Research Progress in Understanding the Molecular Biology of Cordyceps militaris
Abstract
1. Introduction
2. C. militaris Genome Analysis
3. C. militaris Transcriptome Analysis
| Strain | Sample Information | Main Results | Year | Ref. |
|---|---|---|---|---|
| C. militaris TBRC6039 | Mycelia and fruiting bodies | Compared the metabolic functions and regulation of C. militaris relevant to its developmental stages | 2023 | [49] |
| C. militaris YCC | Mycelia from 6 generations of subculture | Compared the changes in C. militaris for six generations during fruiting body growth | 2017 | [36] |
| C. militaris CM10 | Mycelia (cultured under light and dark conditions) | Discovered the genes related to the biosynthesis of C. militaris carotenoid | 2021 | [55] |
| C. militaris Cm01 | Mycelia with different salt treatment | Provided a global transcriptome characterization of the salt treatment adaptation process in C. militaris | 2021 | [51] |
| C. militaris TBRC6039 | Mycelia (cultured under three carbon sources) | Uncovering sucrose- and glucose-mediated changes in the transcriptional regulation of central carbon metabolism in C. militaris | 2018 | [52] |
| C. militaris TBRC7358 | Mycelia (cultured under three carbon sources) | Proposed putative alternative routes in formation of precursors for COR production on xylose | 2020 | [53] |
4. C. militaris Proteome Analysis
5. Genetic Manipulation of C. militaris
6. COR Biosynthesis in C. militaris
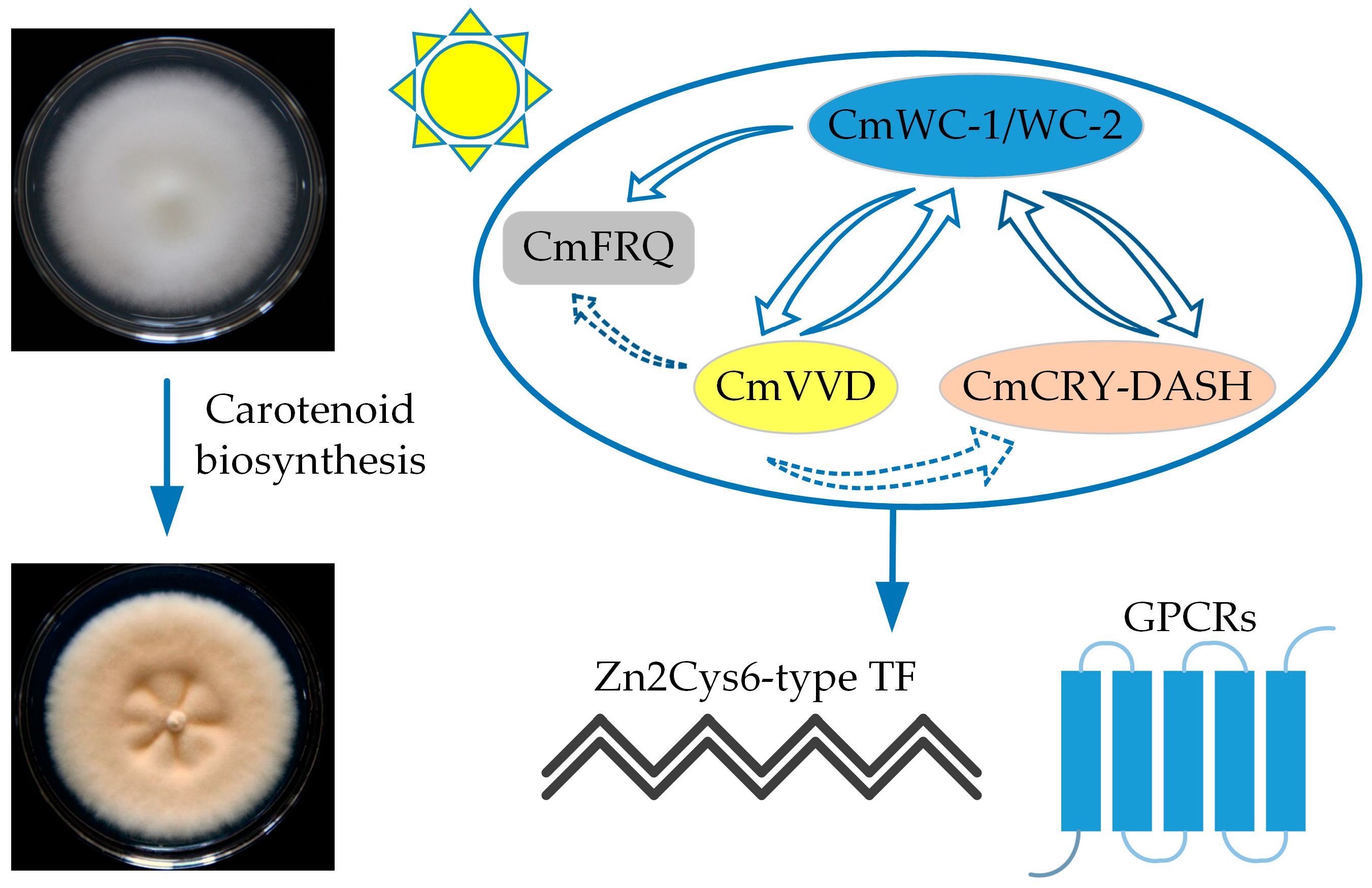
7. Carotenoid Biosynthesis in C. militaris
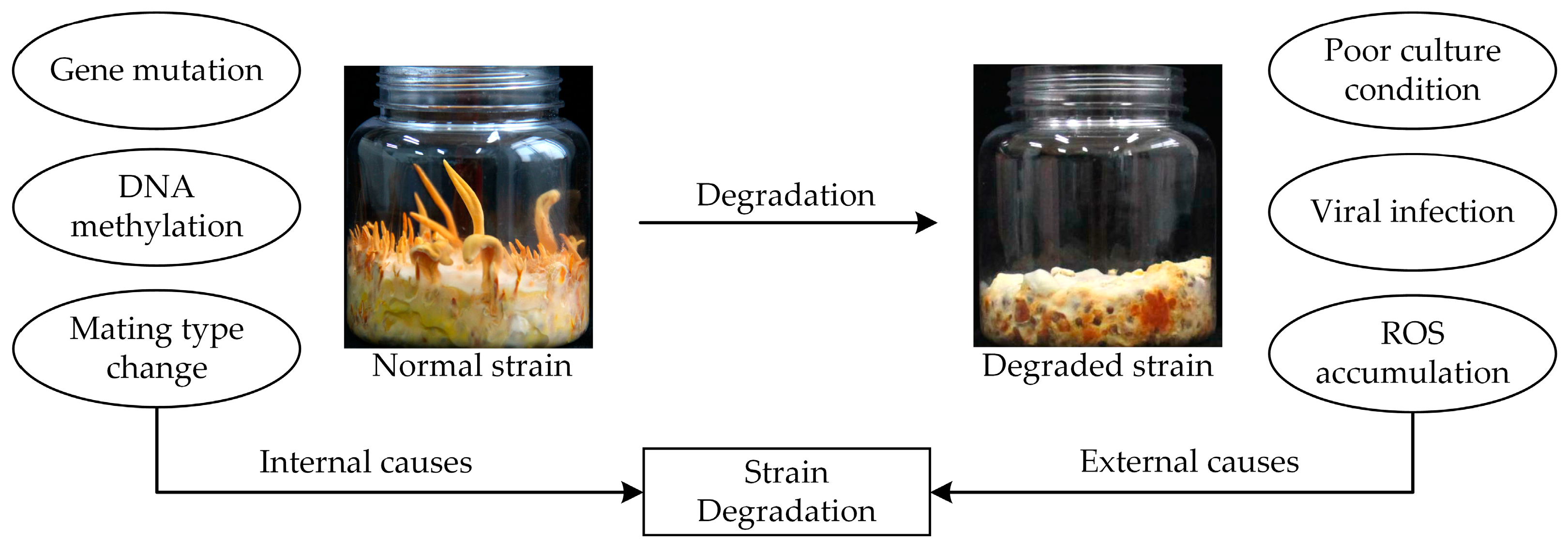
8. C. militaris Strain Degradation Mechanisms
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhou, L.-W.; Yang, Z.-L.; Bau, T.; Li, T.-H.; Dai, Y.-C. Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 2019, 98, 1–76. [Google Scholar] [CrossRef]
- Jiaojiao, Z.; Fen, W.; Kuanbo, L.; Qing, L.; Ying, Y.; Caihong, D. Heat and light stresses affect metabolite production in the fruit body of the medicinal mushroom Cordyceps militaris. Appl. Microbiol. Biotechnol. 2018, 102, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Kunhorm, P.; Chaicharoenaudomrung, N.; Noisa, P. Enrichment of cordycepin for cosmeceutical applications: Culture systems and strategies. Appl. Microbiol. Biotechnol. 2019, 103, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Aung, W.L.; Kyaw, M. Identification and Determination of Secondary Metabolites and Amino Acids in Cordyceps. Partn. Univers. Int. Innov. J. 2023, 1, 251–258. [Google Scholar]
- Song, Q.; Zhu, Z. Using Cordyceps militaris extracellular polysaccharides to prevent Pb 2+-induced liver and kidney toxicity by activating Nrf2 signals and modulating gut microbiota. Food Funct. 2020, 11, 9226–9239. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Shin, H.-S.; Leyva-Gómez, G.; Prado-Audelo, M.L.D.; Cortes, H.; Singh, Y.D.; Panda, M.K.; Mishra, A.P.; Nigam, M.; Saklani, S. Cordyceps spp.: A review on its immune-stimulatory and other biological potentials. Front. Pharmacol. 2021, 11, 2250. [Google Scholar] [CrossRef]
- Van Nguyen, T.; Chumnanpuen, P.; Parunyakul, K.; Srisuksai, K.; Fungfuang, W. A study of the aphrodisiac properties of Cordyceps militaris in streptozotocin-induced diabetic male rats. Vet. World. 2021, 14, 537–544. [Google Scholar] [CrossRef]
- Dong, C.; Guo, S.; Wang, W.; Liu, X. Cordyceps industry in China. Mycology 2015, 6, 121–129. [Google Scholar] [CrossRef]
- Wei, H.-P.; Ye, X.-L.; Chen, Z.; Zhong, Y.-J.; Li, P.-M.; Pu, S.-C.; Li, X.-G. Synthesis and pharmacokinetic evaluation of novel N-acyl-cordycepin derivatives with a normal alkyl chain. Eur. J. Med. Chem. 2009, 44, 665–669. [Google Scholar] [CrossRef]
- Quy, T.N.; Xuan, T.D.; Andriana, Y.; Tran, H.-D.; Khanh, T.D.; Teschke, R. Cordycepin isolated from Cordyceps militaris: Its newly discovered herbicidal property and potential plant-based novel alternative to glyphosate. Molecules 2019, 24, 2901. [Google Scholar] [CrossRef]
- Lan, D.; Thu, N.; Lan, P.; Nha, P.; Tung, B. Cordyceps militaris (L.) link: Chemical bioactive compounds and pharmacological activities. J. Pharm. Nutr. Sci. 2016, 6, 153–159. [Google Scholar] [CrossRef]
- Jędrejko, K.J.; Lazur, J.; Muszyńska, B. Cordyceps militaris: An overview of its chemical constituents in relation to biological activity. Foods 2021, 10, 2634. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.-Y.; Liu, X.-C.; Dong, F.-Y.; Guo, M.-Z.; Wang, X.-T.; Wang, Z.; Zhang, Y.-M. Influence of fermentation conditions on polysaccharide production and the activities of enzymes involved in the polysaccharide synthesis of Cordyceps militaris. Appl. Microbiol. Biotechnol. 2016, 100, 3909–3921. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Xin, X.D.; Weng, Y.J.; Li, S.H.; Jia, J.Q.; Gui, Z.Z. Genotypic analysis of degenerative Cordyceps militaris cultured in the pupa of Bombyx mori. Entomol. Res. 2018, 48, 137–144. [Google Scholar] [CrossRef]
- Wang, F.; Song, X.; Dong, X.; Zhang, J.; Dong, C. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris. Appl. Microbiol. Biotechnol. 2017, 101, 4645–4657. [Google Scholar] [CrossRef] [PubMed]
- Yong, T.; Chen, S.; Xie, Y.; Chen, D.; Su, J.; Shuai, O.; Jiao, C.; Zuo, D. Cordycepin, a characteristic bioactive constituent in Cordyceps militaris, ameliorates hyperuricemia through URAT1 in hyperuricemic mice. Front. Microbiol. 2018, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wei, T.; Lin, Y.; Ye, Z.-W.; Lin, J.-F.; Guo, L.-Q.; Yun, F.; Kang, L. Developing a novel two-stage process for carotenoid production by Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms. 2019, 21, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Lou, Z.; Wang, H.; Chen, C. Antimicrobial effect and proposed action mechanism of cordycepin against Escherichia coli and Bacillus subtilis. J. Microbiol. 2019, 57, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cai, G.; He, Y.; Tong, G. Separation of cordycepin from Cordyceps militaris fermentation supernatant using preparative HPLC and evaluation of its antibacterial activity as an NAD+-dependent DNA ligase inhibitor. Exp. Ther. Med. 2016, 12, 1812–1816. [Google Scholar] [CrossRef]
- Jeong, M.-H.; Lee, C.-M.; Lee, S.-W.; Seo, S.-Y.; Seo, M.-J.; Kang, B.-W.; Jeong, Y.-K.; Choi, Y.-J.; Yang, K.-M.; Jo, W.-S. Cordycepin-enriched Cordyceps militaris induces immunomodulation and tumor growth delay in mouse-derived breast cancer. Oncol. Rep. 2013, 30, 1996–2002. [Google Scholar] [CrossRef]
- Wu, W.-C.; Hsiao, J.-R.; Lian, Y.-Y.; Lin, C.-Y.; Huang, B.-M. The apoptotic effect of cordycepin on human OEC-M1 oral cancer cell line. Cancer Chemother. Pharmacol. 2007, 60, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, Y.; Cui, Y.; Liu, H.; Dong, C.; Sun, Y. Structural characterization, antioxidant and immunomodulatory activities of a neutral polysaccharide from Cordyceps militaris cultivated on hull-less barley. Carbohydr. Polym. 2020, 235, 115969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Gu, S.; Pan, L.; Sun, H.; Gong, E.; Zhu, Z.; Wen, T.; Daba, G.M.; Elkhateeb, W.A. Structure analysis and antioxidant activity of polysaccharide-iron (III) from Cordyceps militaris mycelia. Int. J. Biol. Macromol. 2021, 178, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Won, S.-Y.; Park, E.-H. Anti-inflammatory and related pharmacological activities of cultured mycelia and fruiting bodies of Cordyceps militaris. J. Ethnopharmacol. 2005, 96, 555–561. [Google Scholar] [CrossRef]
- Phull, A.-R.; Ahmed, M.; Park, H.-J. Cordyceps militaris as a bio functional food source: Pharmacological potential, anti-inflammatory actions and related molecular mechanisms. Microorganisms 2022, 10, 405. [Google Scholar] [CrossRef]
- Gao, J.; Lian, Z.-Q.; Zhu, P.; Zhu, H.-B. Lipid-lowering effect of cordycepin (3’-deoxyadenosine) from Cordyceps militaris on hyperlipidemic hamsters and rats. Yao Xue Xue Bao Acta Pharm. Sin. 2011, 46, 669–676. [Google Scholar]
- Li, R.-F.; Zhou, X.-B.; Zhou, H.-X.; Yang, Z.-F.; Jiang, H.-M.; Wu, X.; Li, W.-J.; Qiu, J.-J.; Mi, J.-N.; Chen, M. Novel fatty acid in Cordyceps suppresses influenza A (H1N1) virus-induced proinflammatory response through regulating innate signaling pathways. ACS Omega 2021, 6, 1505–1515. [Google Scholar] [CrossRef]
- Jiang, Y.; Wong, J.; Fu, M.; Ng, T.; Liu, Z.; Wang, C.; Li, N.; Qiao, W.; Wen, T.; Liu, F. Isolation of adenosine, iso-sinensetin and dimethylguanosine with antioxidant and HIV-1 protease inhibiting activities from fruiting bodies of Cordyceps militaris. Phytomedicine 2011, 18, 189–193. [Google Scholar] [CrossRef]
- Panya, A.; Songprakhon, P.; Panwong, S.; Jantakee, K.; Kaewkod, T.; Tragoolpua, Y.; Sawasdee, N.; Lee, V.S.; Nimmanpipug, P.; Yenchitsomanus, P.-T. Cordycepin inhibits virus replication in dengue virus-infected vero cells. Molecules 2021, 26, 3118. [Google Scholar] [CrossRef]
- Ryu, E.; Son, M.; Lee, M.; Lee, K.; Cho, J.Y.; Cho, S.; Lee, S.K.; Lee, Y.M.; Cho, H.; Sung, G.-H. Cordycepin is a novel chemical suppressor of Epstein-Barr virus replication. Oncoscience 2014, 1, 866–881. [Google Scholar] [CrossRef]
- Du, Y.; Yu, J.; Du, L.; Tang, J.; Feng, W.-H. Cordycepin enhances Epstein–Barr virus lytic infection and Epstein–Barr virus-positive tumor treatment efficacy by doxorubicin. Cancer Lett. 2016, 376, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Mori, K.; Satoh, S.; Dansako, H.; Ikeda, M.; Kato, N. Anti-HCV activity of the Chinese medicinal fungus Cordyceps militaris. Biochem. Biophys. Res. Commun. 2014, 447, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K. Cordycepin: A bioactive metabolite of Cordyceps militaris and polyadenylation inhibitor with therapeutic potential against COVID-19. J. Biomol. Struct. Dyn. 2022, 40, 3745–3752. [Google Scholar] [CrossRef] [PubMed]
- Dubhashi, S.; Sinha, S.; Dwivedi, S.; Ghanekar, J.; Kadam, S.; Samant, P.; Datta, V.; Singh, S.; Chaudry, I.H.; Gurmet, P. Early Trends to Show the Efficacy of Cordyceps militaris in Mild to Moderate COVID Inflammation. Cureus 2023, 15, e43731. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.-A.; Lin, T.-H.; Wang, J.-S.; Chen, J.-J.; Hsu, W.-K.; Ying, L.-C.; Liang, Z.-C. The effects of Cordyceps militaris fruiting bodies in micturition and prostate size in benign prostatic hyperplasia patients: A pilot study. Pharmacol. Res.-Mod. Chin. Med. 2022, 4, 100143. [Google Scholar] [CrossRef]
- Yin, J.; Xin, X.; Weng, Y.; Gui, Z. Transcriptome-wide analysis reveals the progress of Cordyceps militaris subculture degeneration. PLoS ONE 2017, 12, e0186279. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-J.; Deng, C.-H.; Zhang, L.-Y.; Hu, K.-H. Molecular analysis and biochemical characteristics of degenerated strains of Cordyceps militaris. Arch. Microbiol. 2017, 199, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Yao, X.; Huang, Y.; Qu, Q.; Shi, X.; Zhang, H.; Shi, X. Identification of cordycepin biosynthesis-related genes through de novo transcriptome assembly and analysis in Cordyceps cicadae. R. Soc. Open Sci. 2018, 5, 181247. [Google Scholar] [CrossRef]
- Kramer, G.J.; Nodwell, J.R. Chromosome level assembly and secondary metabolite potential of the parasitic fungus Cordyceps militaris. BMC Genom. 2017, 18, 912. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Y.; Liu, L.; Feng, J.; Zhang, T.; Qin, S.; Zhao, X.; Wang, C.; Li, D.; Han, W. Study of the whole genome, methylome and transcriptome of Cordyceps militaris. Sci. Rep. 2019, 9, 898. [Google Scholar] [CrossRef]
- Massonnet, M.; Morales-Cruz, A.; Minio, A.; Figueroa-Balderas, R.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Whole-genome resequencing and pan-transcriptome reconstruction highlight the impact of genomic structural variation on secondary metabolite gene clusters in the grapevine esca pathogen Phaeoacremonium minimum. Front. Microbiol. 2018, 9, 1784. [Google Scholar] [CrossRef]
- Brunner-Mendoza, C.; Reyes-Montes, M.d.R.; Moonjely, S.; Bidochka, M.J.; Toriello, C. A review on the genus Metarhizium as an entomopathogenic microbial biocontrol agent with emphasis on its use and utility in Mexico. Biocontrol. Sci. Technol. 2019, 29, 83–102. [Google Scholar] [CrossRef]
- Staats, C.C.; Junges, A.; Guedes, R.L.M.; Thompson, C.E.; de Morais, G.L.; Boldo, J.T.; de Almeida, L.G.P.; Andreis, F.C.; Gerber, A.L.; Sbaraini, N. Comparative genome analysis of entomopathogenic fungi reveals a complex set of secreted proteins. BMC Genom. 2014, 15, 822. [Google Scholar] [CrossRef] [PubMed]
- Qiang, G.; Kai, J.; Sheng-Hua, Y.; Yongjun, Z.; Guohua, X.; Yanfang, S.; Zhibing, D.; Xiao, H.; Xue-Qin, X.; Gang, Z.; et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar]
- Xiao, G.; Ying, S.-H.; Zheng, P.; Wang, Z.-L.; Zhang, S.; Xie, X.-Q.; Shang, Y.; St Leger, R.J.; Zhao, G.-P.; Wang, C. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [PubMed]
- Suparmin, A.; Kato, T.; Dohra, H.; Park, E.Y. Insight into cordycepin biosynthesis of Cordyceps militaris: Comparison between a liquid surface culture and a submerged culture through transcriptomic analysis. PLoS ONE 2017, 12, e0187052. [Google Scholar] [CrossRef] [PubMed]
- Thananusak, R.; Laoteng, K.; Raethong, N.; Koffas, M.; Vongsangnak, W. Dissecting metabolic regulation in mycelial growth and fruiting body developmental stages of Cordyceps militaris through integrative transcriptome analysis. Biotechnol. Bioprocess Eng. 2023, 28, 406–418. [Google Scholar] [CrossRef]
- Qin, P.; Wang, Z.; Lu, D.; Kang, H.; Li, G.; Guo, R.; Zhao, Y.; Han, R.; Ji, B.; Zeng, Y. Neutral lipid content in lipid droplets: Potential biomarker of cordycepin accumulation in cordycepin-producing fungi. Molecules 2019, 24, 3363. [Google Scholar] [CrossRef]
- Lv, G.; Zhu, Y.; Cheng, X.; Cao, Y.; Zeng, B.; Liu, X.; He, B. Transcriptomic Responses of Cordyceps militaris to Salt Treatment during Cordycepins Production. Front. Nutr. 2021, 8, 793795. [Google Scholar] [CrossRef]
- Raethong, N.; Laoteng, K.; Vongsangnak, W. Uncovering global metabolic response to cordycepin production in Cordyceps militaris through transcriptome and genome-scale network-driven analysis. Sci. Rep. 2018, 8, 9250. [Google Scholar] [CrossRef] [PubMed]
- Wongsa, B.; Raethong, N.; Chumnanpuen, P.; Wong-Ekkabut, J.; Laoteng, K.; Vongsangnak, W. Alternative metabolic routes in channeling xylose to cordycepin production of Cordyceps militaris identified by comparative transcriptome analysis. Genomics 2020, 112, 629–636. [Google Scholar] [CrossRef]
- Shendurse, A.; Khedkar, C. Glucose: Properties and analysis. In Encyclopedia of Food and Health; Academic Press: Oxford, UK, 2016; Volume 3, pp. 239–247. [Google Scholar]
- Lou, H.-W.; Zhao, Y.; Tang, H.-B.; Ye, Z.-W.; Wei, T.; Lin, J.-F.; Guo, L.-Q. Transcriptome analysis of Cordyceps militaris reveals genes associated with carotenoid synthesis and identification of the function of the Cmtns gene. Front. Microbiol. 2019, 10, 2105. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yu, G.; Chen, Y.; Jiang, S.; Wang, M.; Jin, Y.; Lan, X.; Liang, Y.; Sun, H. Genome-wide transcriptome and proteome analysis on different developmental stages of Cordyceps militaris. PLoS ONE 2012, 7, e51853. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, G.; Li, C.; Ling, J. Cordycepin and pentostatin biosynthesis gene identified through transcriptome and proteomics analysis of Cordyceps kyushuensis Kob. Microbiol. Res. 2019, 218, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Huang, C.; Cao, L.; Xie, C.; Han, R. Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in medicinal fungus Cordyceps militaris. Fungal Biol. 2011, 115, 265–274. [Google Scholar] [CrossRef]
- He, R.; Zhang, L.; Lan, J.; Mei, S.; Li, Y. Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris. Biology 2022, 11, 1535. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-X.; Xue, L.-N.; Wei, T.; Ye, Z.-W.; Li, X.-H.; Guo, L.-Q.; Lin, J.-F. Enhancement of ergothioneine production by discovering and regulating its metabolic pathway in Cordyceps militaris. Microb. Cell Factories 2022, 21, 169. [Google Scholar] [CrossRef]
- Choi, H.; Park, S.-W.; Oh, J.; Kim, C.S.; Sung, G.-H.; Sang, H. Efficient disruption of CmHk1 using CRISPR/Cas9 ribonucleoprotein delivery in Cordyceps militaris. FEMS Microbiol. Lett. 2023, 370, fnad072. [Google Scholar] [CrossRef]
- Zou, G.; Xiao, M.; Chai, S.; Zhu, Z.; Wang, Y.; Zhou, Z. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents. Microb. Biotechnol. 2021, 14, 2343–2355. [Google Scholar] [CrossRef]
- Cunningham, K.; Manson, W.; Spring, F.; Hutchinson, S. Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature 1950, 166, 949. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Bao, H.; Bau, T. Isolation and identification of one natural pigment from cultured Cordyceps militaris. Mycosystema 2010, 29, 777–781. [Google Scholar]
- Chen, C.; Bao, H.; Bau, T. Chemical composition analysis of cultured Cordyceps militaris. Food Sci. 2013, 34, 36–40. [Google Scholar]
- Dong, J.Z.; Wang, S.H.; Ai, X.R.; Yao, L.; Sun, Z.W.; Lei, C.; Wang, Y.; Wang, Q. Composition and characterization of cordyxanthins from Cordyceps militaris fruit bodies. J. Funct. Foods 2013, 5, 1450–1455. [Google Scholar] [CrossRef]
- Yang, T.; Guo, M.; Yang, H.; Guo, S.; Dong, C. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2016, 100, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, Q.; Zhang, J.; Liu, K.; Li, K.; Liu, G.; Dong, C. Comparative transcriptome analysis between a spontaneous albino mutant and its sibling strain of Cordyceps militaris in response to light stress. Front. Microbiol. 2018, 9, 1237. [Google Scholar] [CrossRef]
- Lian, T.; Dong, C.; Yang, T.; Sun, J. Three types of geranylgeranyl diphosphate synthases from the medicinal caterpillar fungus, Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms. 2014, 16, 115–124. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Yang, Y.; Wang, Y.; Dong, C. CmVVD is involved in fruiting body development and carotenoid production and the transcriptional linkage among three blue-light receptors in edible fungus Cordyceps militaris. Environ. Microbiol. 2020, 22, 466–482. [Google Scholar] [CrossRef]
- Shrestha, B.; Lee, W.-H.; Han, S.-K.; Sung, J.-M. Observations on some of the mycelial growth and pigmentation characteristics of Cordyceps militaris isolates. Mycobiology 2006, 34, 83–91. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limón, M. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Bovier, E.R.; Hammond, B.R. A randomized placebo-controlled study on the effects of lutein and zeaxanthin on visual processing speed in young healthy subjects. Arch. Biochem. Biophys. 2015, 572, 54–57. [Google Scholar] [CrossRef]
- Yang, T.; Dong, C. Photo morphogenesis and photo response of the blue-light receptor gene Cmwc-1 in different strains of Cordyceps militaris. FEMS Microbiol. Lett. 2014, 352, 190–197. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Yu-Xian, W.; Xin-Xin, T.; Yokoyama, W.; Jing, C.; Fang, W.; Cheng, P.; Jin-Lin, G. Overexpression of ribonucleotide reductase small subunit, RNRM, increases cordycepin biosynthesis in transformed Cordyceps militaris. Chin. J. Nat. Med. 2020, 18, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Takano, H. The regulatory mechanism underlying light-inducible production of carotenoids in nonphototrophic bacteria. Biosci. Biotechnol. Biochem. 2016, 80, 1264–1273. [Google Scholar] [CrossRef]
- Chen, A.; Wang, Y.; Shao, Y.; Huang, B. A novel technique for rejuvenation of degenerated caterpillar medicinal mushroom, Cordyceps militaris (Ascomycetes), a valued traditional Chinese medicine. Int. J. Med. Mushrooms. 2017, 19, 87–91. [Google Scholar] [CrossRef]
- Wang, C.; Butt, T.M.; Leger, R.J.S. Colony sectorization of Metarhizium anisopliae is a sign of ageing. Microbiology 2005, 151, 3223–3236. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.-L.; Qiu, X.-H.; Han, R.-C. Identification of the genes involved in the fruiting body production and cordycepin formation of Cordyceps militaris fungus. Mycobiology 2015, 43, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Li, M.; Kang, J.; He, J. A molecular genetic study on fruiting-body formation of Cordyceps militaris. Afr. J. Microbiol. Res. 2012, 6, 5215–5221. [Google Scholar]
- Wang, H.; Wei, J.; Lin, N.; Feng, A.; Chen, M.; Bao, D. Distribution of mating-type genes in fruiting and non-fruiting forms of Cordyceps militaris. Acta Edulis Fungi. 2010, 17, 1–4. [Google Scholar]
- Wang, H.; Cai, T.; Wei, J.; Feng, A.; Lin, N.; Bao, D.-P. Molecular markers to detect the formation of heterokaryon and homokaryon from asexual spores of the caterpillar medicinal mushroom, Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms. 2015, 17, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Zu, Z.; Wang, S.; Zhao, Y.; Fan, W.; Li, T. Integrated enzymes activity and transcriptome reveal the effect of exogenous melatonin on the strain degeneration of Cordyceps militaris. Front. Microbiol. 2023, 14, 1112035. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Yin, J.; Zhang, B.; Li, Z.; Zhao, S.; Gui, Z. Genome-wide analysis of DNA methylation in subcultured Cordyceps militaris. Arch. Microbiol. 2019, 201, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Homolka, L. Preservation of live cultures of Basidiomycetes—Recent methods. Fungal Biol. 2014, 118, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, F.; Liu, K.; Dong, C. Influence of strain preservation methods on fruiting body growth and metabolite production by the medicinal mushroom Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms. 2018, 20, 1003–1011. [Google Scholar] [CrossRef]
- Lin, Q.; Qiu, X.; Zheng, Z.; Xie, C.; Xu, Z.; Han, R. Characteristics of the degenerate strains of Cordyceps militaris. Mycosystema 2010, 29, 670–677. [Google Scholar]
- Hu, X.; Lv, A.; JN, U. Discussion of the degenerate strains of Cordyceps militaris. Edible Fungi China 2015, 34, 1–3. [Google Scholar]
- Wang, Y.-L.; Wang, Z.-X.; Liu, C.; Wang, S.-B.; Huang, B. Genome-wide analysis of DNA methylation in the sexual stage of the insect pathogenic fungus Cordyceps militaris. Fungal Biol. 2015, 119, 1246–1254. [Google Scholar] [CrossRef]
- Zhang, G.; Liang, Y. Improvement of fruiting body production in Cordyceps militaris by molecular assessment. Arch. Microbiol. 2013, 195, 579–585. [Google Scholar] [CrossRef]


| Strain | Size/Mb | Coverage/Fold | Assembly Level | GenBank Assembly Accession | Ref. |
|---|---|---|---|---|---|
| Cm01 | 32.2 | 147× | 33 scaffolds | GCA_000225605 | [38] |
| ATCC34164 | 33.6 | 180× | 7 chromosomes | GCA_008080495 | [41] |
| HN | 32.6 | 300× | 14 contigs | GCA_003332165 | [42] |
| Strain | Size/Mb | Coverage /Fold | %G + C Content | Protein- Coding Genes | NCBI Accession | Ref. |
|---|---|---|---|---|---|---|
| C. militaris Cm01 | 32.2 | 147× | 51.4 | 9684 | AEVU00000000 | [38] |
| M. robertsii ARSEF23 | 39.0 | 100× | 51.5 | 10582 | ADNJ00000000 | [46] |
| M. acridum CQMa102 | 38.1 | 107× | 50.0 | 9849 | ADNI00000000 | [45,47] |
| M. anisopliae E6 | 38.5 | 19× | 51.0 | 10817 | AZNF00000000 | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, G.; Tian, X.; Shang, Y.; Yan, H.; Yao, L.; Hu, Z. Research Progress in Understanding the Molecular Biology of Cordyceps militaris. Fermentation 2024, 10, 167. https://doi.org/10.3390/fermentation10030167
Wang L, Li G, Tian X, Shang Y, Yan H, Yao L, Hu Z. Research Progress in Understanding the Molecular Biology of Cordyceps militaris. Fermentation. 2024; 10(3):167. https://doi.org/10.3390/fermentation10030167
Chicago/Turabian StyleWang, Lihong, Ganghua Li, Xueqin Tian, Yitong Shang, Huanhuan Yan, Lihua Yao, and Zhihong Hu. 2024. "Research Progress in Understanding the Molecular Biology of Cordyceps militaris" Fermentation 10, no. 3: 167. https://doi.org/10.3390/fermentation10030167
APA StyleWang, L., Li, G., Tian, X., Shang, Y., Yan, H., Yao, L., & Hu, Z. (2024). Research Progress in Understanding the Molecular Biology of Cordyceps militaris. Fermentation, 10(3), 167. https://doi.org/10.3390/fermentation10030167





