Functional Study of Different Lignocellulases from Trichoderma guizhouence NJAU4742 in the Synergistic Degradation of Natural Straw
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of the Experimental Materials in This Study
2.2. Cloning and Expression of EGL, BGL, and XYN
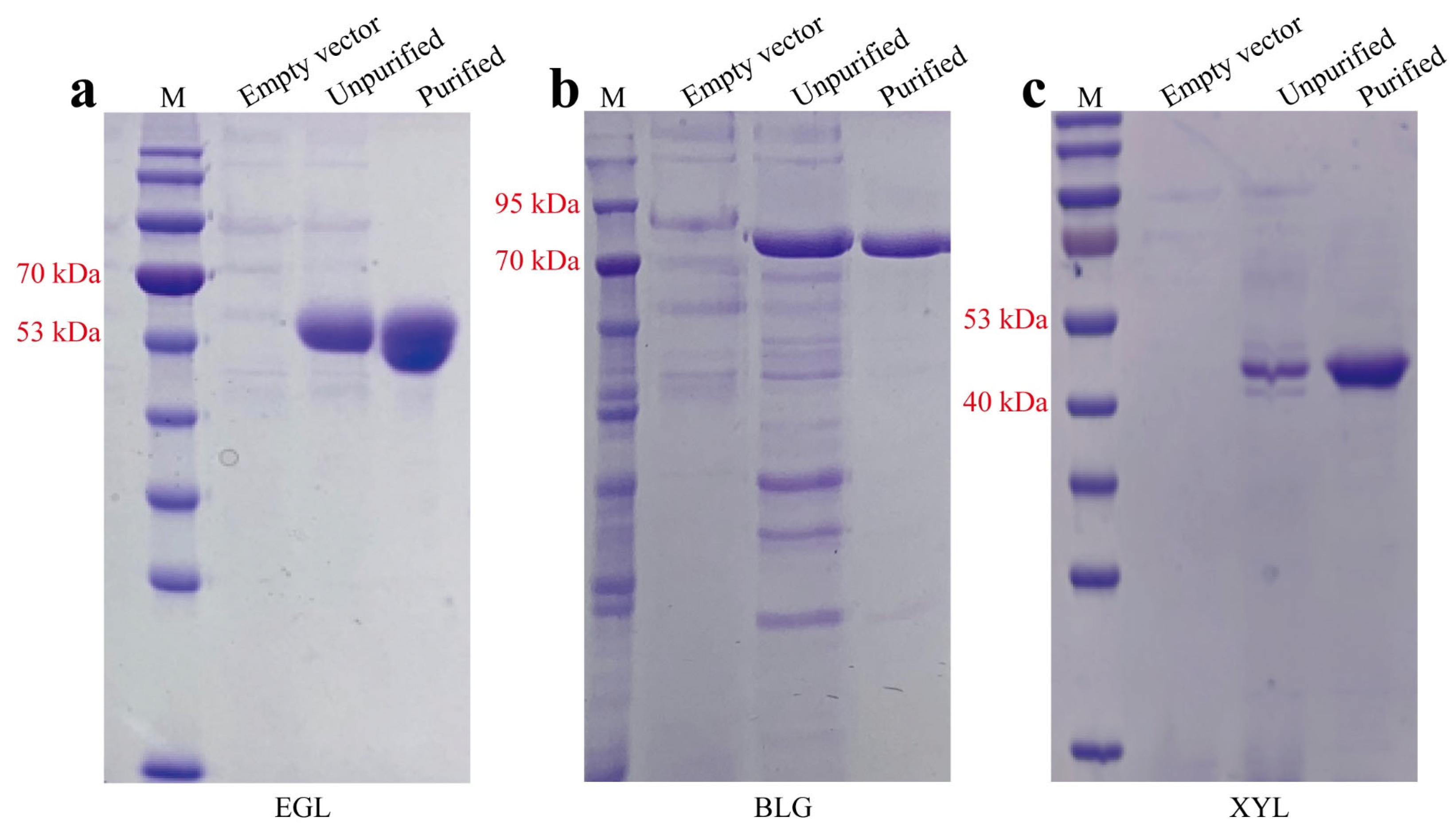
2.3. Purification of EGL, BGL, and XYN
2.4. Enzymatic Property Evaluation
2.5. Enzyme Kinetics Analysis
2.6. Investigation of Synergistic Effects between Different Enzymes
2.7. Statistical Analysis
3. Results and Discussion
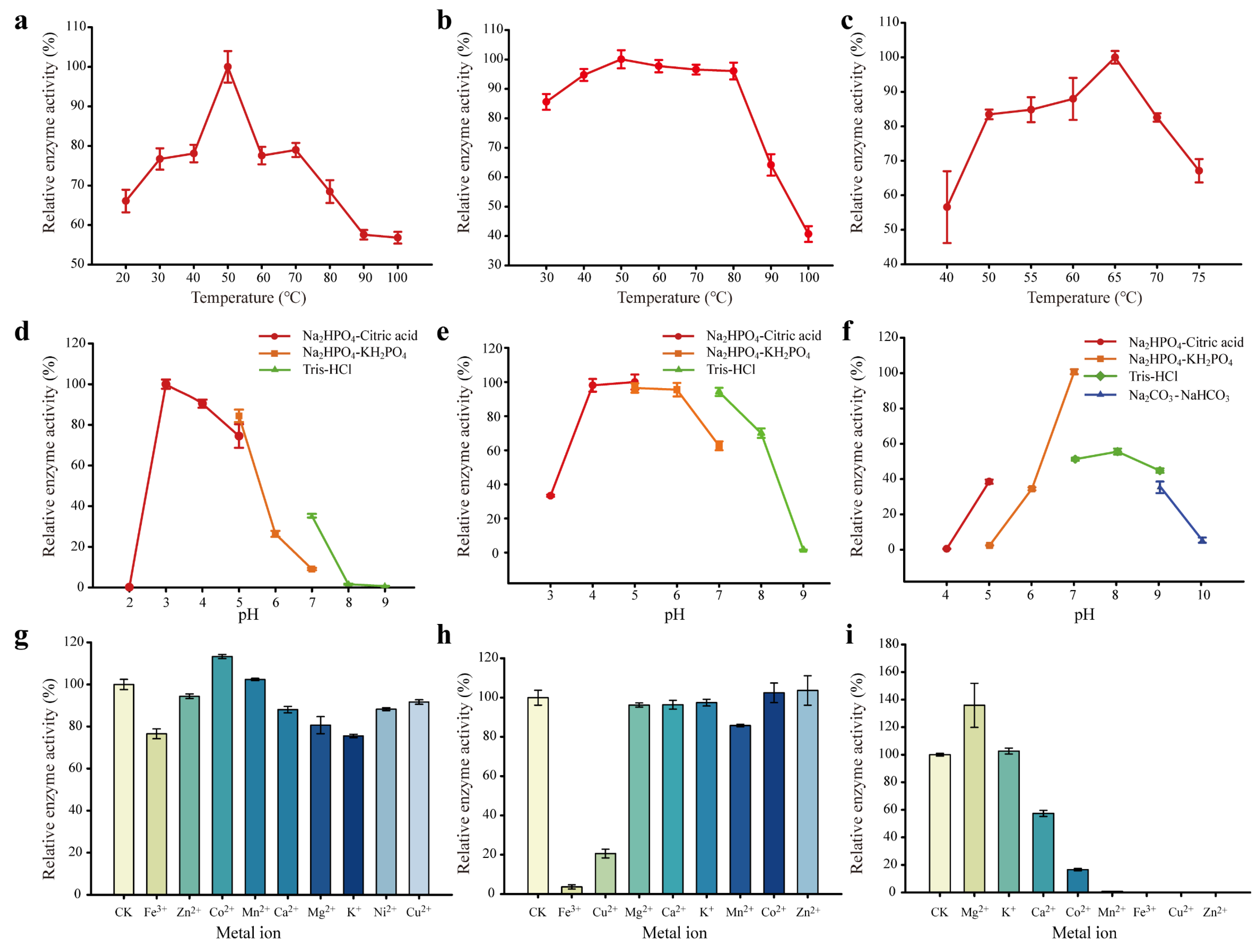
3.1. Enzymatic Property Evaluation of EGL, BGL, and XYN
3.2. Kinetics Analysis of EGL, BGL, and XYN
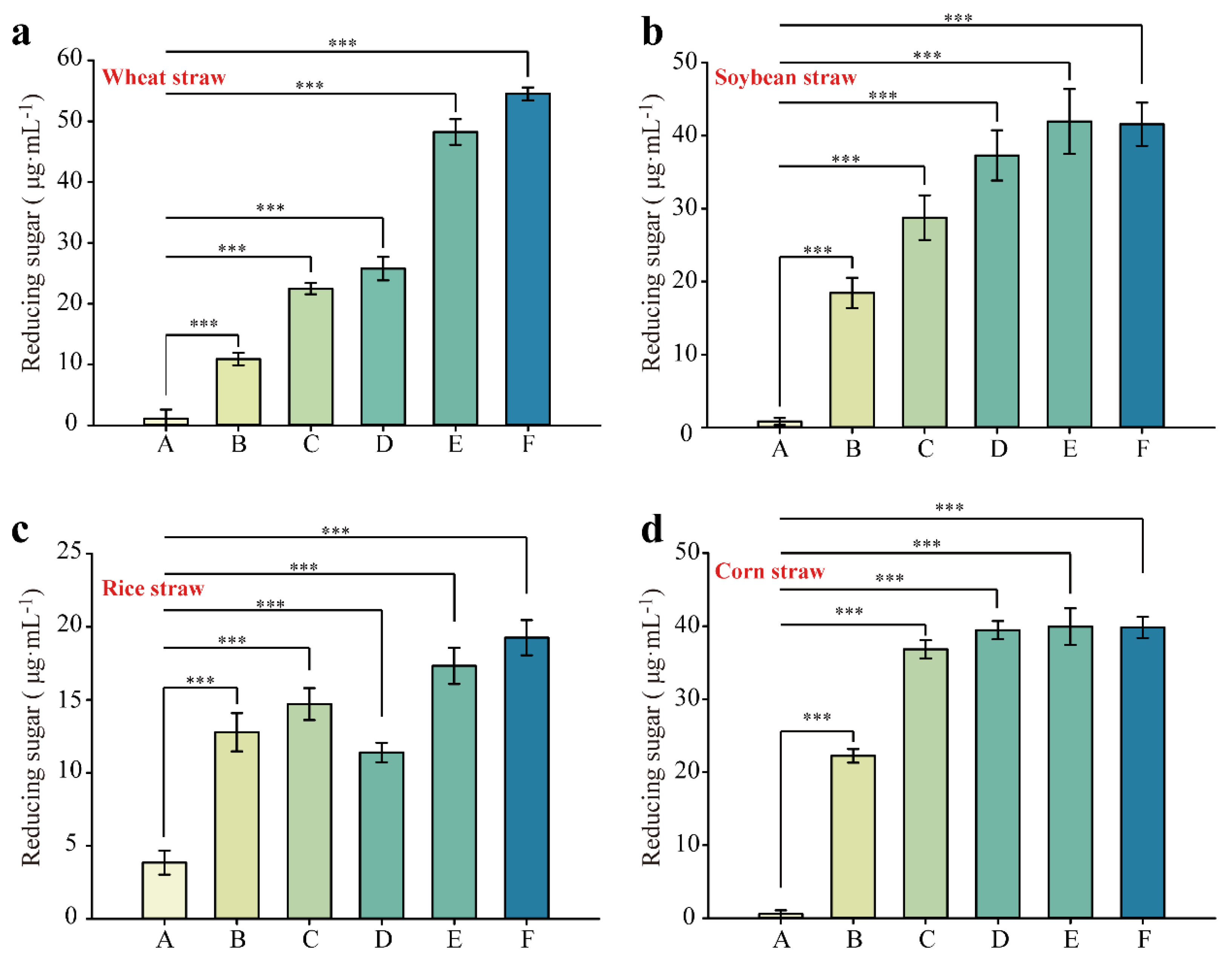
3.3. Synergistic Degradation of Different Natural Substrates by EGL, BGL, and XYN
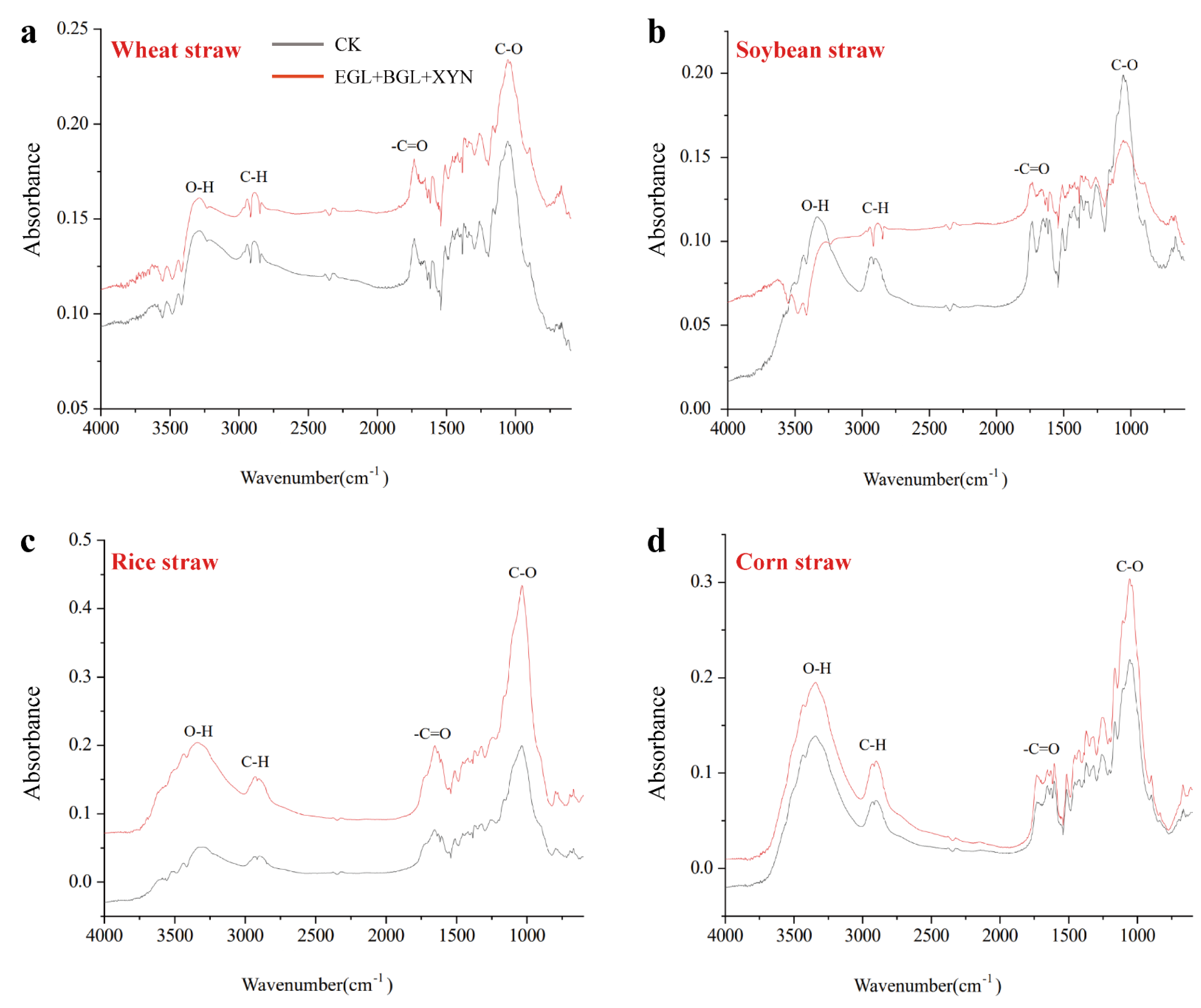
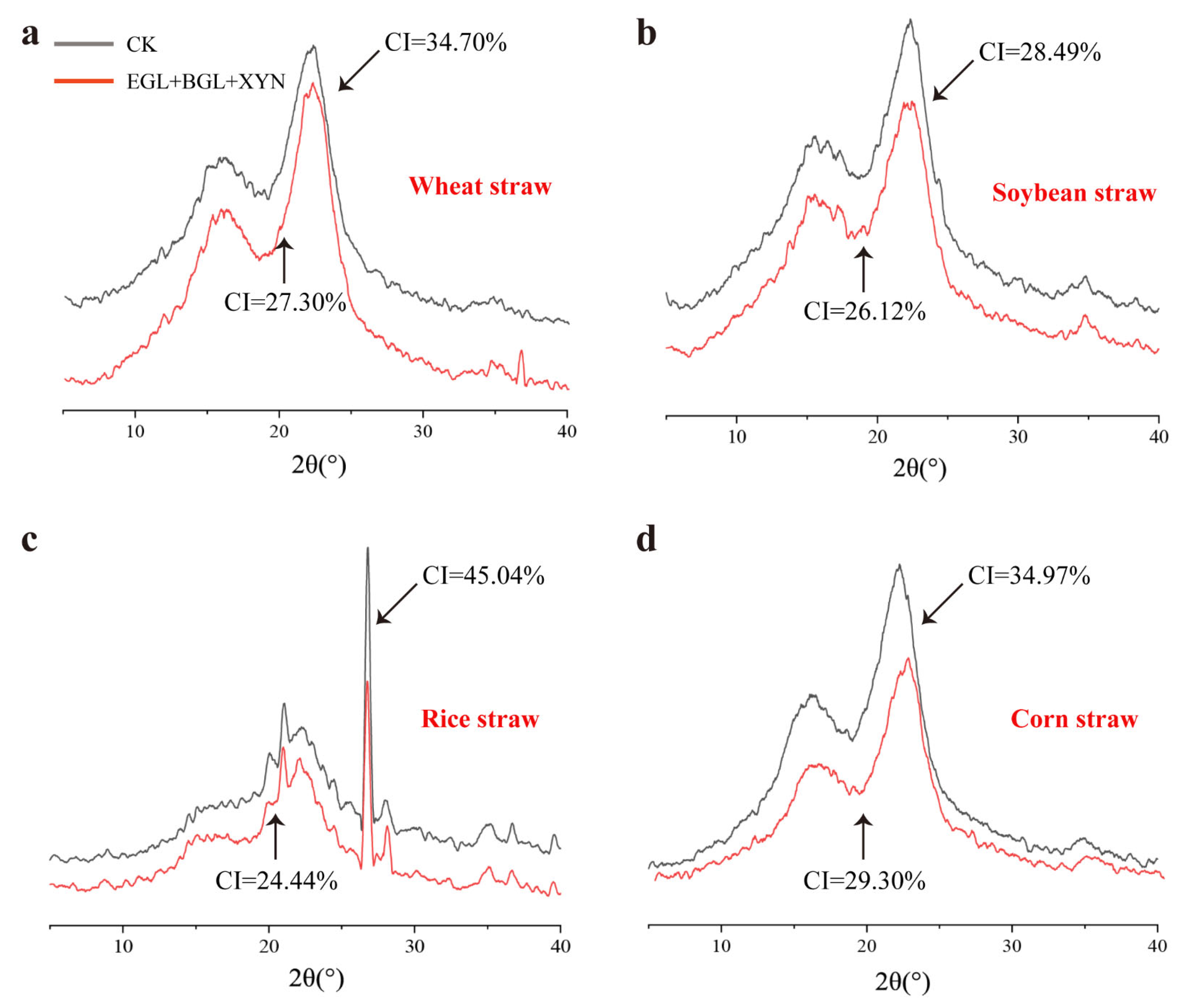
3.4. Evaluation of Functional Groups and Crystallinity of the Degraded Substrate
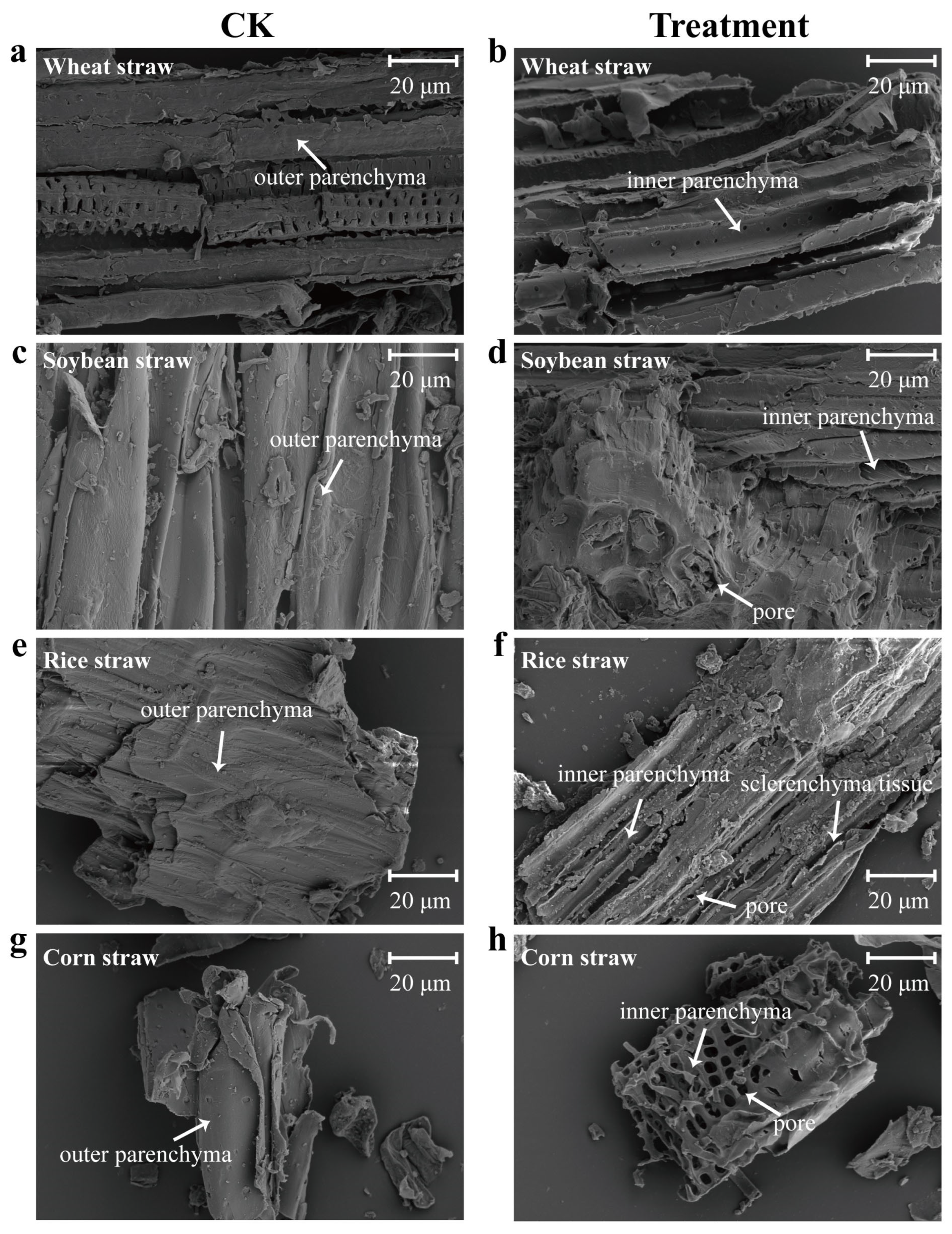
3.5. Scanning Electron Microscopy (SEM) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Definition |
| EGL | Endoglucanase |
| BGL | β-glucosidase |
| XYN | Xylanase |
| CK | Control |
| SEM | Scanning electron microscopy |
| FTIR | Fourier transform infrared spectroscopy |
| XRD | X-ray diffraction |
| CI | Crystallinity index |
References
- Rezai, A.; Van Der Ploeg, F. Abandoning fossil fuel: How fast and how much. Manch. Sch. 2017, 85, e16–e44. [Google Scholar] [CrossRef]
- Lin, B.; Xu, B. How does fossil energy abundance affect China’s economic growth and CO2 emissions? Sci. Total Environ. 2020, 719, 137503. [Google Scholar] [CrossRef] [PubMed]
- Alzoubi, A. Renewable Green hydrogen energy impact on sustainability performance. Int. J. Comput. Inf. Manuf. 2021, 1, 1. [Google Scholar] [CrossRef]
- Malherbe, S.; Cloete, T.E. Lignocellulose biodegradation: Fundamentals and applications. Rev. Environ. Sci. Biotechnol. 2002, 1, 105–114. [Google Scholar] [CrossRef]
- Jurgens, G.; Survase, S.; Berezina, O.; Sklavounos, E.; Linnekoski, J.; Kurkijärvi, A.; Väkevä, M.; van Heiningen, A.; Granström, T. Butanol production from lignocellulosics. Biotechnol. Lett. 2012, 34, 1415–1434. [Google Scholar] [CrossRef] [PubMed]
- Margeot, A.; Hahn-Hagerdal, B.; Edlund, M.; Slade, R.; Monot, F. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 2009, 20, 372–380. [Google Scholar] [CrossRef]
- Hamawand, I.; Seneweera, S.; Kumarasinghe, P.; Bundschuh, J. Nanoparticle technology for separation of cellulose, hemicellulose and lignin nanoparticles from lignocellulose biomass: A short review. Nano Struct. Nano Objects 2020, 24, 100601. [Google Scholar] [CrossRef]
- Sharma, H.K.; Xu, C.; Qin, W. Biological pretreatment of lignocellulosic biomass for biofuels and bioproducts: An overview. Waste Biomass Valorization 2019, 10, 235–251. [Google Scholar] [CrossRef]
- Zupančič, G.D.; Grilc, V. Anaerobic treatment and biogas production from organic waste. In Management of Organic Waste; Institute for Environmental Protection and Sensor: Ljubljana, Slovenia, 2012; p. 2. [Google Scholar]
- Zhang, Y.P. Reviving the carbohydrate economy via multi-product lignocellulose biorefineries. J. Ind. Microbiol. Biotechnol. 2008, 35, 367–375. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Microbial cellulases: Engineering, production and applications. Renew. Sustain. Energy Rev. 2014, 33, 188–203. [Google Scholar] [CrossRef]
- Himmel, M.E.; Ding, S.-Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Chundawat, S.P.; Beckham, G.T.; Himmel, M.E.; Dale, B.E. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Zaldivar, J.; Nielsen, J.; Olsson, L. Fuel ethanol production from lignocellulose: A challenge for metabolic engineering and process integration. Appl. Microbiol. Biotechnol. 2001, 56, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chandra, R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.J.; Lee, H.J.; Choi, I.-G.; Kim, K.H. Synergistic proteins for the enhanced enzymatic hydrolysis of cellulose by cellulase. Appl. Microbiol. Biotechnol. 2014, 98, 8469–8480. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, K.; Dai, L.; Zhang, J.; Fang, X. The structural and biochemical basis for cellulose biodegradation. J. Chem. Technol. Biotechnol. 2013, 88, 491–500. [Google Scholar] [CrossRef]
- Rani, V.; Mohanram, S.; Tiwari, R.; Nain, L.; Arora, A. Beta-glucosidase: Key enzyme in determining efficiency of cellulase and biomass hydrolysis. J. Bioprocess. Biotech. 2014, 5, 197. [Google Scholar]
- Motta, F.; Andrade, C.; Santana, M. A review of xylanase production by the fermentation of xylan: Classification, characterization and applications. Sustain. Degrad. Lignocellul. Biomass Tech. Appl. Commer. 2013, 1, 251–276. [Google Scholar]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Factories 2016, 15, 106. [Google Scholar] [CrossRef]
- Häkkinen, M.; Arvas, M.; Oja, M.; Aro, N.; Penttilä, M.; Saloheimo, M.; Pakula, T.M. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb. Cell Factories 2012, 11, 134. [Google Scholar] [CrossRef]
- Schuster, A.; Schmoll, M. Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 2010, 87, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Qualhato, T.F.; Lopes, F.A.C.; Steindorff, A.S.; Brandao, R.S.; Jesuino, R.S.A.; Ulhoa, C.J. Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: Evaluation of antagonism and hydrolytic enzyme production. Biotechnol. Lett. 2013, 35, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, J.; Guo, C.; Xu, H.; Wang, W.; Yang, M.; Shen, Q.; Zhang, R.; Miao, Y. Exploring the multi-level regulation of lignocellulases in the filamentous fungus Trichoderma guizhouense NJAU4742 from an omics perspective. Microb. Cell Factories 2022, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Kong, Z.; Zhang, X.; Wang, X.; Chai, L.; Liu, D.; Shen, Q. Deciphering the effect of exogenous lignocellulases addition on the composting efficiency and microbial communities. Bioresour. Technol. 2022, 361, 127751. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, J.; Zhao, S.; Zhang, R.; Wang, M.; Miao, Y.; Shen, Y.; Shen, Q. Secretome diversity and quantitative analysis of cellulolytic Aspergillus fumigatus Z5 in the presence of different carbon sources. Biotechnol. Biofuels 2013, 6, 149. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Fan, G.; Yu, T.; Sun, B.; Tang, H.; Teng, C.; Yang, R.; Li, X. Biochemical characteristics of the mutant xylanase T-XynC (122) C (166) and production of xylooligosaccharides from corncobs. Ind. Crop. Prod. 2019, 142, 111848. [Google Scholar] [CrossRef]
- Bansal, P.; Hall, M.; Realff, M.J.; Lee, J.H.; Bommarius, A.S. Modeling cellulase kinetics on lignocellulosic substrates. Biotechnol. Adv. 2009, 27, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Lin, L.; Liu, T.; Ouyang, P.; He, B.; Liu, S. Reducing sugar content in hemicellulose hydrolysate by DNS method: A revisit. J. Biobased Mater. Bioenergy 2008, 2, 156–161. [Google Scholar] [CrossRef]
- Basotra, N.; Kaur, B.; Di Falco, M.; Tsang, A.; Chadha, B.S. Mycothermus thermophilus (Syn. Scytalidium thermophilum): Repertoire of a diverse array of efficient cellulases and hemicellulases in the secretome revealed. Bioresour. Technol. 2016, 222, 413–421. [Google Scholar] [CrossRef]
- Kim, S.B.; Lee, S.J.; Lee, J.H.; Jung, Y.R.; Thapa, L.P.; Kim, J.S.; Um, Y.; Park, C.; Kim, S.W. Pretreatment of rice straw with combined process using dilute sulfuric acid and aqueous ammonia. Biotechnol. Biofuels 2013, 6, 109. [Google Scholar] [CrossRef]
- Segal, L.; Creely, J.J.; Martin Jr, A.; Conrad, C. An empirical method for estimating the degree of crystallinity of native cellulose using the X-ray diffractometer. Text. Res. J. 1959, 29, 786–794. [Google Scholar] [CrossRef]
- McDonald, J.E.; Houghton, J.N.; Rooks, D.J.; Allison, H.E.; McCarthy, A.J. The microbial ecology of anaerobic cellulose degradation in municipal waste landfill sites: Evidence of a role for fibrobacters. Environ. Microbiol. 2012, 14, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- de Cassia Pereira, J.; Giese, E.C.; de Souza Moretti, M.M.; dos Santos Gomes, A.C.; Perrone, O.M.; Boscolo, M.; da Silva, R.; Gomes, E.; Martins, D.A.B. Effect of metal ions, chemical agents and organic compounds on lignocellulolytic enzymes activities. Enzym. Inhib. Act. 2017, 29, 139–164. [Google Scholar]
- Zhou, Y.; Yang, J.; Luo, C.; Yang, B.; Liu, C.; Xu, B. Effect of metal ions and surfactants on the enzymatic hydrolysis of pretreated lignocellulose. BioResources 2019, 14, 1653–1667. [Google Scholar] [CrossRef]
- Corrêa, A.P.F.; Daroit, D.J.; Brandelli, A. Characterization of a keratinase produced by Bacillus sp. P7 isolated from an Amazonian environment. Int. Biodeterior. Biodegrad. 2010, 64, 1–6. [Google Scholar] [CrossRef]
- Andreini, C.; Bertini, I.; Cavallaro, G.; Holliday, G.L.; Thornton, J.M. Metal ions in biological catalysis: From enzyme databases to general principles. J. Biol. Inorg. Chem. 2008, 13, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Beltrame, P.L.; Carniti, P.; Focher, B.; Marzetti, A.; Sarto, V. Enzymatic hydrolysis of cellulosic materials: A kinetic study. Biotechnol. Bioeng. 1984, 26, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, G.; Car, S.; Scott-Craig, J.S.; Borrusch, M.S.; Walton, J.D. Rapid optimization of enzyme mixtures for deconstruction of diverse pretreatment/biomass feedstock combinations. Biotechnol. Biofuels 2010, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Berlin, A.; Maximenko, V.; Gilkes, N.; Saddler, J. Optimization of enzyme complexes for lignocellulose hydrolysis. Biotechnol. Bioeng. 2007, 97, 287–296. [Google Scholar] [CrossRef]
- Arantes, V.; Gourlay, K.; Saddler, J.N. The enzymatic hydrolysis of pretreated pulp fibers predominantly involves “peeling/erosion” modes of action. Biotechnol. Biofuels 2014, 7, 87. [Google Scholar] [CrossRef]
- Hu, J.; Gourlay, K.; Arantes, V.; Van Dyk, J.; Pribowo, A.; Saddler, J.N. The accessible cellulose surface influences cellulase synergism during the hydrolysis of lignocellulosic substrates. ChemSusChem 2015, 8, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-T.; Gao, Y.; Yang, Y.-M.; Xiao, W.-J.; Liu, S.-H.; Xia, W.-C.; Liu, Z.-L.; Yi, L.; Jiang, Z.-B. Synergistic effect of cellulase and xylanase during hydrolysis of natural lignocellulosic substrates. Bioresour. Technol. 2016, 219, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Spiridon, I.; Teaca, C.A.; Bodirlau, R. Structural changes evidenced by FTIR spectroscopy in cellulosic materials after pre-treatment with ionic liquid and enzymatic hydrolysis. BioResources 2011, 6, 400–413. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, Y.; Li, R.; Wu, Y.; Yang, M. A modified kinetic analysis method of cellulose pyrolysis based on TG–FTIR technique. Thermochim. Acta 2018, 665, 20–27. [Google Scholar] [CrossRef]
- Bian, J.; Peng, F.; Peng, X.-P.; Xiao, X.; Peng, P.; Xu, F.; Sun, R.-C. Effect of [Emim] Ac pretreatment on the structure and enzymatic hydrolysis of sugarcane bagasse cellulose. Carbohydr. Polym. 2014, 100, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Yoon, L.W.; Ang, T.N.; Ngoh, G.C.; Chua, A.S.M. Regression analysis on ionic liquid pretreatment of sugarcane bagasse and assessment of structural changes. Biomass Bioenergy 2012, 36, 160–169. [Google Scholar] [CrossRef]
- Dar, M.A.; Shaikh, A.A.; Pawar, K.D.; Pandit, R.S. Exploring the gut of Helicoverpa armigera for cellulose degrading bacteria and evaluation of a potential strain for lignocellulosic biomass deconstruction. Process Biochem. 2018, 73, 142–153. [Google Scholar] [CrossRef]
- Arantes, V.; Saddler, J.N. Cellulose accessibility limits the effectiveness of minimum cellulase loading on the efficient hydrolysis of pretreated lignocellulosic substrates. Biotechnol. Biofuels 2011, 4, 3. [Google Scholar] [CrossRef]
| Material | Product Information and Suppliers | Purpose |
|---|---|---|
| PDA medium | P8931, Solarbio, Shanghai, China | Strain cultivation |
| Tris-HCl-NaCl buffer (pH 8.0) | prepared in the lab | Protein dialysis |
| TEV protease | P8112S, NEB, Ipswich, MA, USA | Protein purification |
| CMC-Na (99%) | 9004, Sigma, St. Louis, MO, USA | Enzyme activity assay |
| p-NPG (98%) | N1252, Sigma, USA | Enzyme activity assay |
| xylan | X8070-5, Sigma, USA | Enzyme activity assay |
| RNeasy® Plant Mini Kit | 74904, Qiagen, Hilden, Germany | RNA extraction |
| PrimeScript RT Reagent Kit | RR036A, Takara, Dalian, China | cDNA synthesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Pei, R.; Wang, J.; Zhou, Y.; Liu, D. Functional Study of Different Lignocellulases from Trichoderma guizhouence NJAU4742 in the Synergistic Degradation of Natural Straw. Fermentation 2024, 10, 230. https://doi.org/10.3390/fermentation10050230
Li T, Pei R, Wang J, Zhou Y, Liu D. Functional Study of Different Lignocellulases from Trichoderma guizhouence NJAU4742 in the Synergistic Degradation of Natural Straw. Fermentation. 2024; 10(5):230. https://doi.org/10.3390/fermentation10050230
Chicago/Turabian StyleLi, Tuo, Ronghua Pei, Jiaguo Wang, Yihao Zhou, and Dongyang Liu. 2024. "Functional Study of Different Lignocellulases from Trichoderma guizhouence NJAU4742 in the Synergistic Degradation of Natural Straw" Fermentation 10, no. 5: 230. https://doi.org/10.3390/fermentation10050230
APA StyleLi, T., Pei, R., Wang, J., Zhou, Y., & Liu, D. (2024). Functional Study of Different Lignocellulases from Trichoderma guizhouence NJAU4742 in the Synergistic Degradation of Natural Straw. Fermentation, 10(5), 230. https://doi.org/10.3390/fermentation10050230







