Ozaki, Saori Ogura, Narandalai Danshiitsoodol, Etsuji Nakashima, and Masanori Sugiyama. 2024. "Characterization of the Gamma-Aminobutyric Acid (GABA) Biosynthetic Gene Cluster in High GABA-Producing Enterococcus avium G-15" Fermentation 10, no. 8: 379. https://doi.org/10.3390/fermentation10080379
APA StyleNoda, M., Ozaki, M., Ogura, S., Danshiitsoodol, N., Nakashima, E., & Sugiyama, M. (2024). Characterization of the Gamma-Aminobutyric Acid (GABA) Biosynthetic Gene Cluster in High GABA-Producing Enterococcus avium G-15. Fermentation, 10(8), 379. https://doi.org/10.3390/fermentation10080379






