Transcriptome Analysis of Sake Yeast in Co-Culture with kuratsuki Kocuria
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture of Sake Yeast and kuratsuki Kocuria
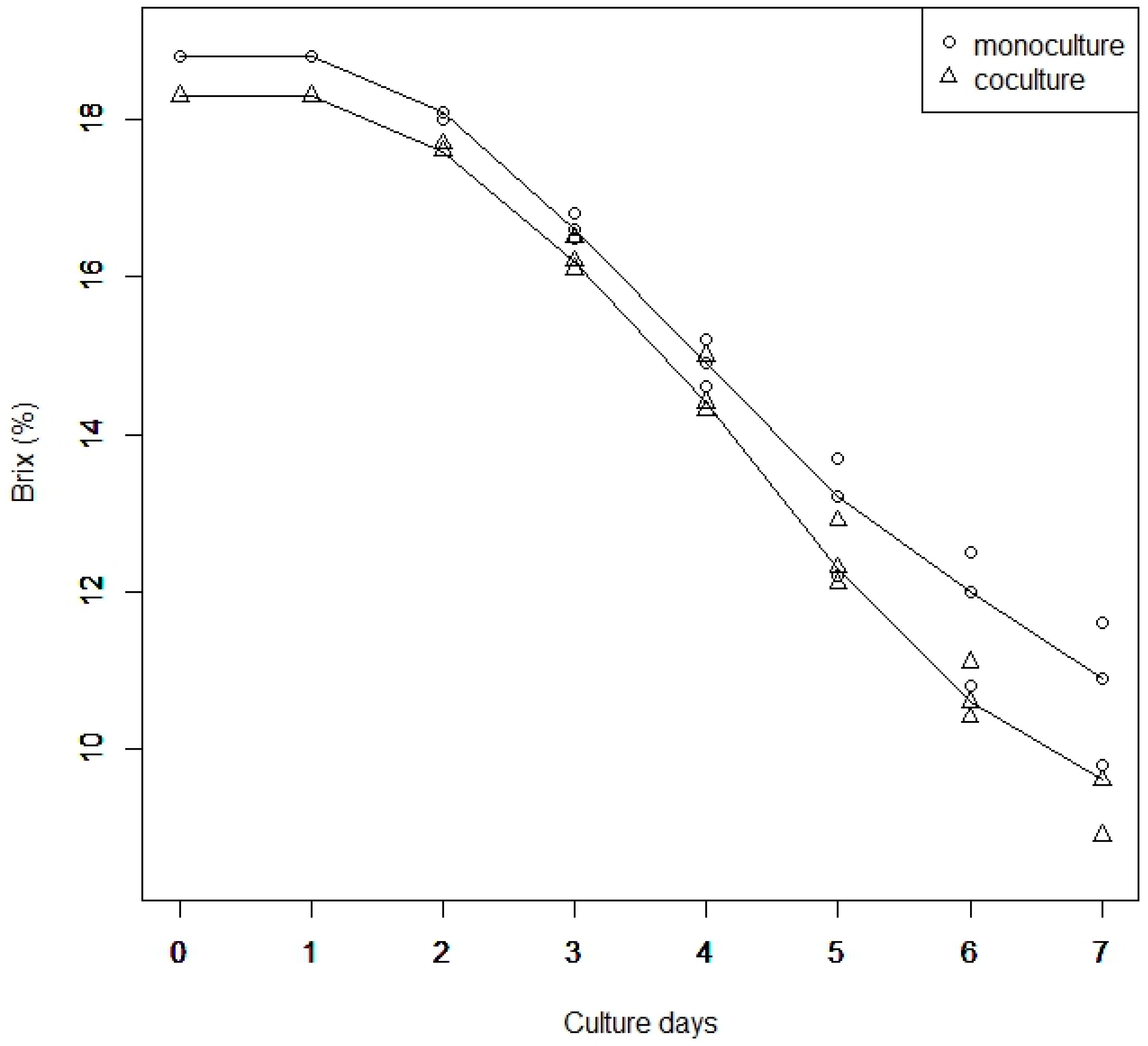
2.2. Measurement of Brix
2.3. RNA Isolation and Sequencing
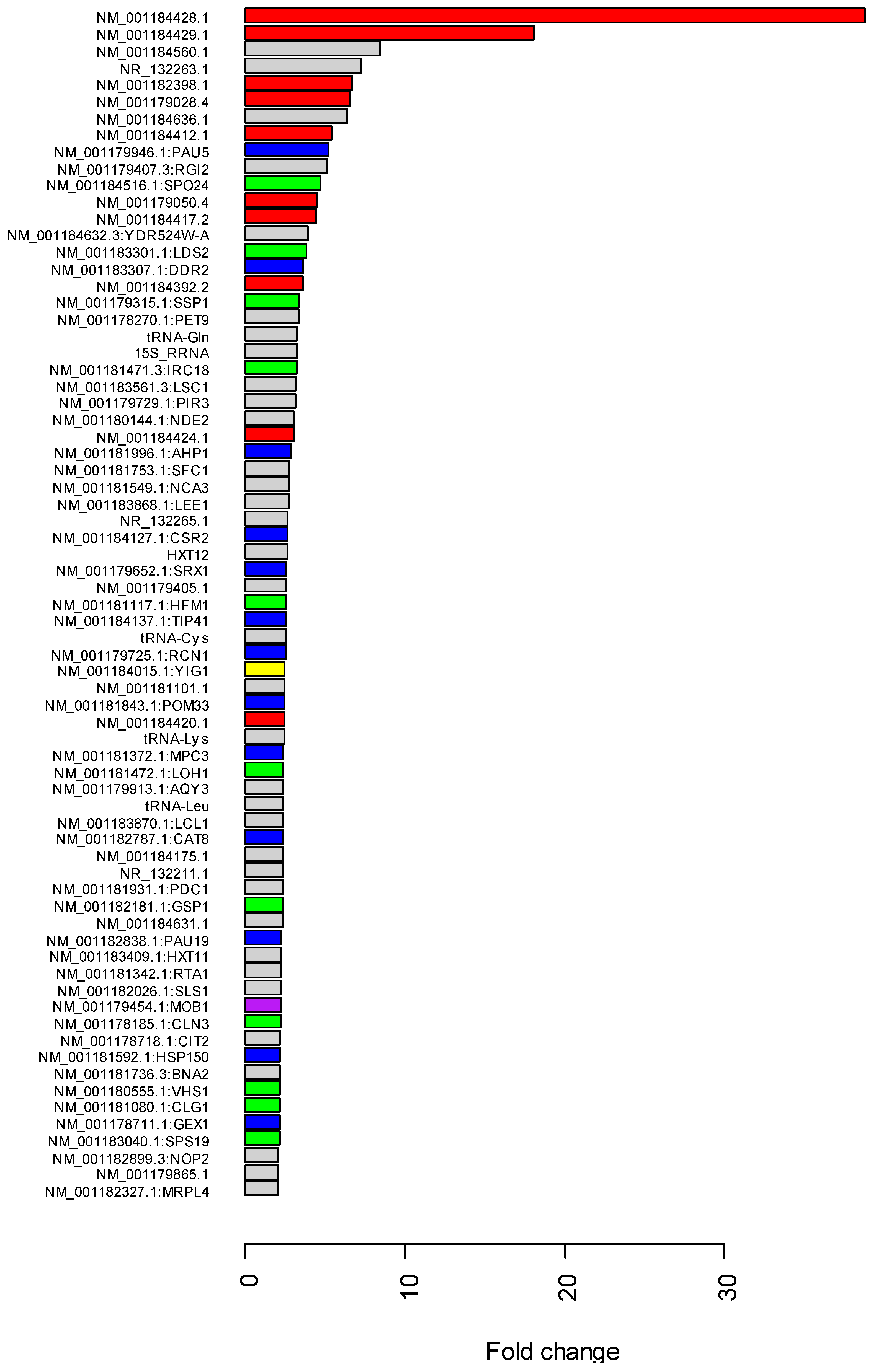
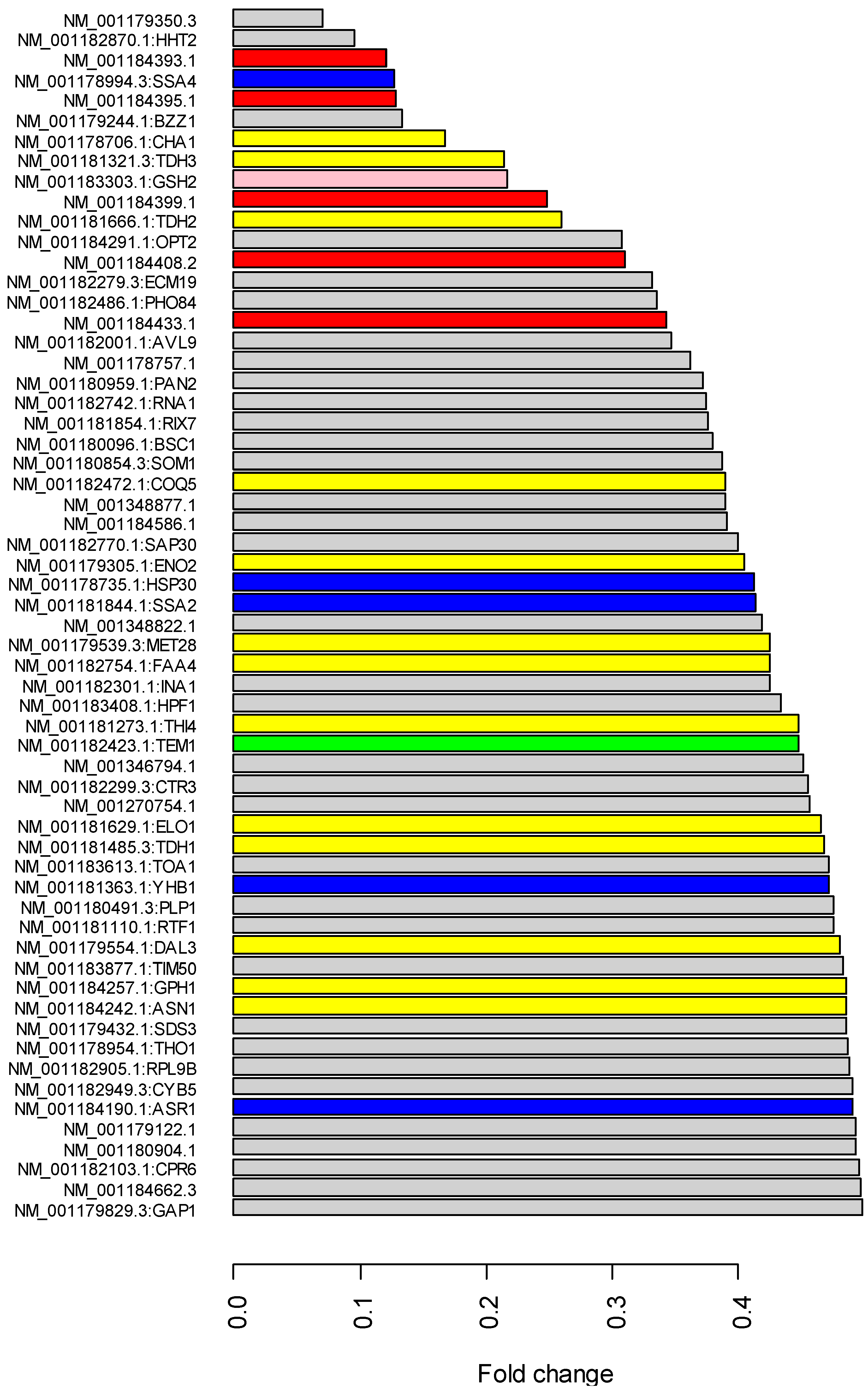
2.4. Gene Expression Comparison
3. Results and Discussion
3.1. Brix of Culture
3.2. RNA Mapping and Gene Expression
3.3. Gene Expression Changes of Retrotransposons and Small Nuclear RNAs in Co-Culture with the kuratsuki Kocuria
3.4. Expression Changes of Stress-Related Genes in Co-Culture with the kuratsuki Kocuria
3.5. Expression Changes of Cell Cycle-Related Genes in Co-Culture with the kuratsuki Kocuria
3.6. Downregulation of Metabolic Genes in Co-Culture with kuratsuki Kocuria
3.7. Expression Changes of Transporter Genes and Mitochondria-Related Genes in Co-Culture with the kuratsuki Kocuria
3.8. Comparison of Gene Expression Changes in Co-Culture with the kuratsuki Kocuria and with Lactic Acid Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonçalves, M.; Pontes, A.; Almeida, P.; Barbosa, R.; Serra, M.; Libkind, D.; Hutzler, M.; Gonçalves, P.; Sampaio, J.P. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 2016, 26, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Kitagaki, H.; Kitamoto, K. Breeding research on sake yeasts in Japan: History, recent technological advances, and future perspectives. Ann. Rev. Food Sci. Technol. 2013, 4, 215–235. [Google Scholar] [CrossRef]
- Akaike, M.; Miyagawa, H.; Kimura, Y.; Terasaki, M.; Kusaba, Y.; Kitagaki, H.; Nishida, H. Chemical and bacterial components in sake and sake production process. Curr. Microbiol. 2020, 77, 632–637. [Google Scholar] [CrossRef]
- Suto, M.; Kawashima, H. Discrimination for sake brewing methods by compound specific isotope analysis and formation mechanism of organic acids in sake. Food Chem. 2022, 381, 132295. [Google Scholar] [CrossRef]
- Maruyama, H.; Yamamiya, T.; Ozawa, A.; Yamazaki, E.; Suzuki, N. Beer brewed with sake yeast strain has unique sake-like flavors. J. Am. Soc. Brew. Chem. 2024, 82, 150–159. [Google Scholar] [CrossRef]
- Yoshimoto, H.; Bogaki, T. Mechanisms of production and control of acetate esters in yeasts. J. Biosci. Bioeng. 2023, 136, 261–269. [Google Scholar] [CrossRef]
- Ohya, Y.; Kashima, M. History, lineage and phenotypic differentiation of sake yeast. Biosci. Biotechnol. Biochem. 2019, 83, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Kumano, M.; Sugimoto, Y.; Ito, M.; Ohashi, M.; Sunada, K.; Takahashi, T.; Yamada, T.; Takagi, H. Metabolic switching of sake yeast by kimoto lactic acid bacteria through the [GAR+] non-genetic element. J. Biosci. Bioeng. 2018, 126, 624–629. [Google Scholar] [CrossRef]
- Watanabe, D.; Takagi, H. Yeast prion-based metabolic reprogramming induced by bacteria in fermented foods. FEMS Yeast Res. 2019, 19, foz061. [Google Scholar] [CrossRef]
- Chen, C.; Xiong, Y.; Xie, Y.; Zhang, H.; Jiang, K.; Pang, X.-N.; Huang, M. Metabolic characteristics of lactic acid bacteria and interaction with yeast isolated from light-flavor Baijiu fermentation. Food Biosci. 2022, 50, 102102. [Google Scholar] [CrossRef]
- Senne de Oliveira Lino, F.; Bajic, D.; Vila, J.C.C.; Sánchez, A.; Sommer, M.O.A. Complex yeast-bacteria interactions affect the yield of industrial ethanol fermentation. Nat. Commun. 2021, 12, 1498. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H. Sake brewing and bacteria inhabiting sake breweries. Front. Microbiol. 2021, 12, 602380. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H. Kuratsuki bacteria and sake making. Biosci. Biotechnol. Biochem. 2024, 88, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, M.; Inoue, A.; Kanamoto, E.; Yoshida, S.; Yamada, M.; Toda, H.; Nishida, H. Co-cultivation of sake yeast and Kocuria isolates from the sake brewing process. FEMS Microbiol. Lett. 2021, 368, fnab053. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, M.; Kimura, Y.; Yamada, M.; Nishida, H. Genomic information of Kocuria isolates from sake brewing process. AIMS Microbiol. 2021, 7, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Kanamoto, E.; Terashima, K.; Shiraki, Y.; Nishida, H. Diversity of Bacillus isolates from the sake brewing process at a sake brewery. Microorganisms 2021, 9, 1760. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, M.; Nishida, H. Bacterial DNA diversity among clear and cloudy sakes, and sake-kasu. Open Bioinform. J. 2020, 13, 74–82. [Google Scholar] [CrossRef]
- Yazaki, A.; Nishida, H. Effect of kuratsuki Kocuria on sake brewing in different koji conditions. FEMS Microbiol. Lett. 2023, 370, fnad020. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, A.; Nishida, H. Effect of kuratsuki Kocuria on sake’s taste varies depending on the sake yeast strain used in sake brewing. Arch. Microbiol. 2023, 205, 290. [Google Scholar] [CrossRef]
- Saito, M.; Nishida, H. Molecular hydrogen treatment of sake yeast and kuratsuki bacteria affects sake taste. Fermentation 2023, 9, 516. [Google Scholar] [CrossRef]
- Kobayashi, K.; Nishida, H. Effect of kuratsuki Bacillus and Priestia on taste of sake. Appl. Microbiol. 2024, 4, 147–161. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCure, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Rolfe, M.; Spanos, A.; Banks, G. Induction of yeast Ty element transcription by ultraviolet light. Nature 1986, 319, 339–340. [Google Scholar] [CrossRef]
- Bradshaw, V.A.; McEntee, K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol. Genet. Genom. 1989, 218, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Sacerdot, C.; Mercier, G.; Todeschini, A.L.; Dutreix, M.; Springer, M.; Lesage, P. Impact of ionizing radiation on the life cycle of Saccharomyces cerevisiae Ty1 retrotransposon. Yeast 2005, 22, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, A.L.; Morillon, A.; Springer, M.; Lesage, P. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005, 25, 7459–7472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stanley, D.; Fraser, S.; Stanley, G.A.; Chambers, P.J. Retrotransposon expression in ethanol-stressed Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010, 87, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987, 6, 4169–4175. [Google Scholar] [CrossRef] [PubMed]
- de Beus, E.; Brockenbrough, J.S.; Hong, B.; Aris, J.P. Yeast NOP2 encodes an essential nuclear protein with homology to a human proliferation marker. J. Cell Biol. 1994, 127, 1799–1813. [Google Scholar] [CrossRef]
- Lee, J.; Spector, D.; Godon, C.; Labarre, J.; Toledano, M.B. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem. 1999, 274, 4537–4544. [Google Scholar] [CrossRef]
- Hedges, D.; Proft, M.; Entian, K.-D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Haurie, V.; Perrot, M.; Mini, T.; Jenö, P.; Sagliocco, F.; Boucherie, H. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Marks, V.D.; Ho Sui, S.J.; Erasmus, D.; van der Merwe, G.K.; Brumm, J.; Wasserman, W.W.; Bryan, J.; van Vuuren, H.J. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 2008, 8, 35–52. [Google Scholar] [CrossRef] [PubMed]
- McClanahan, T.; McEntee, K. DNA damage and heat shock dually regulate genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1986, 6, 90–96. [Google Scholar] [PubMed]
- Treger, J.M.; Magee, T.R.; McEntee, K. Functional analysis of the stress response element and its role in the multistress response of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1998, 243, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Dhaoui, M.; Auchère, F.; Blaiseau, P.-L.; Lesuisse, E.; Landoulsi, A.; Camadro, J.-M.; Haguenauer-Tsapis, R.; Belgareh-Touzé, N. Gex1 is a yeast glutathione exchanger that interferes with pH and redox homeostasis. Mol. Biol. Cell 2011, 22, 2054–2067. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Simonen, M.; Uimari, A.; Teesalu, T.; Makarow, M. Dual regulation by heat and nutrient stress of the yeast HPS150 gene encoding a secretory glycoprotein. Mol. Genet. Genom. 1993, 239, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Ezaki, B.; Gardner, R.C.; Ezaki, Y.; Kondo, H.; Matsumoto, H. Protective roles of two aluminum (Al)-induced genes, HSP150 and SED1 of Saccharomyces cerevisiae, in Al and oxidative stresses. FEMS Microbiol. Lett. 1998, 159, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Luca, F.C.; Mody, M.; Kurischko, C.; Roof, D.M.; Giddings, T.H.; Winey, M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 2001, 21, 6972–6983. [Google Scholar] [CrossRef]
- Hergovich, A. MOB control: Reviewing a conserved family of kinase regulators. Cell Signal. 2011, 23, 1433–1440. [Google Scholar] [CrossRef]
- Timón-Gómez, A.; Proft, M.; Pascual-Ahuir, A. Differential regulation of mitochondrial pyruvate carrier genes modulates respiratory capacity and stress tolerance in yeast. PLoS ONE 2013, 8, e79405. [Google Scholar] [CrossRef] [PubMed]
- Bender, T.; Pena, G.; Martinou, J.-C. Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. EMBO J. 2015, 34, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; van Vuuren, H.J.J. Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology 2009, 155, 4036–4049. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; van Vuuren, H.J.J. Stress-induced production, processing and stability of a seripauperin protein, Pau5p, in Saccharomyces cerevisiae. FEMS Yeast Res. 2008, 8, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Chadrin, A.; Hess, B.; Roman, M.S.; Gatti, X.; Lombard, B.; Loew, D.; Barral, Y.; Palancade, B.; Doye, V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 2010, 189, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Floch, A.G.; Tareste, D.; Fuchs, P.F.; Chadrin, A.; Naciri, I.; Léger, T.; Schlenstedt, G.; Palancade, B.; Doye, V. Nuclear pore targeting of the yeast Pom33 nucleoporin depends on karyopherin and lipid binding. J. Cell Sci. 2015, 128, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Biteau, B.; Labarre, J.; Toledano, M.B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 2003, 425, 980–984. [Google Scholar] [CrossRef]
- Jacinto, E.; Guo, B.; Arndt, K.; Schmelzle, T.; Hall, M. TIP41 interacts with TAP42 and negatively regulated the TOR signaling pathway. Mol. Cell 2001, 8, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Barker, S.L.; Boone, C.; Measday, V. Identification of RCN1 and RSA3 as ethanol-tolerant genes in Saccharomyces cerevisiae using a high copy barcoded library. FEMS Yeast Res. 2012, 12, 48–60. [Google Scholar] [CrossRef]
- Tkach, J.M.; Yimit, A.; Lee, A.Y.; Riffle, M.; Costanzo, M.; Jaschob, D.; Hendry, J.A.; Ou, J.; Moffat, J.; Boone, C.; et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012, 14, 966–976. [Google Scholar] [CrossRef]
- Bhagwat, M.; Nagar, S.; Kaur, P.; Mehta, R.; Vancurova, I.; Vancura, A. Replication stress inhibits synthesis of histone mRNAs in yeast by removing Spt10p and Spt21p from the histone promoters. J. Biol. Chem. 2021, 297, 101246. [Google Scholar] [CrossRef]
- Stolinski, L.A.; Eisenmann, D.M.; Arndt, K.M. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 4490–4500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.W.; Iratni, R.; Erdjument-Bromage, H.; Tempst, P.; Hampsey, M.; Reinberg, D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1998, 1, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Lechner, T.; Carrozza, M.J.; Yu, Y.; Grant, P.A.; Eberharter, A.; Vannier, D.; Brosch, G.; Stillman, D.J.; Shore, D.; Workman, J.L. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3-Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 2000, 275, 40961–40966. [Google Scholar] [CrossRef] [PubMed]
- Betz, C.; Schlenstedt, G.; Bailer, S.M. Asr1p, a novel yeast ring/PHD finger protein, signals alcohol stress to the nucleus. J. Biol. Chem. 2004, 279, 28174–28181. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Sugiyama, K.; Izawa, S.; Kimura, A. Molecular identification of glutathione synthetase (GSH2) gene from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1998, 1395, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Morano, K.A.; Grant, C.M.; Scott Moye-Rowley, W. The response to heat shoch and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef] [PubMed]
- Panaretou, B.; Piper, P.W. The plasma membrane of yeast acquires a novel heat-shock protein (hsp30) and displays a decline in proton-pumping ATPase levels in response to both heat shock and the entry to stationary phase. Eur. J. Biochem. 1992, 206, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Seymour, I.J.; Piper, P.W. Stress induction of HSP30, the plasma membrane heat shock protein gene of Saccharomyces cerevisiae, appears not to use known stress-regulated transcription factors. Microbiology 1999, 145, 231–239. [Google Scholar] [CrossRef]
- Werner-Washburne, M.; Stone, D.E.; Craig, E.A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 2568–2577. [Google Scholar]
- Zhao, X.J.; Raitt, D.; Burke, P.V.; Clewell, A.S.; Kwast, K.E.; Poyton, R.O. Function and expression of flavohemoglobin in Saccharomyces cerevisiae. Evidence for a role in the oxidative stress response. J. Biol. Chem. 1996, 271, 25131–25138. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kolodner, R.D. Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol. Cell. Biol. 2002, 22, 3281–3291. [Google Scholar] [CrossRef] [PubMed]
- Suda, Y.; Rodringuez, R.K.; Coluccio, A.E.; Neiman, A.M. A screen for spore wall permeability mutants identifies a secreted protease required for proper spore wall assembly. PLoS ONE 2009, 4, e7184. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.P.-C.; Kim, C.; Smith, S.O.; Neiman, A.M. A highly redundant gene network controls assembly of the outer spore wall in S. cerevisiae. PLoS Genet. 2013, 9, e1003700. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.P.; Nelson, Z.W.; Hetrick, E.D.; Gottschling, D.E. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics 2008, 179, 1179–1195. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, S.; Guisbert, K.S.K.; Sontheimer, E.J. SPO24 is a transcriptionally dynamic, small ORF-encoding locus required for efficient sporulation in Saccharomyces cerevisiae. PLoS ONE 2014, 9, e105058. [Google Scholar] [CrossRef] [PubMed]
- Gurvitz, A.; Rottensteiner, H.; Kilpeläinen, S.H.; Hartig, A.; Hiltunen, J.K.; Binder, M.; Dawes, I.W.; Hamilton, B. The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-CoA reductase is encoded by the oleate-inducible gene SPS19. J. Biol. Chem. 1997, 272, 22140–22147. [Google Scholar] [CrossRef] [PubMed]
- Nag, D.K.; Koonce, M.P.; Axelrod, J. SSP1, a gene necessary for proper completion of meiotic divisions and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 7029–7039. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Wickner, R.B. CLG1, a new cyclin-like gene of Saccharomyces cerevisiae. Yeast 1993, 9, 929–931. [Google Scholar] [CrossRef]
- Measday, V.; Moore, L.; Retnakaran, R.; Lee, J.; Donoviel, M.; Neiman, A.M.; Andrews, B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 1997, 17, 1212–1223. [Google Scholar] [CrossRef]
- Shi, L.; Tu, B.P. Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2013, 110, 7318–7323. [Google Scholar] [CrossRef]
- Belhumeur, P.; Lee, A.; Tam, R.; DiPaolo, T.; Fortin, N.; Clark, M.W. GSP1 and GSP2, genetic suppressors of the prp20-1 mutant in Saccharomyces cerevisiae: GTP-binding proteins involved in the maintenance of nuclear organization. Mol. Cell. Biol. 1993, 13, 2152–2161. [Google Scholar]
- Muñoz, I.; Simón, E.; Casals, N.; Clotet, J.; Ariño, J. Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast 2003, 20, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Shirayama, M.; Matsui, Y.; Toh-e, A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 1994, 14, 7476–7482. [Google Scholar] [PubMed]
- Colomina, N.; Garí, E.; Gallego, C.; Herrero, E.; Aldea, M. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 1999, 18, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, N.; Abe, K.; Koshika, Y.; Iwano, K. Cln3 blocks IME1 transcription and the Ime1-Ume6 interaction to cause the sporulation incompetence in a sake yeast, Kokai no. 7. J. Biosci. Bioeng. 2010, 110, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dang, V.D.; Valens, M.; Bolotin-Fukuhara, M.; Daignan-Fornier, B. Cloning of the ASN1 and ASN2 genes encoding asparagine synthetases in Saccharomyces cerevisiae: Differential regulation by the CCAAT-box-binding factor. Mol. Microbiol. 1996, 22, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.G.; Kielland-Brandt, M.C.; Nilsson-Tillgren, T.; Bornaes, C.; Holmberg, S. Molecular genetics of serine and threonine catabolism in Saccharomyces cerevisiae. Genetics 1988, 119, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Barkovich, R.J.; Shtanko, A.; Shepherd, J.A.; Lee, P.T.; Myles, D.C.; Tzagoloff, A.; Clarke, C.F. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 1997, 272, 9182–9188. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.S.; Genbauffe, F.S.; Cooper, T.G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol. Cell. Biol. 1985, 5, 2279–2288. [Google Scholar]
- Schneiter, R.; Tatzer, V.; Gogg, G.; Leitner, E.; Kohlwein, S.D. Elo1p-dependent carboxy-terminal elongation of C14:1Δ9 to C16:1Δ11 Fatty Acids in Saccharomyces cerevisiae. J. Bacteriol. 2000, 182, 3655–3660. [Google Scholar] [CrossRef]
- McAlister, L.; Holland, M.J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J. Biol. Chem. 1982, 257, 7181–7188. [Google Scholar] [CrossRef]
- Ashrafi, K.; Farazi, T.A.; Gordon, J.I. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 1998, 273, 25864–25874. [Google Scholar] [CrossRef]
- Hwang, P.K.; Tugendreich, S.; Fletterick, R.J. Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989, 9, 1659–1666. [Google Scholar]
- Kuras, L.; Cherest, H.; Surdin-Kerjan, Y.; Thomas, D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996, 15, 2519–2529. [Google Scholar] [CrossRef]
- McAlister, L.; Holland, M.J. Isolation and characterization of yeast strains carrying mutations in the glyceraldehyde-3-phosphate dehydrogenase genes. J. Biol. Chem. 1985, 260, 15013–15018. [Google Scholar] [CrossRef]
- Stanley, D.; Chambers, P.J.; Stanley, G.A.; Borneman, A.; Fraser, S. Transcriptional changes associated with ethanol tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010, 88, 231–239. [Google Scholar] [CrossRef]
- Praekelt, U.M.; Byrne, K.L.; Meacock, P.A. Regulation of THI4 (MOL1), a thiamine-biosynthetic gene of Saccharomyces cerevisiae. Yeast 1994, 10, 481–490. [Google Scholar] [CrossRef]
- Nourani, A.; Wesolowski-Louvel, M.; Delaveau, T.; Jacq, C.; Delahodde, A. Multiple-drug-resistance phenomenon in the yeast Saccharomyces cerevisiae: Involvement of two hexose transporters. Mol. Cell. Biol. 1997, 17, 5453–5460. [Google Scholar] [CrossRef] [PubMed]
- Pena, M.M.; Puig, S.; Thiele, D.J. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 2000, 275, 33244–33251. [Google Scholar] [CrossRef] [PubMed]
- Wiles, A.M.; Cai, H.; Naider, F.; Becker, J.M. Nutrient regulation of oligopeptide transport in Saccharomyces cerevisiae. Microbiology 2006, 152, 3133–3145. [Google Scholar] [CrossRef]
- Bun-Ya, M.; Nishimura, M.; Harashima, S.; Oshima, Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 1991, 11, 3229–3238. [Google Scholar] [CrossRef]
- Kim, K.-S.; Rosenkrantz, M.S.; Guarente, L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol. Cell. Biol. 1986, 6, 1936–1942. [Google Scholar]
- Przybyla-Zawislak, B.; Dennis, R.A.; Zakharkin, S.O.; McCammon, M.T. Genes of succinyl-CoA ligase from Saccharomyces cerevisiae. Eur. J. Biochem. 1998, 258, 736–743. [Google Scholar] [CrossRef]
- Graack, H.R.; Grohmann, L.; Kitakawa, M.; Goldschmidt-Reisin, S. Gene MRP-L4, encoding mitochondrial ribosomal protein YmL4, is indispensable for proper non-respiratory cell functions in yeast. Gene 1995, 152, 107–112. [Google Scholar] [CrossRef]
- Pélissier, P.; Camougrand, N.; Velours, G.; Guérin, M. NCA3, a nuclear gene involved in the mitochondrial expression of subunits 6 and 8 of the Fo-F1 ATP synthase of S. cerevisiae. Curr. Genet. 1995, 27, 409–416. [Google Scholar] [CrossRef]
- Luttik, M.A.H.; Overkamp, K.M.; Kötter, P.; de Vries, S.; van Dijken, J.P.; Pronk, J.T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 1998, 273, 24529–24534. [Google Scholar] [CrossRef]
- Adrian, G.S.; McCammon, M.T.; Montgomery, D.L.; Douglas, M.G. Sequences required for delivery and localization of the ADP/ATP translocator to the mitochondrial inner membrane. Mol. Cell. Biol. 1986, 6, 626–634. [Google Scholar]
- Palmieri, L.; Lasorsa, F.M.; Vozza, A.; Agrimi, G.; Fiermonte, G.; Runswick, M.J.; Walker, J.E.; Palmieri, F. Identification and functions of new transporters in yeast mitochondria. Biochim. Biophys. Acta 2000, 1459, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.C.; Rodeheffer, M.S.; Wearn, C.M.; Shadel, G.S. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics 2002, 160, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Esser, K.; Pratje, E.; Michaelis, G. SOM1, a small new gene required for mitochondrial inner membrane peptidase function in Saccharomyces cerevisiae. Mol. Genet. Genom. 1996, 252, 437–445. [Google Scholar]
- Geissler, A.; Chacinska, A.; Truscott, K.N.; Wiedemann, N.; Brandner, K.; Sickmann, A.; Meyer, H.E.; Meisinger, C.; Pfanner, N.; Rehling, P. The mitochondrial presequence translocase: An essential role of Tim50 in directing preproteins to the import channel. Cell 2002, 111, 507–518. [Google Scholar] [CrossRef]
- Kellermann, E.; Seeboth, P.G.; Hollenberg, C.P. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res. 1986, 14, 8963–8977. [Google Scholar] [CrossRef]
- Mendes, F.; Sieuwerts, S.; de Hulster, E.; Almering, M.J.H.; Luttik, M.A.H.; Pronk, J.T.; Smid, E.J.; Bron, P.A.; Daran-Lapujade, P. Transcriptome-based characterization of interactions between Saccharomyces cerevisiae and Lactobacillus delbrueckii subsp. bulgaricus in lactose-grown chemostat cocultures. Appl. Environ. Microbiol. 2013, 79, 5949–5961. [Google Scholar] [CrossRef]
- Fu, D.; Beeler, T.J.; Dunn, T.M. Sequence, mapping and disruption of CCC2, a gene that cross-complements the Ca2+-sensitive phenotype of csg1 mutants and encodes a P-type ATPase belonging to the Cu2+-ATPase subfamily. Yeast 1995, 11, 283–292. [Google Scholar] [CrossRef]
- Delling, U.; Raymond, M.; Schurr, E. Identification of Saccharomyces cerevisiae genes conferring resistance to quinoline ring-containing antimalarial drugs. Antimicrob. Agents Chemother. 1998, 42, 1034–1041. [Google Scholar] [CrossRef]
- Mendizabal, I.; Rios, G.; Mulet, J.M.; Serrano, R.; de Larrinoa, I.F. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 1998, 425, 323–328. [Google Scholar] [CrossRef]
- Dancis, A.; Haile, D.; Yuan, D.S.; Klausner, R.D. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 1994, 269, 25660–25667. [Google Scholar] [CrossRef]
- Saerens, S.M.G.; Verstrepen, K.J.; Van Laere, S.D.M.; Voet, A.R.D.; Dan Dijck, P.; Delvaux, F.R.; Thevelein, J.M. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 2006, 281, 4446–4456. [Google Scholar] [CrossRef]
- Protchenko, O.; Ferea, T.; Rashford, J.; Tiedeman, J.; Brown, P.O.; Botstein, D.; Philpott, C.C. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 49244–49250. [Google Scholar] [CrossRef]
- van Bakel, H.; Strengman, E.; Wijmenga, C.; Holstege, F.C.P. Gene expression profiling and phenotype analyses of S. cerevisiae in response to changing copper revels six genes with new roles in copper and iron metabolism. Physiol. Genom. 2005, 22, 356–367. [Google Scholar] [CrossRef]
- Protchenko, O.; Philpott, C.C. Regulation of intracellular heme levels by HMX1, a homologue of hem oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 36582–36587. [Google Scholar] [CrossRef]
- Lesuisse, E.; Simon-Casteras, M.; Labbe, P. Siderophore-mediated iron uptake in Saccharomyces cerevisiae: The SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 1998, 144, 3455–3462. [Google Scholar] [CrossRef]
- Ferreira, C.; van Voorst, F.; Martins, A.; Neves, L.; Oliveira, R.; Kielland-Brandt, M.C.; Lucas, C.; Brandt, A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 2068–2076. [Google Scholar] [CrossRef]
- Martínez-Pastor, M.; Vergara, S.V.; Puig, S.; Thiele, D.J. Negative feedback regulation of the yeast CTH1 and CTH2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation. Mol. Cell. Biol. 2013, 33, 2178–2187. [Google Scholar] [CrossRef]
- Russell, D.W.; Smith, M.; Williamson, V.M.; Young, E.T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J. Biol. Chem. 1983, 258, 2674–2682. [Google Scholar] [CrossRef]
- Guaragnella, N.; Butow, R.A. ATO3 encoding a putative outward ammonium transporter is an RTG-independent retrograde responsive gene regulated by GCN4 and the Ssy1-Ptr3-Ssy5 amino acid sensor system. J. Biol. Chem. 2003, 278, 45882–45887. [Google Scholar] [CrossRef]
- Deschamps, J.; Wiame, J.M. Mating-type effect on cis mutations leading to constitutivity of ornithine transaminase in diploid cells of Saccharomyces cerevisiae. Genetics 1979, 92, 749–758. [Google Scholar] [CrossRef]
- Reddy, V.S.; Singh, A.K.; Rajasekharan, R. The Saccharomyces cerevisiae PHM8 gene encodes a soluble magnesium-dependent lysophosphatidic acid phosphatase. J. Biol. Chem. 2008, 283, 8846–8854. [Google Scholar] [CrossRef]
- Smith, F.W.; Hawkesford, M.J.; Prosser, I.M.; Clarkson, D.T. Isolation of a cDNA from Saccharomyces cerevisiae that encodes a high affinity sulphate transporter at the plasma membrane. Mol. Genet. Genom. 1995, 247, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Mai, B.; Breeden, L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol. 1997, 17, 6491–6501. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, T.-Y.; Liu, G.; Ye, Y.; Soteyome, T.; Seneviratne, G.; Xiao, G.; Xu, Z.; Kjellerup, B.V. Microbial interaction between Lactiplantibacillus plantarum and Saccharomyces cerevisiae: Transcriptome level mechanism of cell-cell antagonism. Microbiol. Spectr. 2022, 10, e0143322. [Google Scholar] [CrossRef] [PubMed]
- Molin, M.; Norbeck, J.; Blomberg, A. Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J. Biol. Chem. 2003, 278, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Matsuda, T.; Sugiyama, K.; Izawa, S.; Kimura, A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 27002–27009. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.F.; Vos, M.H.; Lindquist, S. Identification of SSF1, CNS1, and HCH1 as multicopy suppressors of a Saccharomyces cerevisiae Hsp90 loss-of-function mutation. Proc. Natl. Acad. Sci. USA 1999, 96, 1409–1414. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, Y.; Taulien, J.; Borkovich, K.A.; Lindquist, S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992, 11, 2357–2364. [Google Scholar] [CrossRef]
- Viswanathan, M.; Muthukumar, G.; Cong, Y.S.; Lenard, J. Seripauperins of Saccharomyces cerevisiae: A new multiple family encoding serine-poor relatives of serine-rich proteins. Gene 1994, 148, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Wolfger, H.; Mamnun, Y.M.; Kuchler, K. The yeast Pdr15p ATP-binding cassette (ABC) protein is a general stress response factor implicated in cellular detoxification. J. Biol. Chem. 2004, 279, 11593–11599. [Google Scholar] [CrossRef] [PubMed]
- Kihara, A.; Igarashi, Y. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. J. Biol. Chem. 2002, 277, 30048–30054. [Google Scholar] [CrossRef]
- Kim, B.; Lee, Y.; Choi, H.; Huh, W.-K. The trehalose-6-phosphate phosphatase Tps2 regulates ATG8 transcription and autophagy in Saccharomyces cerevisiae. Autophagy 2021, 17, 1013–1027. [Google Scholar] [CrossRef]
- Machado, A.K.; Morgan, B.A.; Merrill, G.F. Thioredoxin reductase-dependent inhibition of MCB cell cycle box activity in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 17045–17054. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Scott, J.D.; McMullen, B.; Hurwitz, M.; Krebs, E.G.; Wigler, M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 1371–1377. [Google Scholar]
- Ni, L.; Snyder, M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 2147–2170. [Google Scholar] [CrossRef] [PubMed]
- Borkovich, K.A.; Farrelly, F.W.; Finkelstein, D.B.; Taulien, J.; Lindquist, S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 1989, 9, 3919–3930. [Google Scholar] [PubMed]
- Kolodrubetz, D.; Rykowski, M.C.; Grunstein, M. Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes. Proc. Natl. Acad. Sci. USA 1982, 79, 7814–7818. [Google Scholar] [CrossRef] [PubMed]
- Sideri, T.C.; Willetts, S.A.; Avery, S.V. Methionine sulphoxide reductases protect iron-sulphur clusters from oxidative inactivation in yeast. Microbiology 2009, 155, 612–623. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khan, M.M.; Lee, S.; Couoh-Cardel, S.; Oot, R.A.; Kim, H.; Wilkens, S.; Roh, S.-H. Oxidative stress protein Oxr1 promotes V-ATPase holoenzyme disassembly in catalytic activity-independent manner. EMBO J. 2022, 41, e109360. [Google Scholar] [CrossRef] [PubMed]
- Nittis, T.; George, G.N.; Winge, D.R. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 2001, 276, 42520–42526. [Google Scholar]
- Muller, E.G. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem. 1991, 266, 9194–9202. [Google Scholar] [CrossRef]
| Genbank ID | Gene | Product | Length (nt) | No. of Mapped Reads | RPKM | Ratio | ||
|---|---|---|---|---|---|---|---|---|
| Monoculture | Coculture | Mono | Co | Co/Mono | ||||
| NM_001182870.1 | HHT2 | histone H3 | 411 | 36894 | 4354 | 3749.46 | 358.96 | 0.096 |
| NM_001178358.1 | HHT1 | histone H3 | 411 | 2419 | 1624 | 245.84 | 133.89 | 0.545 |
| NM_001178243.1 | HTA2 | histone H2A | 399 | 2201 | 1517 | 230.41 | 128.83 | 0.559 |
| NM_001178357.1 | HHF1 | histone H4 | 312 | 1616 | 1169 | 216.34 | 126.96 | 0.587 |
| NM_001180533.3 | HTA1 | histone H2A | 399 | 4178 | 3769 | 437.37 | 320.08 | 0.732 |
| NM_001178242.1 | HTB2 | histone H2B | 396 | 829 | 749 | 87.44 | 64.09 | 0.733 |
| NM_001182869.1 | HHF2 | histone H4 | 312 | 3556 | 3677 | 476.06 | 399.34 | 0.839 |
| NM_001183941.1 | HHO1 | histone H1 | 777 | 936 | 1194 | 50.32 | 52.07 | 1.035 |
| NM_001183266.1 | HTZ1 | histone H2AZ | 405 | 445 | 588 | 45.89 | 49.20 | 1.072 |
| NM_001180532.3 | HTB1 | histone H2B | 396 | 600 | 855 | 63.29 | 73.16 | 1.156 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, K.; Nishida, H. Transcriptome Analysis of Sake Yeast in Co-Culture with kuratsuki Kocuria. Fermentation 2024, 10, 249. https://doi.org/10.3390/fermentation10050249
Kobayashi K, Nishida H. Transcriptome Analysis of Sake Yeast in Co-Culture with kuratsuki Kocuria. Fermentation. 2024; 10(5):249. https://doi.org/10.3390/fermentation10050249
Chicago/Turabian StyleKobayashi, Karin, and Hiromi Nishida. 2024. "Transcriptome Analysis of Sake Yeast in Co-Culture with kuratsuki Kocuria" Fermentation 10, no. 5: 249. https://doi.org/10.3390/fermentation10050249
APA StyleKobayashi, K., & Nishida, H. (2024). Transcriptome Analysis of Sake Yeast in Co-Culture with kuratsuki Kocuria. Fermentation, 10(5), 249. https://doi.org/10.3390/fermentation10050249






