Distinct Short-Term Response of Intracellular Amino Acids in Saccharomyces cerevisiae and Pichia pastoris to Oxidative and Reductive Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Organisms and Media
2.2. Chemostat Cultivation and Application of Oxidative Stress
2.3. Rapid Sampling, Quenching, and Extraction of Intracellular Metabolites
2.4. Analytical Methods
2.5. Elemental Balances, Statistical Evaluations
3. Results and Discussion
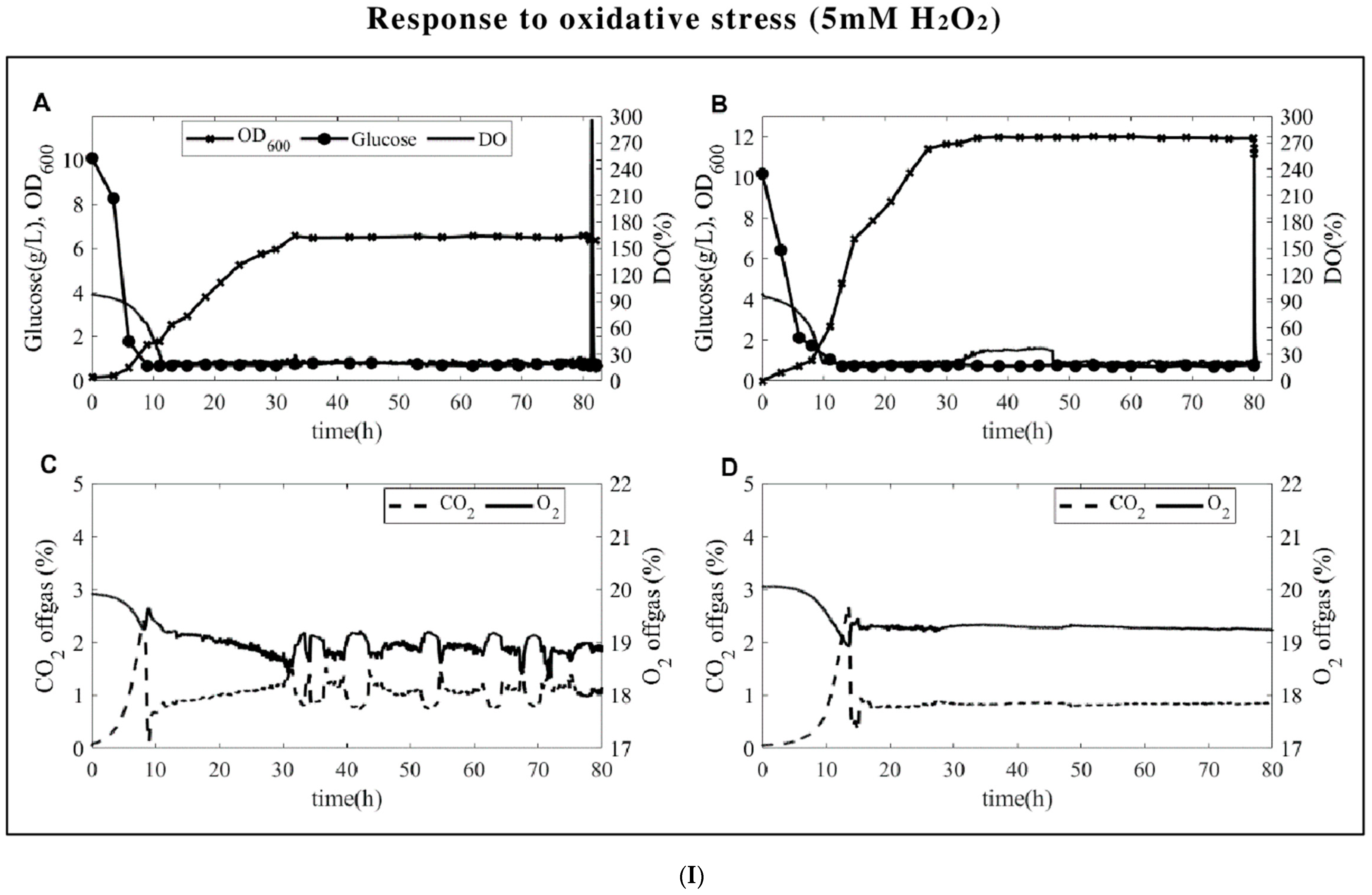
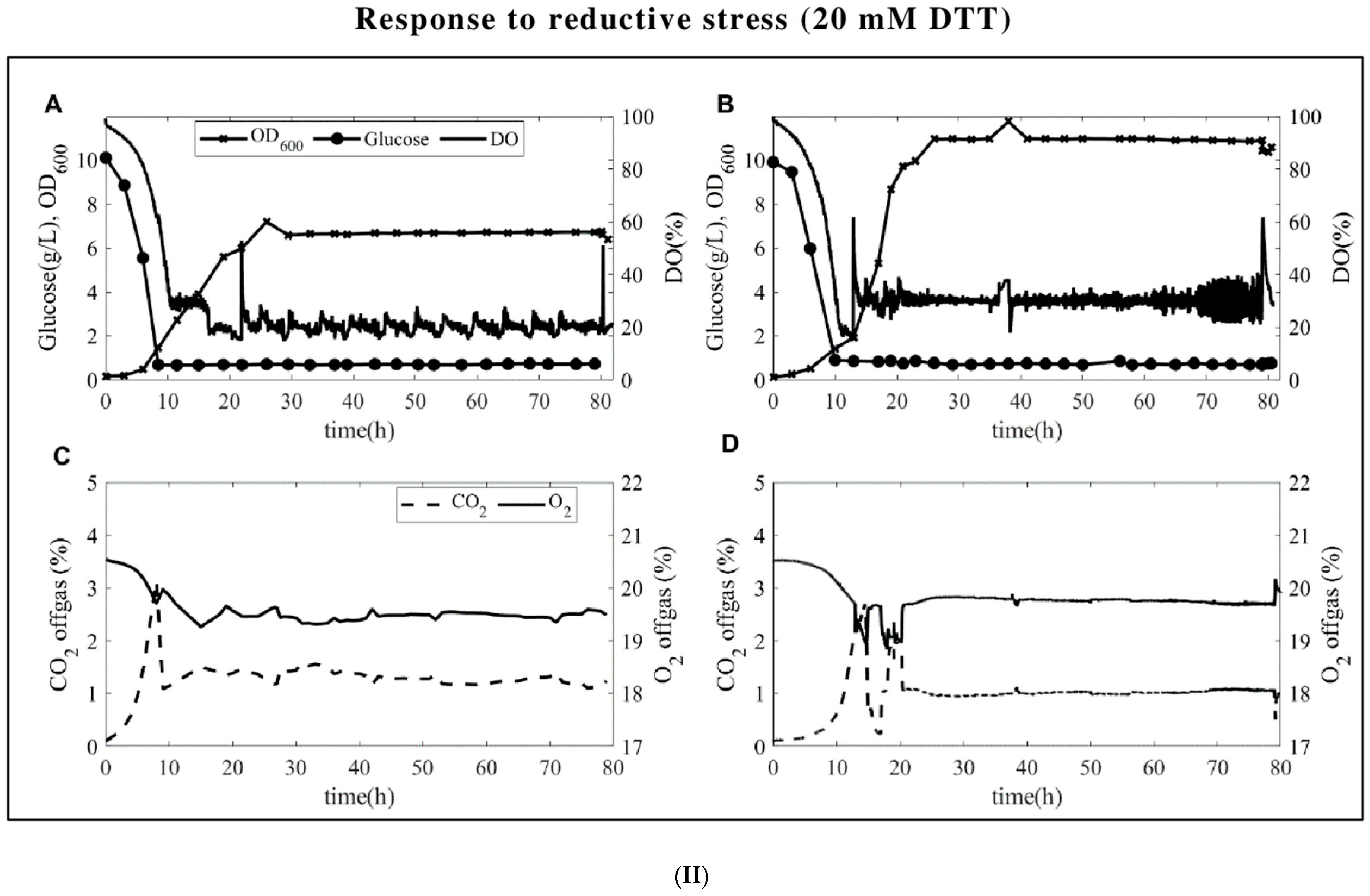
3.1. Fermentation Profiles
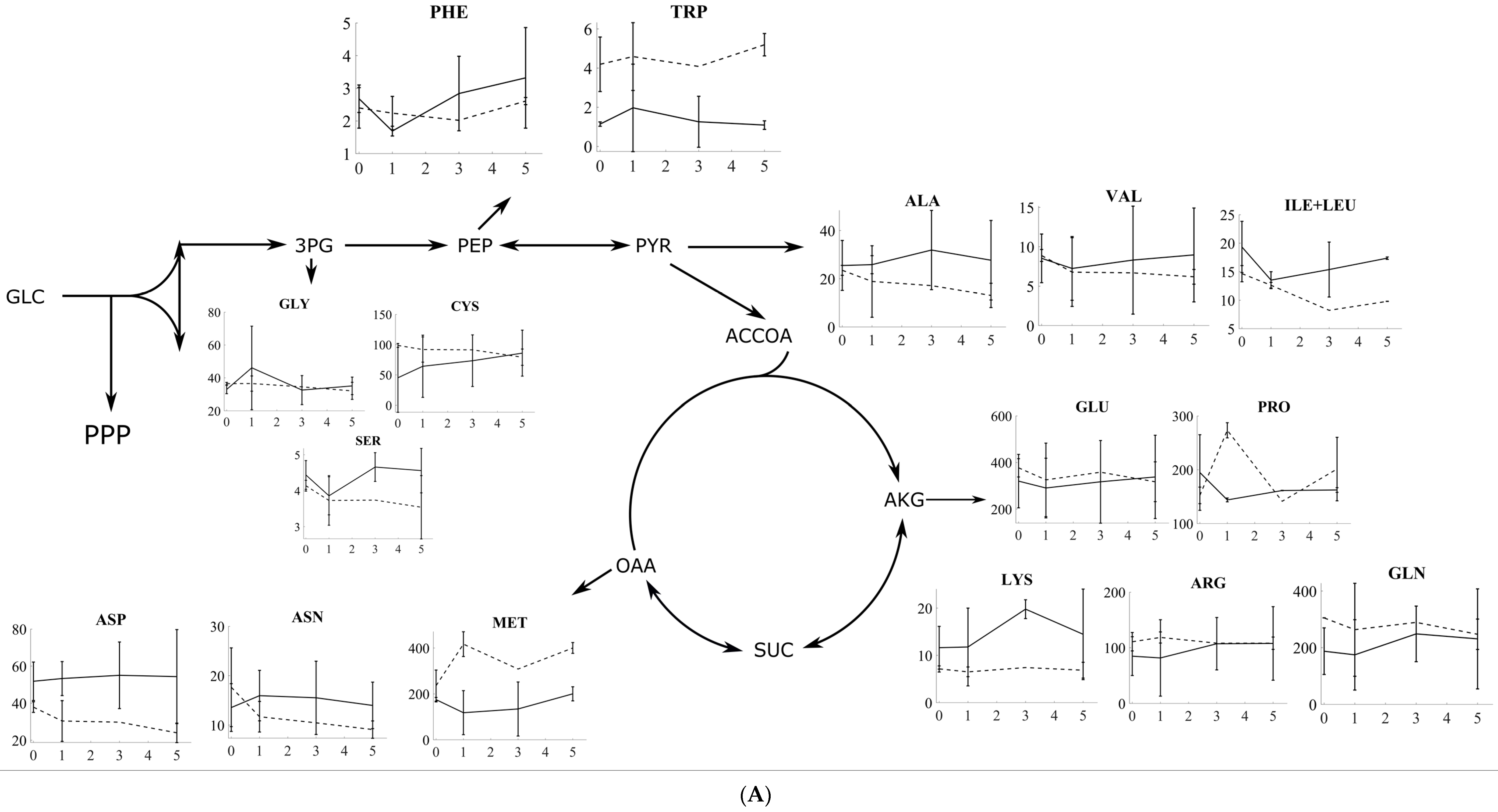
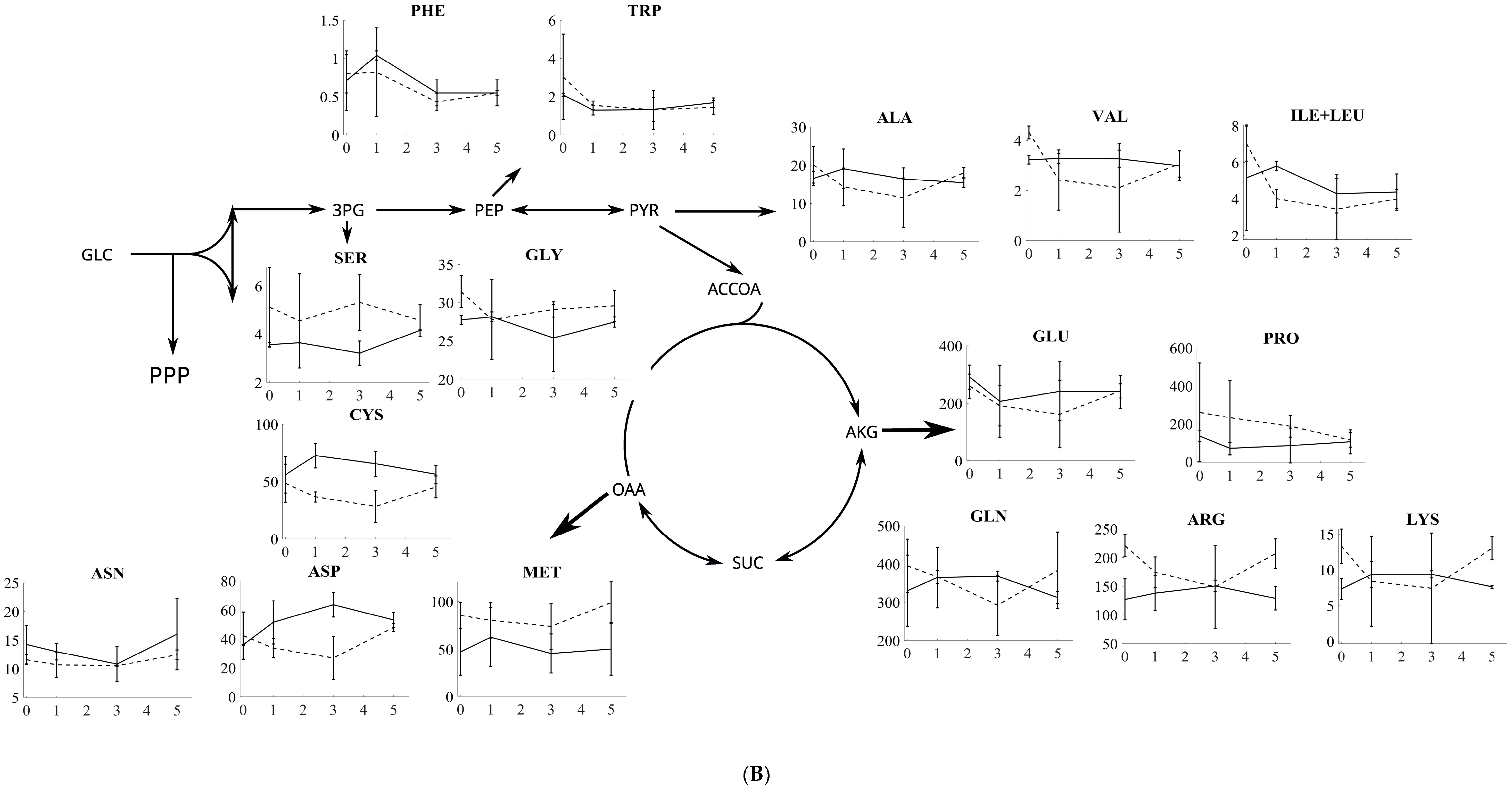
3.2. Amino Acid Profiling of S. cerevisiae and P. pastoris after Oxidative and Reductive Stress
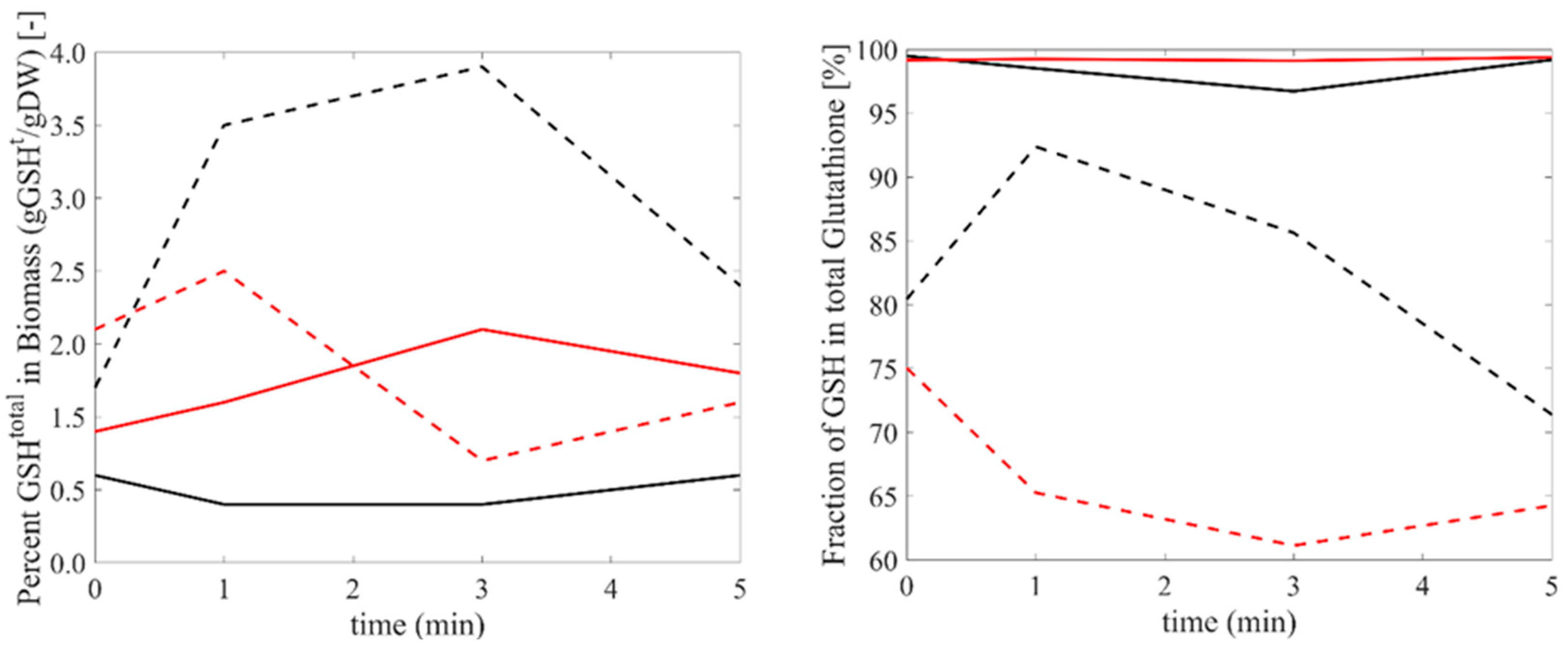
3.3. Glutathione Response to Oxidative and Reductive Stress
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mashego, M.R.; van Gulik, W.M.; Vinke, J.L.; Visser, D.; Heijnen, J.J. In Vivo Kinetics with Rapid Perturbation Experiments in Saccharomyces Cerevisiae Using a Second-Generation BioScope. Metab. Eng. 2006, 8, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Canelas, A.B.; Ten Pierick, A.; Ras, C.; Seifar, R.M.; Van Dam, J.C.; Van Gulik, W.M.; Heijnen, J.J. Quantitative Evaluation of Intracellular Metabolite Extraction Techniques for Yeast Metabolomics. Anal. Chem. 2009, 81, 7379–7389. [Google Scholar] [CrossRef] [PubMed]
- Nikerel, I.E.; Canelas, A.B.; Jol, S.J.; Verheijen, P.J.T.; Heijnen, J.J. Construction of Kinetic Models for Metabolic Reaction Networks: Lessons Learned in Analysing Short-Term Stimulus Response Data. Math. Comput. Model. Dyn. Syst. 2011, 17, 243–260. [Google Scholar] [CrossRef]
- Nikerel, I.E.; Verheijen, P.J.T.; van Gulik, W.M.; Heijnen, J.J. Model-Based Design of Superior Cell Factory: An Illustrative Example of Penicillium Chrysogenum. In Systems Metabolic Engineering; Springer: Dordrecht, The Netherlands, 2012; pp. 221–270. [Google Scholar]
- Nikerel, I.E. Managing Complexity of Cellular Systems: Theoretical Tools for Dynamic Modeling of Metabolic Reaction Networks. Ph.D. Thesis, Delft University of Technology, Delft, The Netherlands, 2009; 231p. [Google Scholar]
- Kresnowati, M.T.A.P.; Suarez-Mendez, C.M.; van Winden, W.A.; van Gulik, W.M.; Heijnen, J.J. Quantitative Physiological Study of the Fast Dynamics in the Intracellular pH of Saccharomyces Cerevisiae in Response to Glucose and Ethanol Pulses. Metab. Eng. 2008, 10, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Baumann, K.; Carnicer, M.; Dragosits, M.; Graf, A.B.; Stadlmann, J.; Jouhten, P.; Maaheimo, H.; Gasser, B.; Albiol, J.; Mattanovich, D.; et al. A Multi-Level Study of Recombinant Pichia Pastoris in Different Oxygen Conditions. BMC Syst. Biol. 2010, 4, 141. [Google Scholar] [CrossRef]
- Diezmann, S. Oxidative Stress Response and Adaptation to H2O2 in the Model Eukaryote Saccharomyces Cerevisiae and Its Human Pathogenic Relatives Candida Albicans and Candida Glabrata. Fungal Biol. Rev. 2014, 28, 126–136. [Google Scholar] [CrossRef]
- Korge, P.; Calmettes, G.; Weiss, J.N. Increased Reactive Oxygen Species Production during Reductive Stress: The Roles of Mitochondrial Glutathione and Thioredoxin Reductases. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 514–525. [Google Scholar] [CrossRef]
- Gasch, A.; Spellman, P. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Kurylenko, O.O.; Dmytruk, K.V.; Sibirny, A. Glutathione Metabolism in Yeasts and Construction of the Advanced Producers of This Tripeptide; Springer: Cham, Switzerland, 2019; ISBN 9783030211103. [Google Scholar]
- Chen, Y.; Yang, X.; Zhang, S.; Wang, X.; Guo, C.; Guo, X.; Xiao, D. Development of Saccharomyces Cerevisiae Producing Higher Levels of Sulfur Dioxide and Glutathione to Improve Beer Flavor Stability. Appl. Biochem. Biotechnol. 2012, 166, 402–413. [Google Scholar] [CrossRef]
- Schmacht, M.; Lorenz, E.; Senz, M. Microbial Production of Glutathione. World J. Microbiol. Biotechnol. 2017, 33, 106. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.X.; He, R.Z.; Xu, Y.; Yu, X.W. Oxidative Stress Tolerance Contributes to Heterologous Protein Production in Pichia Pastoris. Biotechnol. Biofuels 2021, 14, 160. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Cheng, J.H.; Sun, D.W. Metabolomic Analyses on Microbial Primary and Secondary Oxidative Stress Responses. Compr. Rev. Food Sci. Food Saf. 2021, 20, 5675–5697. [Google Scholar] [CrossRef] [PubMed]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; Van Dijken, J.P. Effect of Benzoic Acid on Metabolic Fluxes in Yeasts: A Continuous-culture Study on the Regulation of Respiration and Alcoholic Fermentation. Yeast 1992, 8, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Lange, H.C.; Heijnen, J.J. Statistical Reconciliation of the Elemental and Molecular Biomass Composition of Saccharomyces Cerevisiae. Biotechnol. Bioeng. 2001, 75, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Lange, H.C.; Eman, M.; Van Zuijlen, G.; Visser, D.; Van Dam, J.C.; Frank, J.; De Teixeira Mattos, M.J.; Heijnen, J.J. Improved Rapid Sampling for in Vivo Kinetics of Intracellular Metabolites in Saccharomyces Cerevisiae. Biotechnol. Bioeng. 2001, 75, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Pudlik, A.M.; Lolkema, J.S. Rerouting Citrate Metabolism in Lactococcus Lactis to Citrate-Driven Transamination. Appl. Environ. Microbiol. 2012, 78, 6665–6673. [Google Scholar] [CrossRef]
- Rollini, M.; Manzoni, M. Influence of Different Fermentation Parameters on Glutathione Volumetric Productivity by Saccharomyces Cerevisiae. Process Biochem. 2006, 41, 1501–1505. [Google Scholar] [CrossRef]
- Carnicer, M.; Ten Pierick, A.; Van Dam, J.; Heijnen, J.J.; Albiol, J.; Van Gulik, W.; Ferrer, P. Quantitative Metabolomics Analysis of Amino Acid Metabolism in Recombinant Pichia Pastoris under Different Oxygen Availability Conditions. Microb. Cell Fact. 2012, 11, 83. [Google Scholar] [CrossRef]
- van der Hijden, R.T.J.M.; Heijnen, J.J.; Hellinga, C.; Romein, B.; Luyben, K.C.A.M. Linear Constrain Relations in Biochemical Reaction Systems: I. Classification of the Calculability and the Balanceability of Conversion Rates. Biotechnol. Bioeng. 1994, 43, 3–10. [Google Scholar] [CrossRef]
- Trotter, E.W.; Grant, C.M. Thioredoxins Are Required for Protection against a Reductive Stress in the Yeast Saccharomyces Cerevisiae. Mol. Microbiol. 2002, 46, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Netto, L.E.S.; Stadtman, E.R. The Iron-Catalyzed Oxidation of Dithiothreitol Is a Biphasic Process: Hydrogen Peroxide Is Involved in the Initiation of a Free Radical Chain of Reactions. Arch. Biochem. Biophys. 1996, 333, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, J.; Liu, J.; Han, B. Transcriptome Analysis Reveals the Oxidative Stress Response in Saccharomyces Cerevisiae. RSC Adv. 2015, 5, 22923–22934. [Google Scholar] [CrossRef]
- Sha, W.; Martins, A.M.; Laubenbacher, R.; Mendes, P.; Shulaev, V. The Genome-Wide Early Temporal Response of Saccharomyces Cerevisiae to Oxidative Stress Induced by Cumene Hydroperoxide. PLoS ONE 2013, 8, e74939. [Google Scholar] [CrossRef]
- Christodoulou, D.; Kuehne, A.; Estermann, A.; Fuhrer, T.; Lang, P.; Sauer, U. Reserve Flux Capacity in the Pentose Phosphate Pathway by NADPH Binding Is Conserved across Kingdoms. iScience 2019, 19, 1133–1144. [Google Scholar] [CrossRef]
- Wang, B.; Liang, G.; Zhou, Q.; Xie, J.; Mo, Y. Combined Amino Acids Modulation with H2O2 Stress for Glutathione Overproduction in Candida Utilis. Afr. J. Biotechnol. 2010, 9, 5399–5406. [Google Scholar]
- Wei, G.Y.; Wang, D.H.; Chen, J. Overproduction of Glutathione by L-Cysteine Addition and a Temperature-Shift Strategy. Biotechnol. Bioprocess. Eng. 2008, 13, 347–353. [Google Scholar] [CrossRef]
- Wang, Z.; Tan, T.; Song, J. Effect of Amino Acids Addition and Feedback Control Strategies on the High-Cell-Density Cultivation of Saccharomyces Cerevisiae for Glutathione Production. Process Biochem. 2007, 42, 108–111. [Google Scholar] [CrossRef]
- Wen, S.; Zhang, T.; Tan, T. Maximizing Production of Glutathione by Amino Acid Modulation and High-Cell-Density Fed-Batch Culture of Saccharomyces Cerevisiae. Process Biochem. 2006, 41, 2424–2428. [Google Scholar] [CrossRef]
- Liang, G.B.; Du, G.C.; Chen, J. A Novel Strategy of Enhanced Glutathione Production in High Cell Density Cultivation of Candida Utilis-Cysteine Addition Combined with Dissolved Oxygen Controlling. Enzym. Microb. Technol. 2008, 42, 284–289. [Google Scholar] [CrossRef]
| Measured (Unbalanced) Specific Rates | ||||||
|---|---|---|---|---|---|---|
| - | - | C-Recovery | DoR-Recovery | |||
| S. cerevisiae | 1.046 ± 0.001 | 3.847 ± 0.004 | 2.450 ± 0.022 | 2.274 ± 0.029 | 100 ± 0.00 | 100 ± 0.01 |
| P. pastoris | 0.848 ± 0.010 | 3.713 ± 0.019 | 1.633 ± 0.081 | 1.438 ± 0.167 | 104 ± 0.01 | 104 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Şirin Kaya, B.; Nikerel, E. Distinct Short-Term Response of Intracellular Amino Acids in Saccharomyces cerevisiae and Pichia pastoris to Oxidative and Reductive Stress. Fermentation 2024, 10, 166. https://doi.org/10.3390/fermentation10030166
Şirin Kaya B, Nikerel E. Distinct Short-Term Response of Intracellular Amino Acids in Saccharomyces cerevisiae and Pichia pastoris to Oxidative and Reductive Stress. Fermentation. 2024; 10(3):166. https://doi.org/10.3390/fermentation10030166
Chicago/Turabian StyleŞirin Kaya, Burcu, and Emrah Nikerel. 2024. "Distinct Short-Term Response of Intracellular Amino Acids in Saccharomyces cerevisiae and Pichia pastoris to Oxidative and Reductive Stress" Fermentation 10, no. 3: 166. https://doi.org/10.3390/fermentation10030166
APA StyleŞirin Kaya, B., & Nikerel, E. (2024). Distinct Short-Term Response of Intracellular Amino Acids in Saccharomyces cerevisiae and Pichia pastoris to Oxidative and Reductive Stress. Fermentation, 10(3), 166. https://doi.org/10.3390/fermentation10030166






