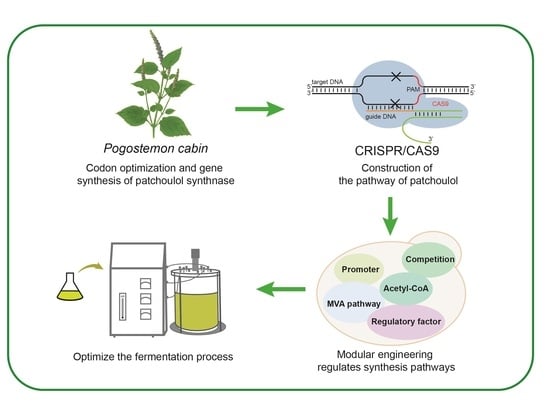
Metabolic Engineering for Efficient Synthesis of Patchoulol in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Plasmids Construction
2.3. Yeast Transformation and Strains Construction
2.4. Cultivation in Shaking Flask
2.5. Extraction and Detection Methods
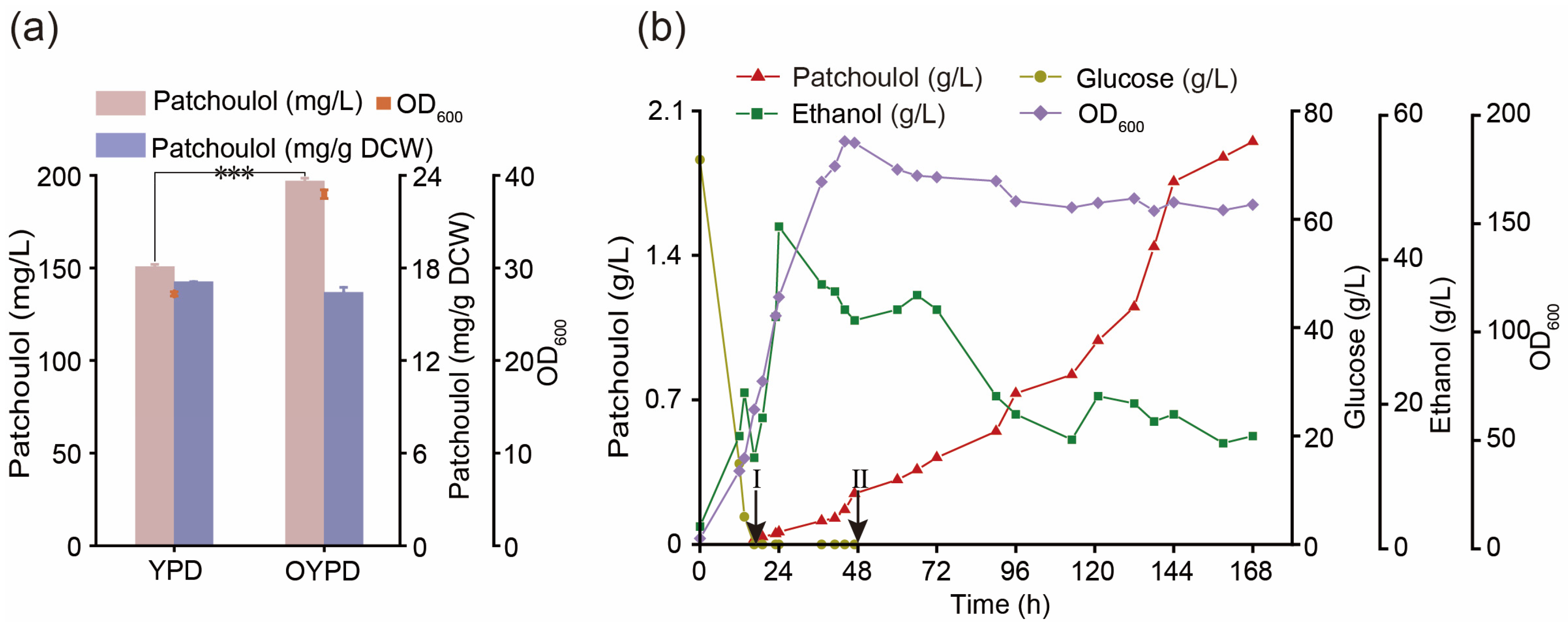
2.6. Fed-Batch Fermentation
- Supplementary medium Ⅰ for the first stage (1 L): 800 g glucose, 300 g yeast extract.
- Supplementary medium Ⅱ for the second stage (1 L): 60% ethanol.
- Metal ion solution A (1 L): 5.75 g ZnSO4∙7H2O, 0.32 g MnC12∙4H2O, 0.47 g CoC12∙6H2O, 0.48 g Na2MoO4∙2H2O, 2.9 g CaC12∙2H2O, 2.8 g FeSO4∙7H2O, 80 mL 0.5 M EDTA, Adjust pH = 8.0.
- Vitamin solution B (1 L): 0.05 g biotin, 1 g calcium pantothenate, l g nicotinic acid, 25 g myo-inositol, 1 g thiamine HCl, 1 g pyridoxal HCl, 0.02 g p-aminobenzoic acid.
3. Results and Discussion
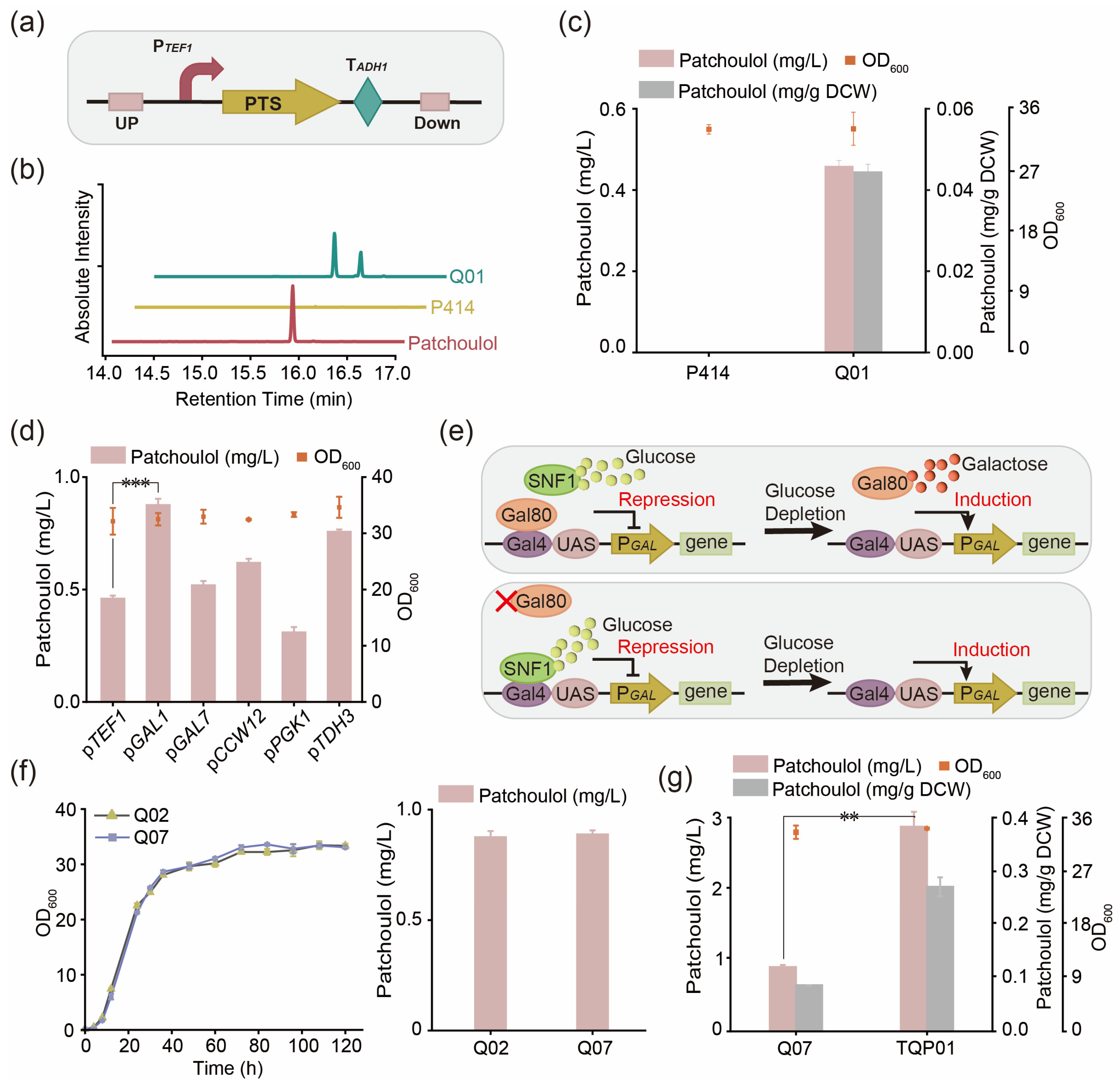
3.1. Construction of Synthetic Pathway of Patchoulol in Saccharomyces cerevisiae
3.2. Promoter Screening for Rational Control of Patchoulol Biosynthesis
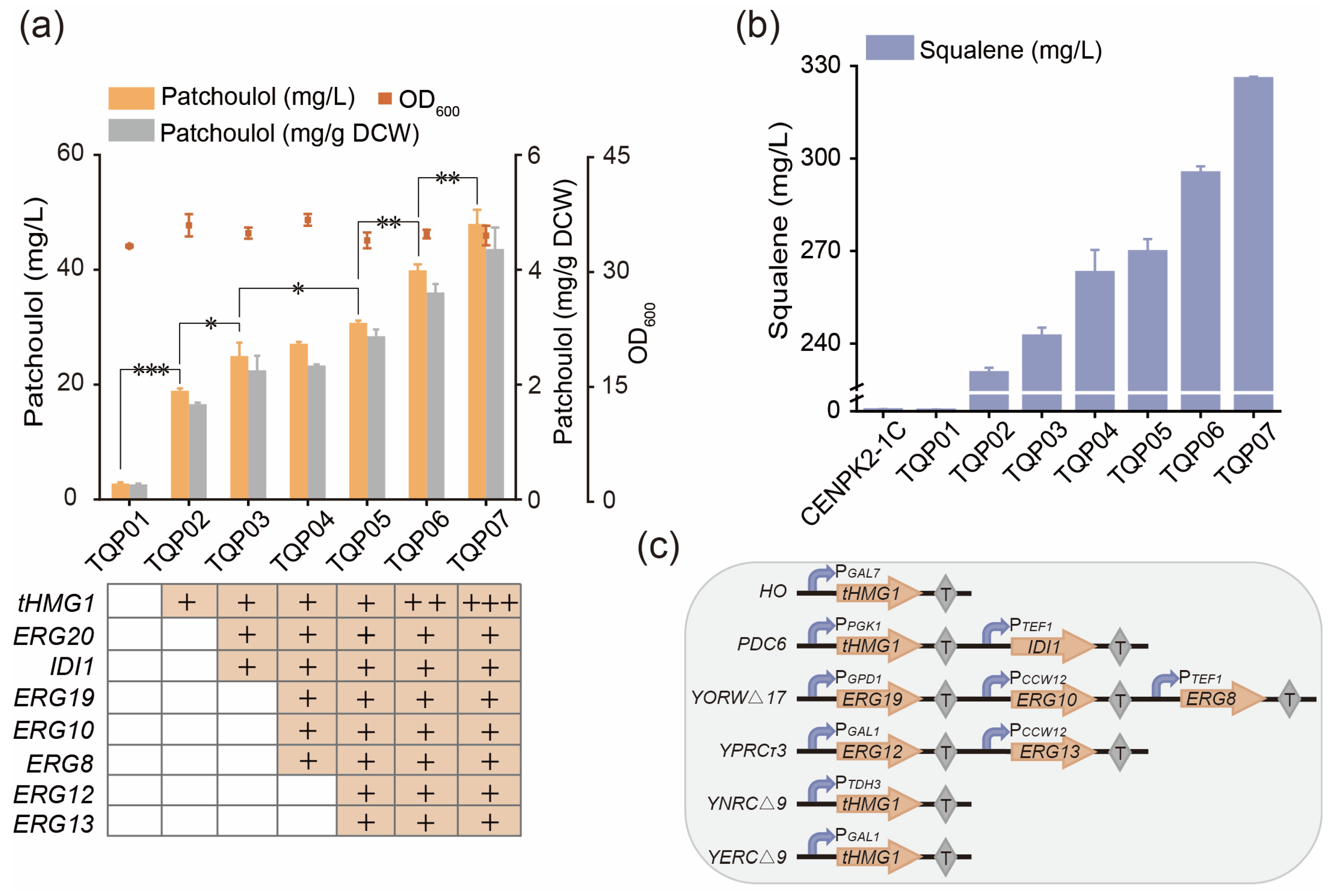
3.3. Strengthen the MVA Pathway in Saccharomyces cerevisiae
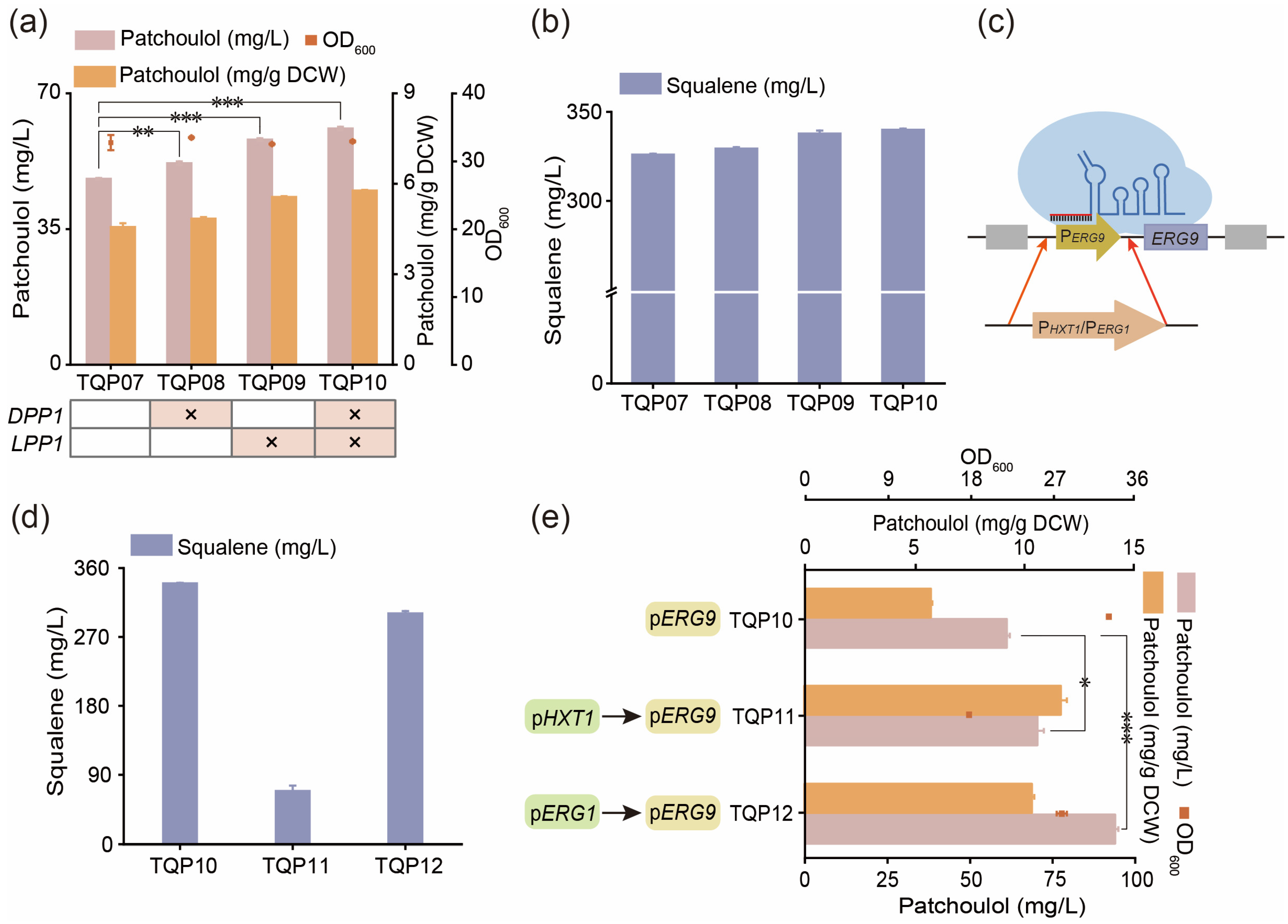
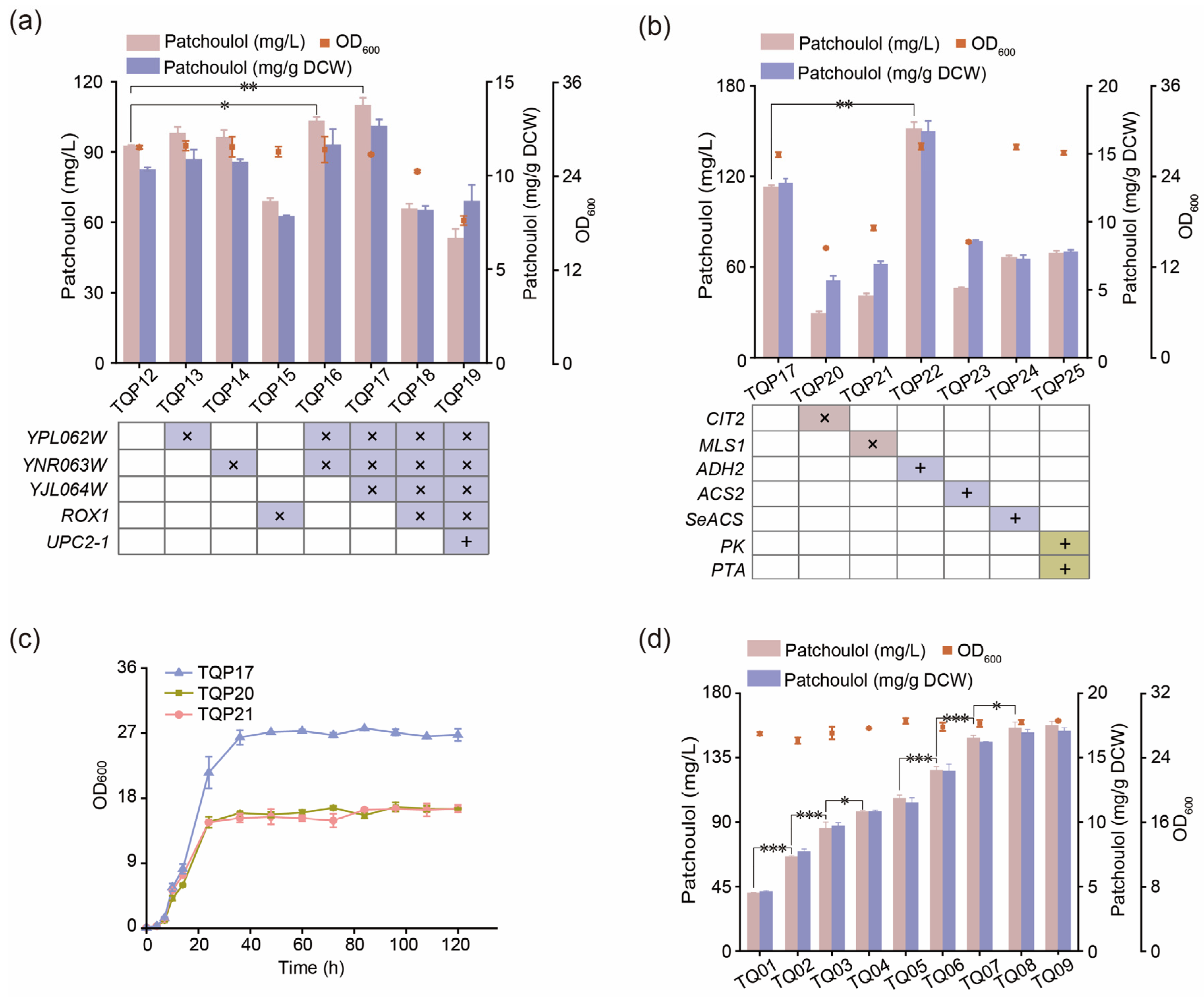
3.4. Reduce the Consumption of Precursor FPP by Balancing Competition Pathway
3.5. Improving the Production of Patchoulol by Modifying Transcription Regulatory Factors
3.6. Regulating the Acetyl-CoA Pathway to Promote the Synthesis of Patchoulol
3.7. Optimize the Fermentation Process to Improve the Production of Patchoulol
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Wu, Y.G.; Xu, Y.; Zhang, J.F.; Song, X.Q.; Zhu, G.P.; Hu, X.W. Dynamic accumulation of sesquiterpenes in essential oil of Pogostemon cablin. Rev. Bras. Farmacogn. 2014, 24, 626–634. [Google Scholar] [CrossRef][Green Version]
- Aguilar, F.; Ekramzadeh, K.; Scheper, T.; Beutel, S. Whole-cell production of patchouli oil sesquiterpenes in Escherichia coli: Metabolic engineering and fermentation optimization in solid-liquid phase partitioning cultivation. Acs Omega 2020, 5, 32436–32446. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Lee, J.; Smolensky, D.; Lee, S.H. Potential benefits of patchouli alcohol in prevention of human diseases: A mechanistic review. Int. Immunopharmacol. 2020, 89, 107056. [Google Scholar] [CrossRef]
- Swamy, M.K.; Sinniah, U.R. Patchouli (Pogostemon cablin Benth.): Botany, agrotechnology and biotechnological aspects. Ind. Crops Prod. 2016, 87, 161–176. [Google Scholar] [CrossRef]
- Mitsui, R.; Nishikawa, R.; Yamada, R.; Matsumoto, T.; Ogino, H. Construction of yeast producing patchoulol by global metabolic engineering strategy. Biotechnol. Bioeng. 2020, 117, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lin, Y.C.; Guo, J.J.; Du, M.M.; Tao, X.; Gao, B.; Zhao, M.; Ma, Y.; Wang, F.Q.; Wei, D.Z. High-level production of sesquiterpene patchoulol in Saccharomyces cerevisiae. ACS Synth. Biol. 2021, 10, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.H.; Liu, J.H.; Liu, Y.F.; Li, J.H.; Du, G.C.; Lv, X.Q.; Liu, L. Metabolic engineering of Saccharomyces cerevisiae for efficient retinol synthesis. J. Fungi 2023, 9, 512. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, M.A.; Maury, J.; Moller, K.; Nielsen, K.F.; Schalk, M.; Clark, A.; Nielsen, J. Production of plant sesquiterpenes in Saccharomyces cerevisiae: Effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol. Bioeng. 2008, 99, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Liu, M.; Li, Z.H.; Tao, X.; Wei, D.Z.; Wang, F.Q. Significantly enhanced production of patchoulol in metabolically engineered Saccharomyces cerevisiae. J. Agric. Food Chem. 2019, 67, 8590–8598. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Zhang, Y.P.; Nielsen, J.; Liu, Z.H. Production of β-carotene in Saccharomyces cerevisiae through altering yeast lipid metabolism. Biotechnol. Bioeng. 2021, 118, 2043–2052. [Google Scholar] [CrossRef]
- Luo, G.J.; Lin, Y.; Chen, S.T.; Xiao, R.M.; Zhang, J.X.; Li, C.; Sinskey, A.J.; Ye, L.; Liang, S.L. Overproduction of patchoulol in metabolically engineered Komagataella phaffii. J. Agric. Food Chem. 2023, 71, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Y.; Han, L.; Wang, Q.; Liu, H.; Cheng, P.; Li, R.; Guo, X.; Zhou, Z. Enhancement of patchoulol production in Escherichia coli via multiple engineering strategies. J. Agric. Food Chem. 2021, 69, 7572–7580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, W.; Wei, W.; Xia, L.; Gao, S.; Zeng, W.; Liu, S.; Zhou, J. Regulation of ethanol assimilation for efficient accumulation of squalene in Saccharomyces cerevisiae. J. Agric. Food Chem. 2023, 71, 6389–6397. [Google Scholar] [CrossRef]
- Xia, L.; Lv, Y.B.; Liu, S.; Yu, S.Q.; Zeng, W.Z.; Zhou, J.W. Enhancing squalene production in Saccharomyces cerevisiae by metabolic engineering and random mutagenesis. Front. Chem. Sci. Eng. 2022, 3, 790261. [Google Scholar] [CrossRef]
- Rasool, A.; Ahmed, M.S.; Li, C. Overproduction of squalene synergistically downregulates ethanol production in Saccharomyces cerevisiae. Chem. Eng. Sci. 2016, 152, 370–380. [Google Scholar] [CrossRef]
- Li, T.; Liu, G.-S.; Zhou, W.; Jiang, M.; Ren, Y.-H.; Tao, X.-Y.; Liu, M.; Zhao, M.; Wang, F.-Q.; Gao, B.; et al. Metabolic engineering of Saccharomyces cerevisiae to overproduce squalene. J. Agric. Food Chem. 2020, 68, 2132–2138. [Google Scholar] [CrossRef]
- Xiong, L.; Zeng, Y.; Tang, R.Q.; Alper, H.S.; Bai, F.W.; Zhao, X.Q. Condition-specific promoter activities in Saccharomyces cerevisiae. Microb. Cell Factories 2018, 17, 58. [Google Scholar] [CrossRef]
- Tang, H.T.; Wu, Y.L.; Deng, J.L.; Chen, N.Z.; Zheng, Z.H.; Wei, Y.J.; Luo, X.Z.; Keasling, J.D. Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 2020, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Kim, J.S. Engineering of GAL1 promoter-driven expression system with artificial transcription factors. Biochem. Biophys. Res. Commun. 2006, 351, 412–417. [Google Scholar] [CrossRef]
- Wang, F.; Lv, X.M.; Xei, W.P.; Zhou, P.P.; Zhu, Y.Q.; Yao, Z.; Yang, C.C.; Yang, X.H.; Ye, L.D.; Yu, H.W. Combining Gal4p-mediated expression enhancement and directed evolution of isoprene synthase to improve isoprene production in Saccharomyces cerevisiae. Metab. Eng. 2017, 39, 257–266. [Google Scholar] [CrossRef]
- Promdonkoy, P.; Sornlek, W.; Preechakul, T.; Tanapongpipat, S.; Runguphan, W. Metabolic engineering of Saccharomyces cerevisiae for production of fragrant terpenoids from agarwood and sandalwood. Fermentation 2022, 8, 429. [Google Scholar] [CrossRef]
- Zha, W.L.; An, T.Y.; Li, T.; Zhu, J.X.; Gao, K.; Sun, Z.J.; Xu, W.D.; Lin, P.C.; Zi, J.C. Reconstruction of the biosynthetic pathway of santalols under control of the GAL regulatory system in yeast. ACS Synth. Biol. 2020, 9, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Westfall, P.J.; Pitera, D.J.; Lenihan, J.R.; Eng, D.; Woolard, F.X.; Regentin, R.; Horning, T.; Tsuruta, H.; Melis, D.J.; Owens, A.; et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. USA 2012, 109, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.P.; Ye, L.D.; Lv, X.M.; Xu, H.M.; Yu, H.W. Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab. Eng. 2015, 28, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, A.; Chen, X.M.; Rush, J.; Horazdovsky, B.; Waechter, C.J.; Carman, G.M.; Sternweis, P.C. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 14831–14837. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, W.; Liang, C.; Zhu, L.; Li, Y.; Mo, Q.; Xu, S.; Chu, A.; Zhang, L.; Ding, Z.; et al. Overproduction of α-farnesene in Saccharomyces cerevisiae by farnesene synthase screening and metabolic engineering. J. Agric. Food Chem. 2021, 69, 3103–3113. [Google Scholar] [CrossRef] [PubMed]
- Scalcinati, G.; Knuf, C.; Partow, S.; Chen, Y.; Maury, J.; Schalk, M.; Daviet, L.; Nielsen, J.; Siewers, V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene α-santalene in a fed-batch mode. Metab. Eng. 2012, 14, 91–103. [Google Scholar] [CrossRef]
- Özaydin, B.; Burd, H.; Lee, T.S.; Keasling, J.D. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab. Eng. 2013, 15, 174–183. [Google Scholar] [CrossRef]
- Trikka, F.A.; Nikolaidis, A.; Athanasakoglou, A.; Andreadelli, A.; Ignea, C.; Kotta, K.; Argiriou, A.; Kampranis, S.C.; Makris, A.M. Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microb. Cell Factories 2015, 14, 60. [Google Scholar] [CrossRef]
- Cao, X.; Yu, W.; Chen, Y.; Yang, S.; Zhao, Z.K.; Nielsen, J.; Luan, H.; Zhou, Y.J. Engineering yeast for high-level production of diterpenoid sclareol. Metab. Eng. 2022, 75, 19–28. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Liu, M.; Qu, J.Z.; Yao, M.D.; Li, B.; Ding, M.Z.; Liu, H.; Xiao, W.H.; Yuan, Y.J. Primary and secondary metabolic effects of a key gene deletion (delta YPL062W) in metabolically engineered terpenoid-producing Saccharomyces cerevisiae. Appl. Environ. Microb. 2019, 85, e01990-18. [Google Scholar] [CrossRef] [PubMed]
- Scalcinati, G.; Partow, S.; Siewers, V.; Schalk, M.; Daviet, L.; Nielsen, J. Combined metabolic engineering of precursor and co-factor supply to increase α-santalene production by Saccharomyces cerevisiae. Microb. Cell Factories 2012, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Jordá, T.; Martínez-Martín, A.; Martínez-Pastor, M.T.; Puig, S. Modulation of yeast Erg1 expression and terbinafine susceptibility by iron bioavailability. Microb. Biotechnol. 2022, 15, 2705–2716. [Google Scholar] [CrossRef] [PubMed]
- Leak, F.W.; Tove, S.; Parks, L.W. In yeast, upc2-1 confers a decrease in tolerance to LiCl and NaCl, which can be suppressed by the P-type ATPase encoded by ENA2. DNA Cell Biol. 1999, 18, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Daviet, L.; Schalk, M.; Siewers, V.; Nielsen, J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab. Eng. 2013, 15, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; DaSilva, N.A. Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast 2005, 22, 431–440. [Google Scholar]
- Zhang, X.; Liu, X.; Meng, Y.; Zhang, L.; Qiao, J.; Zhao, G.-R. Combinatorial engineering of Saccharomyces cerevisiae for improving limonene production. Biochem. Eng. J. 2021, 176, 108155. [Google Scholar] [CrossRef]
- Qin, N.; Li, L.Y.; Ji, X.; Li, X.W.; Zhang, Y.M.; Larsson, C.; Chen, Y.; Nielsen, J.; Liu, Z.H. Rewiring central carbon metabolism ensures increased provision of acetyl-CoA and NADPH required for 3-OH-propionic acid production. ACS Synth. Biol. 2020, 9, 3236–3244. [Google Scholar] [CrossRef]
- Zhao, F.L.; Du, Y.H.; Bai, P.; Liu, J.J.; Lu, W.Y.; Yuan, Y.J. Enhancing Saccharomyces cerevisiae reactive oxygen species and ethanol stress tolerance for high-level production of protopanoxadiol. Bioresour. Technol. 2017, 227, 308–316. [Google Scholar] [CrossRef]

| Names | Description | Reference |
|---|---|---|
| pESC-URA | 2 μ, URA3, Amp | This Lab |
| p414-TEF1p-Cas9-CYC1t | Cas9 expression vector | This Lab |
| p426-SNR52p-gRNA.CAN1.Y-SUP4t | gRNA expression vector | This Lab |
| YEplac181 | 2 μ, LEU2 | This Lab |
| YEplac181-PTS | PTEF1-PTS-TADH1 cassette in YEplac181 | GENEWIZ |
| pESC-PTS | PGAL1-PTS-TCYC1 cassette in pESC-URA | This work |
| p426-YMRWΔ15 | gRNA expressing vector carrying YMRWΔ15 crRNA sequence | This work |
| p426-GAL80 | gRNA expressing vector carrying GAL80 crRNA sequence | This work |
| p426-HO | gRNA expressing vector carrying HO crRNA sequence | This work |
| p426-PDC6 | gRNA expressing vector carrying PDC6 crRNA sequence | This work |
| p426-YORWΔ17 | gRNA expressing vector carrying YORWΔ17 crRNA sequence | This work |
| p426-YPRCτ3 | gRNA expressing vector carrying YPRCτ3 crRNA sequence | This work |
| p426-YNRCΔ9 | gRNA expressing vector carrying YNRCΔ9 crRNA sequence | This work |
| p426-YERCΔ8 | gRNA expressing vector carrying YERCΔ8 crRNA sequence | This work |
| p426-DPP1 | gRNA expressing vector carrying DPP1 crRNA sequence | This work |
| p426-LPP1 | gRNA expressing vector carrying LPP1 crRNA sequence | This work |
| p426-YPL062W | gRNA expressing vector carrying YPL062W crRNA sequence | This work |
| p426-YNR063W | gRNA expressing vector carrying YNR063W crRNA sequence | This work |
| p426-ROX1 | gRNA expressing vector carrying ROX1 crRNA sequence | This work |
| p426 -YJL064W | gRNA expressing vector carrying YJL064W crRNA sequence | This work |
| p426-CIT2 | gRNA expressing vector carrying CIT2 crRNA sequence | This work |
| p426-MLS1 | gRNA expressing vector carrying MLS1 crRNA sequence | This work |
| p426-XI-3 | gRNA expressing vector carrying XI-3 crRNA sequence | This work |
| p426-YORWΔ22 | gRNA expressing vector carrying YORWΔ22 crRNA sequence | This work |
| p426-YPRCΔ15 | gRNA expressing vector carrying YPRCΔ15 crRNA sequence | This work |
| p426-NDT80 | gRNA expressing vector carrying NDT80 crRNA sequence | This work |
| p426-X-2 | gRNA expressing vector carrying X-2 crRNA sequence | This work |
| p426-YHRCΔ14 | gRNA expressing vector carrying YHRCΔ14 crRNA sequence | This work |
| p426-XI-2 | gRNA expressing vector carrying XI-2 crRNA sequence | This work |
| p426-X-4 | gRNA expressing vector carrying X-4 crRNA sequence | This work |
| p426-XI-1 | gRNA expressing vector carrying XI-1 crRNA sequence | This work |
| p426-YARCΔ8 | gRNA expressing vector carrying YARCΔ8 crRNA sequence | This work |
| Names | Description | Reference |
|---|---|---|
| S. cerevisiae CEN.PK2-1C | Mata; his3Δ1; leu2-3-112; ura3-52; trp1-289; MAL2-8c; SUC2 | This Lab |
| P414 | CEN.PK2-1C, contains p414-TEF1p-Cas9-CYC1t plasmid | This work |
| Q01 | CEN.PK2-1C, YMRWΔ15:: PTEF1-PTS-TADH1 | This work |
| Q02 | CEN.PK2-1C, YMRWΔ15:: PGAL1-PTS-TADH1 | This work |
| Q03 | CEN.PK2-1C, YMRWΔ15:: PGAL7-PTS-TADH1 | This work |
| Q04 | CEN.PK2-1C, YMRWΔ15:: PCCW12-PTS-TADH1 | This work |
| Q05 | CEN.PK2-1C, YMRWΔ15:: PPGK1-PTS-TADH1 | This work |
| Q06 | CEN.PK2-1C, YMRWΔ15:: PTDH3-PTS-TADH1 | This work |
| Q07 | Q02, ΔGAL80 | This work |
| TQP01 | Q07, pESC-PTS | This work |
| TQP02 | TQP01, ΔHO:: PGAL7–tHMG1-TCYC1 | This work |
| TQP03 | TQP02, ΔPDC6:: TGPM1-ERG20-PPGK1-PTEF1-IDI1-TACT1 | This work |
| TQP04 | TQP03, YORWΔ17:: PGPD1-ERG19-TPGK1-PCCW12-ERG10-TADH1-PTEF1-ERG8-TCYC1 | This work |
| TQP05 | TQP04, ΔYPRCτ3:: PTDH3-ERG12-TPGK1-PCCW12-ERG13-TADH1 | This work |
| TQP06 | TQP05, YNRCΔ9:: PTDH3–tHMG1-TCYC1 | This work |
| TQP07 | TQP06, YERCΔ8:: PGAL1–tHMG1-TCYC1 | This work |
| TQP08 | TQP07, ΔDPP1 | This work |
| TQP09 | TQP07, ΔLPP1 | This work |
| TQP10 | TQP08, ΔLPP1 | This work |
| TQP11 | TQP10, PERG9→PHXT1 | This work |
| TQP12 | TQP10, PERG9→PERG1 | This work |
| TQP13 | TQP12, ΔYPL062W | This work |
| TQP14 | TQP12, ΔYNR063W | This work |
| TQP15 | TQP12, ΔROX1 | This work |
| TQP16 | TQP13, ΔYNR063W | This work |
| TQP17 | TQP16, ΔYJL064W | This work |
| TQP18 | TQP17, ΔROX1 | This work |
| TQP19 | TQP17, ΔROX1:: PGAL1–UPC2-1-TCYC1 | This work |
| TQP20 | TQP17, ΔCIT2 | This work |
| TQP21 | TQP17, ΔMLS1 | This work |
| TQP22 | TQP17, ΔXI-3:: PTDH3-ADH2-TADH1 | This work |
| TQP23 | TQP17, YORWΔ22:: PTDH3-ACS2-TADH1 | This work |
| TQP24 | TQP17, YPRCΔ15:: PTEF1-SeACS-TCYC1 | This work |
| TQP25 | TQP17, ΔNDT80:: PGAL1-PK-TPGK1–TCYC1-PTA-PTDH3 | This work |
| Q22 | TQP22, discard the PESC-PTS plasmid | This work |
| TQ01 | Q22, ΔX-2:: PGAL1-PTS-TCYC1 | This work |
| TQ02 | TQ01, YHRCΔ14:: PGAL1-PTS-TCYC1 | This work |
| TQ03 | TQ02, ΔXI-2:: PGAL1-PTS-TCYC1 | This work |
| TQ04 | TQ03, ΔX-4:: PGAL1-PTS-TCYC1 | This work |
| TQ05 | TQ04, YPRCΔ15::PGAL1-PTS-TCYC1 | This work |
| TQ06 | TQ05, YORWΔ22:: PGAL1-PTS-TCYC1 | This work |
| TQ07 | TQ06, ΔNDT80:: PGAL1-PTS-TCYC1 | This work |
| TQ08 | TQ07, ΔXI-1:: PGAL1-PTS-TCYC1 | This work |
| TQ09 | TQ08, YARCΔ8:: PGAL1-PTS-TCYC1 | This work |
| E. coli DH5α | The strain for plasmid construction | This Lab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Q.; Du, G.; Chen, J.; Zhang, J.; Peng, Z. Metabolic Engineering for Efficient Synthesis of Patchoulol in Saccharomyces cerevisiae. Fermentation 2024, 10, 211. https://doi.org/10.3390/fermentation10040211
Tao Q, Du G, Chen J, Zhang J, Peng Z. Metabolic Engineering for Efficient Synthesis of Patchoulol in Saccharomyces cerevisiae. Fermentation. 2024; 10(4):211. https://doi.org/10.3390/fermentation10040211
Chicago/Turabian StyleTao, Qiu, Guocheng Du, Jian Chen, Juan Zhang, and Zheng Peng. 2024. "Metabolic Engineering for Efficient Synthesis of Patchoulol in Saccharomyces cerevisiae" Fermentation 10, no. 4: 211. https://doi.org/10.3390/fermentation10040211
APA StyleTao, Q., Du, G., Chen, J., Zhang, J., & Peng, Z. (2024). Metabolic Engineering for Efficient Synthesis of Patchoulol in Saccharomyces cerevisiae. Fermentation, 10(4), 211. https://doi.org/10.3390/fermentation10040211