Abstract
Leghemoglobin (LegH) is a plant-derived hemoglobin that can be used as a food additive to confer red color and meat flavor to plant-based meat products. Although LegH has been expressed in Saccharomyces cerevisiae, the productivity is low at the shaking-flask level, and the downstream process of purification is complicated. Herein, the intracellular expression of LegH reached 151.2 mg/L through initial promoter modification. Then, the fermentation strategy was optimized, and the titer of LegH reached 544.8 mg/L (5.2 mg/L/OD600 per unit yield) in the two-stage fed-batch fermentation in a 5-L fermenter. After the modification of signal peptide and knockout of proteases, the secretory expression of LegH was achieved in recombinant S. cerevisiae, and the final secretory titer of LegH reached 88.5 mg/L at the 5-L fermenter level. Based on the results of this study, the secreted LegH can be widely applied in the fields of food processing and biocatalysis in the future.
1. Introduction
Leghemoglobin (LegH) is a hemoprotein produced by legumes infected with Rhizobium bacteria [1]. Due to the biochemical properties of LegH, including the transport and storage of oxygen, and the red color of legume nodules, it was applied in the field of food processing [2,3]. In 2016, Impossible Food™ developed LegH as a food additive to simulate the meaty color and flavor and obtained the permission from the United States Food and Drug Administration (FDA) for the production of commercial Impossible Burger™ hamburgers [4].
Currently, LegH is mainly extracted from soybeans, but the preparation procedure is complex and time-consuming due to the low concentration of LegH in soybeans [5]. With the development of synthetic biology, the utilization of microbial fermentation to produce LegH has attracted widespread attention. Although the titer of LegH synthesized by E. coli could reach 170 mg/L at the shaking-flask level [6], the complicated renaturing steps of inclusion bodies and the risk of endotoxins limited the food-grade production and application on a large scale [7,8]. Recently, the hemoglobin produced in C. glutamicum is up to approximately 20% of total protein [9], but the yield is not high and the process of downstream purification is complicated. In addition, the highest yield of LegH in microorganisms has been achieved in the methanol-induced K. phaffii, reaching up to 3.5 g/L, including both the target protein and its degradation products [10]. However, the application of recombinant proteins from K. phaffii in food and pharmaceutical fields requires detailed scrutiny, and the strict purification and safety assessments are demanding. Moreover, the food additive “LegH Prep”, which is synthesized in K. phaffii, only has a purity of 65% [2].
Due to its clear genetic background, fast growth, ease of cultivation, and safety, S. cerevisiae has been extensively utilized as a host for synthetic biology [11]. Moreover, S. cerevisiae has shown great advantages in the synthesis of pharmaceutical proteins and has a promising potential for the large-scale production of many other high-value-added proteins in the future [12]. In a previous study, LegH was synthesized in S. cerevisiae with a titer up to 124.3 ± 6.6 mg/L at the shaking-flask level [13]; its productivity is low and not suitable for industrial large-scale production. Obviously, secretory expression can simplify the downstream steps of purification and eliminate the residual proteins from the host. Currently, the secretory expression of LegH has not been achieved in S. cerevisiae.
It has been found that the upstream activator sequence (UAS) works cooperatively with the core promoter, and the strength of the synthetic promoter constructed by replacing the core promoter region of the PGAL1 is two-fold higher than that of the original promoter [14]. In addition, an appropriate increase in the number of UAS elements can enhance the strength of the promoters [15,16]. In the GAL regulatory system, the PGAL1 promoter is regulated by the transcriptional activator Gal4p and the transcriptional repressor Gal80p. It has been proven that the knockout of GAL80 can improve the expressional intensity of the PGAL1 promoter [17,18].
As for the secretory expression in S. cerevisiae, 11 pre-peptides with high secretion efficiency have been mined [19]. In a previous study, it was shown that the combination of different pro-peptides and pre-peptides can produce hybrid signal peptides with different secretion efficiencies [14]. The secretion of human hemoglobin has been achieved by knocking out the proteases Pep4p and Vps10p in S. cerevisiae, but the expressional level is extremely low, and the target protein can only be detected after the supernatant is concentrated 360-fold [20]. Therefore, to improve the secretory production of heterologous proteins, it is not only necessary to find the appropriate secretion signal peptide but also to maintain the stability of target proteins by knocking out special proteases related to degradation.
In this study, we started with the strain GAL1-LegH (CEN.PK2-1C strain harboring plasmid pESC-PGAL1-LegH) [13] and then optimized the elements in the PGAL1 promoter. To reduce the reliance of the expression on galactose and improve the intensity of PGAL1, the gene GAL80 was knocked out. Next, the fermentation conditions were optimized at the shaking-flask level, and a feasible two-stage fed-batch strategy for high-cell-density fermentation was developed to efficiently synthesize LegH at the 5-L fermenter level. In the following study, to simplify the purification procedure, an appropriate protein secretory signal peptide for LegH was identified, and the associated proteases were knocked out in the new engineered recombinant S. cerevisiae strain. Finally, based on all the effective strategies, the highest secretory titer of LegH was achieved for S. cerevisiae on a large scale.
2. Materials and Methods
2.1. Strains and Culture Conditions
E. coli DH5α was used as the cloning host and cultivated at 37 °C and 220 rpm in Luria–Bertani (LB) medium with 100 mg/L of ampicillin or 50 mg/L of kanamycin. LegH was expressed in the S. cerevisiae CEN.PK2-1C strain, the genotypes of which are provided in Table S1 for its derivative strains. YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) was used to cultivate S. cerevisiae without special needs. Synthetic dropout medium (YNB medium: 6.7 g/L of yeast nitrogen base without amino acids, 20 g/L of glucose, 50 mg/L of leucine, 50 mg/L of histidine, and 50 mg/L of tryptophan) was used to cultivate recombinant S. cerevisiae strains harboring the pESC-URA plasmids. YPD-F medium derived by adding 1 g of 5-fluoroorotic to YPD medium was used to remove the plasmid in the recombinant S. cerevisiae strains.
2.2. The Construction of Recombinant Plasmids
The information for all plasmids constructed in this study is detailed in Table S1. All of the primers used to construct recombinant plasmids are listed in Table S2, the elements of the promoters are listed in Table S3, and the hybrid signal peptides are listed in Table S4. Using the genome of S. cerevisiae CEN.PK2-1C as a template, the sequence of the target promoter element was amplified via PCR, and the vector framework was amplified by using pESC-PGAL1-LegH as a template. When designing the primers for the signal peptide recombinant plasmid, the signal peptide sequences were inserted into the 5’ ends of the upstream and downstream primers. Then, pESC-PGAL1-LegH was used as the template to amplify the linear fragment that replaced the original signal peptide. The purified DNA fragments were assembled via Gibson assembly [21] to obtain the target recombinant plasmids.
To construct plasmids for LegH expression with one to four tandem repeats of UASGAL1 elements to the core promoter, the UASGAL1 fragment was obtained from pESC-PGAL1-LegH using primers, followed by cutting the pESC-PGAL1-LegH at the restriction endonuclease site EcoR I, and a long fragment containing an overlap to the UASGAL1 fragment was obtained after incubation for 1 h at 37 °C. The plasmids that randomly assemble different numbers of UASGAL1 fragments were then obtained by assembling these two fragments via Gibson assembly [21].
2.3. The Construction of Engineered Strains
Applying CRISPR/Cas9 system for gene editing, the pML104 plasmid was used to knock down the GAL80, MCA1, PEP4, and VPS10 genes [22]. The sgRNA targeting sequences of target genes were designed according to CHOPCHOP (http://chopchop.cbu.uib.no/ (accessed on August 9, 2023)). The S. cerevisiae S288C genome database from SGD (https://www.yeastgenome.org/ (accessed on October 24, 2022)) served as the basis for the creation of homologous fragments. Using the S. cerevisiae genome as a template, the upstream and downstream homology arms of target genes were amplified and assembled. The purified fusion DNA fragments of target genes and the pML104 plasmid containing the sgRNA sequence of target genes were transformed into S. cerevisiae strains. The donors were introduced into S. cerevisiae via the LiAc/SS carrier DNA/PEG method [23].
2.4. The Culture Conditions for LegH Expression at the Shaking-Flask Level
The engineered strain was cultivated on YNB plates at 30 °C for 2 days, and single colonies were then selected and inoculated into 3 mL of YNB medium, which was then shaken for 18 h at 220 rpm. After that, the seed solution was moved to a 250 mL shaking-flask with 50 mL of YPD medium at an inoculum volume of 1%. And the flask was then incubated for 90 h at 30 °C at 220 rpm. Next, 2% Galactose was added as the inducer for LegH expression at 18 h, and 2% glucose was added for the GAL80 knockout strain.
YPD medium, Li medium [24], Whang J medium [25], and Van Hoek medium [26] were selected for the single-factor optimization of the medium. Different carbon sources (glucose, glycerol, ethanol, and sorbitol) and nitrogen sources (peptone, corn steep liquor, beef extract, and ammonium sulfate) were selected for the single-factor optimization of media composition, all of which were added at a final concentration of 20 g/L. Furthermore, to determine the appropriate initial addition amount, several concentrations of hemin (2.5, 5.0, 10.0, and 20.0 mg/L) were added to the modified YPD medium. The single-factor optimizations of the fermentation temperature were conducted at 25 °C, 30 °C, and 35 °C.
2.5. The Culture Conditions for LegH Expression at the 5-L Fermenter Level
The engineered strain was cultivated on YNB plates at 30 °C for 2 days, and single colonies were then selected and inoculated into 3 mL of YNB medium, which was then shaken for 18 h at 220 rpm. Then, the primary seed cultures were inoculated at 1% inoculum into three 250 mL shaking-flasks containing 100 mL of YNB medium and incubated at 30 °C and 220 rpm for 18 h.
The secondary seed culture (300 mL) was inoculated into a 5 L bioreactor (T&J Bioengineering, Shanghai, China) containing 2.7 L of YPD medium. The temperature was controlled at 30 °C, and the pH was maintained at 5.5 using 50% NH4OH. The dissolved oxygen (DO) was maintained at 30% of air saturation by varying the agitation speed from 300 to 600 rpm, while the airflow rate was fixed at 3 L/min. The final concentrations of 20.0 g/L of inducer (glucose or galactose) and 5 mg/L of hemin were added to the medium at 18 h.
To achieve high-cell-density fermentation, a two-stage feeding strategy was used. In order to maintain rapid cell development, 500 g/L of glucose and 120 g/L of yeast extract were fed in the beginning. In order to support efficient LegH expression, the solution was changed to 300 g/L glucose and 32 mg/L hemin during the second stage.
2.6. The Methods for LegH Purification
LegH was purified using His-Affinity (Ni-NTA-agarose) gravity columns. Yeast cells were collected and resuspended in 50 mL of 20 mM phosphate-buffered saline buffer (PBS, pH 7.4). The cells were crushed using a high-pressure cell crusher instrument (Union Biotech, Shanghai, China) at 4 °C using 700–800 bar. After breaking, the broken cells were collected for centrifugation, and the supernatant obtained from centrifugation was used as the crude protein solution. For secreted LegH, the fermentation supernatant is the crude protein solution. The washing buffer and elution buffer used were 50 mM imidazole in 20 mM PBS buffer and 1 M imidazole in 20 mM PBS buffer, respectively.
2.7. Analytical Methods
The cell density was measured at 600 nm using a spectrophotometer. The concentrations of glucose and ethanol were analyzed using an M100 biosensor analyzer (Sieman Bio-Medical Solutions Company, Shenzhen, China). The nucleic acid fragments were separated using a DYY-6C electrophoresis apparatus (Beijing Liuyi Electrophoresis Factory, Beijing, China). The expression of LegH was detected using a protein electrophoresis instrument (Thermo Fisher, Waltham, MA, USA). The concentration of proteins was measured using the Bradford protein assay kit (Beyotime Biotech, Shanghai, China). The software of Image Lab was used to analyze the percentage of target bands. All data were collected from three independent clones, and significance analysis was performed using GraphPad Prism8.
3. Results and Discussion
3.1. The Optimization of PGAL1 Promoter Elements
In previous research, the original PGAL1 promoter was used to induce the expression of LegH [13]. However, the expressional level of target proteins can be further enhanced via the modification of the PGAL1 promoter [14,18]. To improve the expression of LegH, the elements of the PGAL1 promoter were modified to enhance its strength (Figure 1A). The CRMGAL1-CORETEF1-LegH, CRMGAL1-CORETDH3-LegH, and CRMGAL1-COREGAL7-LegH strains with different hybrid promoters were constructed by replacing the PGAL1 core promoter with CORETEF1/CORETDH3/COREGAL7 (the core promoter of the PTEF1/PTDH3/PGAL7). However, the titers of LegH were significantly decreased by 77.6% in the CRMGAL1-CORETEF1-LegH strain (29.0 mg/L), 29.0% in the CRMGAL1-CORETDH3-LegH strain (91.8 mg/L), and 41.8% in the CRMGAL1-COREGAL7-LegH strain (75.2 mg/L), respectively (Figure 1B). Thus, the increase in the UAS numbers was alternatively attempted [15,27]. One to three tandem repeats of UASGAL1 were fused to the original PGAL1 promoter with one UASGAL1, and three strains were generated (GAL1-UAS × 2-LegH strain, GAL1-UAS × 3-LegH strain, and GAL1-UAS × 4-LegH strain) (Figure 1C). Unfortunately, the results were also not positive (Figure 1D). Thus, the original PGAL1 promoter was employed to control the expression of LegH.
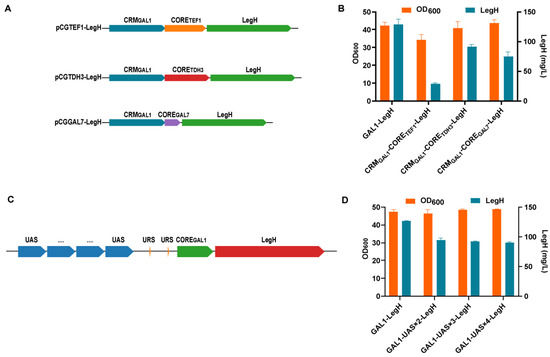
Figure 1.
The engineered strains with different hybrid promoters. (A) The structure of the hybrid promoter. CRM: upstream cis-regulatory module; CORE: core promoter sequence. (B) The expressional level of LegH in recombinant strains with different core regions. (C) A schematic diagram of the heterozygous promoters. (D) The expression of LegH in recombinant strains with different tandem repeats of UASGAL1.
Besides the modification of the PGAL1 promoter, knocking out the GAL80 gene to remove the inhibition of Gal4p (the transcriptional activator of the PGAL1 promoter) is a useful strategy to increase the strength of PGAL1 [17,28]. Although the titer of LegH decreased by 36.5% in the GAL80 knockout strain in the previous study [13], the LegH expression was elevated by 20.3% (151.2 mg/L) after the knockout of the GAL80 gene (GAL1-LegH-ΔGAL80 strain) in this study (Figure 2). This result may be due to the addition of 2% glucose during the fermentation process, and this strategy was applied to obtain a higher titer of LegH.
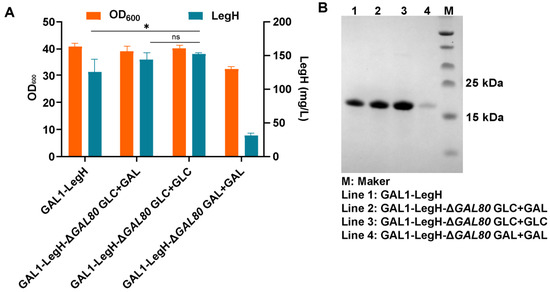
Figure 2.
The expression of LegH in GAL80 knockout strains: (A) the titer of LegH under different addition of carbon sources; (B) the results of SDS-PAGE. “*” p < 0.05 vs. compared group; “ns” stands for “not significant”.
In the previous study, the secretory expression of β-glucosidase under the control of PGAL1 in the GAL80 deletion strain in the presence of galactose was slightly higher than that under glucose growth conditions [18]. Therefore, to explore the effect of galactose on LegH expression, three different combinations of carbon sources were performed: (i) GAL1-LegH-ΔGAL80 GLC+GAL—2% glucose, 2% galactose (143.0 mg/L); (ii) GAL1-LegH-ΔGAL80 GLC+GLC—2% glucose, 2% glucose (151.2 mg/L); (iii) GAL1-LegH-ΔGAL80 GAL+GAL—2% galactose, 2% galactose (31.7 mg/L) (the former carbon source was added at the beginning of fermentation, and the latter carbon source was added at 18 h of fermentation) (Figure 2). The results showed that the expression of LegH (GAL1-LegH-ΔGAL80 strain) increased by 13.8% compared with the control (GAL1-LegH strain, 125.7 mg/L) when galactose and glucose were mixed (2% glucose + 2% galactose). When only glucose was used as a carbon source (2% glucose + 2% glucose), the expression of LegH (GAL1-LegH-ΔGAL80 strain) increased by 20.3%, while when only galactose was used as a carbon source (2% galactose + 2% galactose), the expression of LegH (GAL1-LegH-ΔGAL80 strain) decreased by 74.8%. Thus, we finally chose the carbon source addition of 2% glucose + 2% glucose for fermentation.
3.2. The Optimization of Fermentation Conditions at the Shaking-Flask Level
The fermentation medium was optimized based on the previously selected carbon source. At first, YPD medium, Li medium (trace-metal solution) [24], Whang J medium (a mixture of organic and inorganic nitrogen sources) [25], and van Hoek medium (inorganic salt medium with trace elements) [26] were chosen for the expression of LegH. According to the results, the highest titer of LegH (149.2 mg/L) was obtained in the YPD medium, while 99.6 mg/L and 112.2 mg/L were obtained in the Li medium and Whang J medium, respectively. In contrast, only 8.7 mg/L was produced in the van Hoek medium (Figure 3A). Therefore, the components of the YPD medium were further optimized. Firstly, four carbon sources, glucose, glycerol, ethanol and sorbitol, were selected for the single-factor optimization, and the concentrations of all carbon sources were set at 20 g/L. The results showed that there were certain effects of different carbon sources on the expression of LegH, and when glucose was used as the carbon source, the expression of LegH could reach up to 150.06 mg/L (Figure 3B). Next, based on the optimal carbon source, single-factor optimization was performed for peptone, corn steep liquor, beef extract, and (NH4)2SO4, and the concentrations of all nitrogen sources were set at 20 g/L. The results showed that the organic nitrogen sources were more effective at promoting the growth of the strain and the expression of target proteins compared to the inorganic nitrogen sources. Among all the tested organic nitrogen sources, the higher expressional level of LegH (150.63 mg/L) was achieved by peptone (Figure 3C).
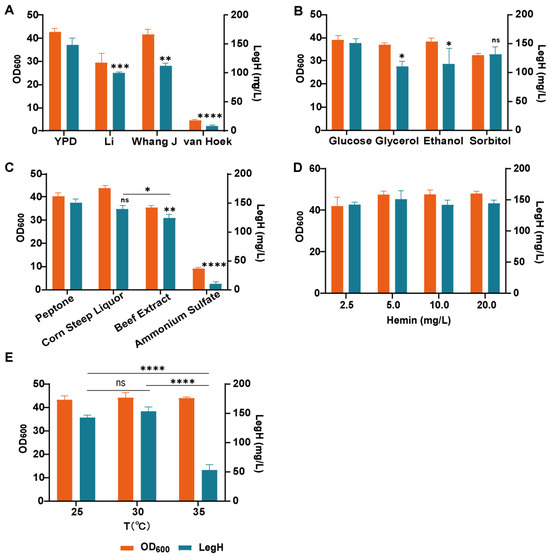
Figure 3.
The optimization of LegH expression at the shaking-flask level: (A) the effects of different media on LegH expression; (B) the effects of different carbon sources on LegH expression; (C) the effects of different nitrogen sources on LegH expression; (D) the effects of different hemin concentrations on LegH expression; (E) LegH was expressed at different temperatures. “*” p < 0.05 vs. compared group, “**” p < 0.01 vs. compared group, “***” p < 0.001 vs. compared group, and “****” p < 0.0001 vs. compared group; “ns” stands for “not significant”.
Hemin is an essential co-factor in the composition of heme-binding proteins. To efficiently synthesize LegH, it is necessary to add hemin to alleviate the insufficient intracellular hemin supply. The concentration of hemin addition was performed via single-factor optimization based on the optimal carbon and nitrogen sources. It can be seen that the concentrations of hemin (2.5, 5.0, 10.0, and 20.0 mg/L) had little effect on the expression of LegH (Figure 3D). Hence, considering the cost of supplementing hemin for large-scale industrial production, it is appropriate to control the concentration of hemin at 2.5 mg/L. Finally, the fermentation temperatures (25, 30, and 35 °C) were optimized. The expression of LegH decreased significantly at 35 °C, which showed that the higher temperature was unfavorable to the expression of LegH. There was no significant difference in the expression of LegH between 25 °C and 35 °C (Figure 3E). In order to reduce the use of condensed water in the fermenter, the fermentation temperature was controlled at 30 °C. Finally, the highest titer of LegH (153.3 mg/L) was achieved in modified YPD medium (20.0 g/L glucose, 20.0 g/L peptone, 10.0 g/L yeast extract, and 2.5 mg/L hemin) at 30 °C.
3.3. Intracellular Expression of LegH in High-Cell-Density Fermentation
Based on the optimized conditions at the shaking-flask level, batch fermentation was performed first. During batch fermentation, it can be seen that LegH started to be expressed after 12 h and reached the highest titer of 340.8 mg/L (4.8 mg/L/OD600) at 48 h (Figure 4A,B). To further increase the titer of LegH, fed-batch fermentations were carried out. DO-STAT, pH-STAT, and exponential feeding strategies were used, but it was found that high-cell-density fermentation [29] could not be achieved (Figures S1–S3). Thus, we adopted a two-stage fed-batch fermentation system to achieve high-cell-density fermentation.
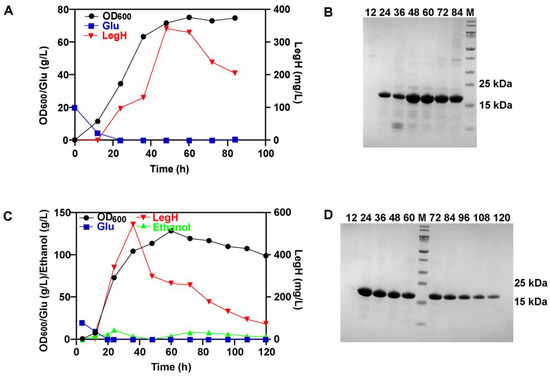
Figure 4.
The efficient synthesis of LegH in a 5-L fermenter: (A) the LegH obtained via batch fermentation; (B) the results of SDS-PAGE for batch fermentation; (C) the LegH obtained via fed-batch fermentation through two-stage fermentation; (D) the results of SDS-PAGE for fed-batch fermentation.
In the first stage, a feeding solution containing 500.0 g/L glucose and 120.0 g/L yeast extract was fed to the bioreactor at a constant rate (8.0 mL/h). During this stage, the GAL1-LegH-ΔGAL80 strain grew rapidly. After glucose consumption, the second stage started, and another feeding solution II containing 300.0 g/L glucose and 32.0 mg/L hemin was fed to the bioreactor using an index feeding strategy. During the second stage, the GAL1-LegH-ΔGAL80 strain gradually utilized ethanol instead of glucose, and LegH began to be expressed due to the derepression of the PGAL1 promoter by glucose. The results showed that the expression of LegH reached the highest titer of 544.8 mg/L at 36 h (5.2 mg/L/OD600) (Figure 4C,D). Based on this two-stage fed-batch strategy, high-cell-density fermentation was achieved, and the highest OD600 could reach 128.7 [29].
3.4. The Optimization of Signal Peptides for the Secretory Expression of LegH
The secretory expression is preferred for the industrial production of recombinant proteins because it simplifies the downstream process of purification. S. cerevisiae can only secrete a small amount of its own proteins during fermentation [30], and many signal peptides can be used to secrete recombinant proteins [31,32]. Therefore, it is possible to establish the strategy of LegH secretory expression through the rational design of signal peptides in S. cerevisiae. Firstly, the commercially applied MFα signal peptide was selected to realize the secretory expression of LegH, and the titer of secreted LegH could reach 15.7 mg/L (Figure 5A). Then, to obtain the optimal signal peptide for LegH secretory expression, three different strategies were selected to modify the MFα signal peptide: (i) the truncated or extended pro-peptide of α-factor signal peptide (the truncated or extended pro-peptides are determined by the secondary structure predicted by Jpred and the tertiary interactions predicted by the knob-socket model [14]) fused with the pre-Ost1 signal peptide or pre-α factor signal peptide; (ii) single-amino-acid substitutions located within the MFα signal peptide and the single-amino-acid substitutions obtained via directed evolution [33]; (iii) the pro-peptide of α-factor signal peptide fused with endogenous signal pre-peptides with strong secretory ability from S. cerevisiae [19].
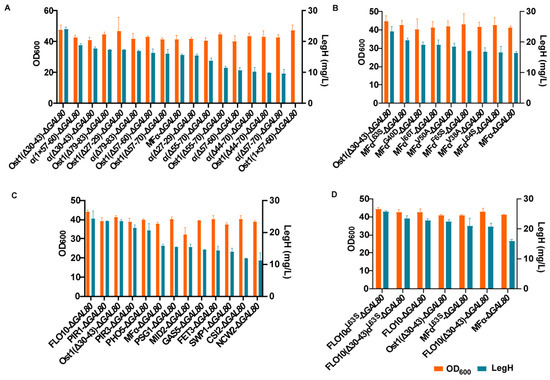
Figure 5.
Screening the suitable secreted signal peptides for LegH expression: (A) the secreted titer of LegH in the recombinant strains with different heterozygous signal peptides; (B) the secreted titer of LegH in recombinant strains with different modifications in the MF-α signal peptide; (C) the secreted titer of LegH in recombinant strains with different signal peptides obtained via the hybridization of endogenous pre-peptide with pro-α; (D) the secreted titer of LegH in recombinant strains combining the optimal pre-peptide and pro-peptide.
In strategy (i), the Ost1(Δ30-43)-ΔGAL80 strain (24.0 mg/L) had the highest secreted titer of LegH of all the constructed strains, which was 1.53-fold higher than the secreted titer obtained for the MFα-ΔGAL80 strain (Figure 5A). In strategy (ii), the secretion titer of LegH was 1.25-fold higher in the MFαL63S-ΔGAL80 strain (20.7 mg/L) than the titer in the MFα-ΔGAL80 strain (Figure 5B). Of the 11 hybrid signal peptides constructed via strategy (iii), the highest secreted titer of LegH was obtained for the FLO10-ΔGAL80 strain (24.4 mg/L), which was 1.54-fold higher than the titer for the MFα-ΔGAL80 strain (Figure 5C). In addition, the FLO10(Δ30-43)-ΔGAL80 strain (deletion of 44 to 70 residues in the pro-peptide of α-factor signal peptide) was constructed because the pro-α(Δ30-43) showed better secretory effect fused with both pre-Ost1 and pre-α. And the FLO10αL63S-ΔGAL80 strain was constructed by the FLO10-ΔGAL80 strain with the L63S single mutation in the pro-α. Finally, the FLO10(Δ30-43)αL63S-ΔGAL80 strain was constructed based on the FLO10(Δ30-43)-ΔGAL80 strain with the L63S single mutation in the pro-α.
After comparing the secretory effect in FLO10(Δ30-43)-ΔGAL80, FLO10αL63S-ΔGAL80, and FLO10(Δ30-43)αL63S-ΔGAL80 strains with the effect in Ost1(Δ30-43)-ΔGAL80, MFαL63S-ΔGAL80, and FLO10-ΔGAL80 strains, the results revealed that the highest secreted titer was achieved in the FLO10αL63S-ΔGAL80 strain (25.8 mg/L), which was 1.62-fold higher than that of the MFα-ΔGAL80 strain (Figure 5D). Furthermore, through the comparison of four engineered strains (FLO10-ΔGAL80, FLO10αL63S-ΔGAL80, FLO10(Δ30-43)-ΔGAL80 and FLO10(Δ30-43)αL63S-ΔGAL80 strains), it was found that the hybrid signal peptide obtained via the fusion of pro-α(Δ30-43) with pre-FLO10 had a negative effect on the secretory expression of LegH, whereas the L63S mutation partially rescued some of the inhibition caused by pro-α(Δ30-43).
3.5. Knockout of Degradation-Related Proteases to Increase LegH Expression
In previous studies, it was found that LegH was severely degraded during the secretion of K. phaffii [10], and the knockout of PEP4 reduced the degradation of porcine myoglobin secreted by the engineered K. phaffii [34]. Similarly, the biosynthesis of human hemoglobin could be elevated after PEP4 knockout in engineered S. cerevisiae [20]. Additionally, the knockout of VPS10 prevented the targeting of heterologous proteins to vacuoles [35], and knockout of MCA1 (YCA1) increased the aggregation of foreign proteins [36].
To reduce the degradation of secreted LegH, the α-ΔGAL80-ΔPEP4, α-ΔGAL80-ΔVPS10, and α-ΔGAL80-ΔMCA1 strains were constructed via the single knockdown of the PEP4, VPS10, and MCA1 (YCA1) genes, respectively. The secretory expression of LegH was significantly higher in the α-ΔGAL80-ΔPEP4 strain (24.8 mg/L), which was 1.69-fold higher than in the MFα-ΔGAL80 strain (Figure 6A). As the degradation of human hemoglobin could be further reduced by the double-knockout of the PEP4 and VPS10 genes [20], the α-ΔGAL80-ΔPEP4-ΔVPS10 strain was constructed (Figure 6B). The result demonstrates that knocking out VPS10 and PEP4 at the same time has no synergistic effect on the secreted expression of LegH, and the negative effect of VPS10 knockdown may be attributed to the increasing extracellular levels of carboxypeptidase Y (CPY) and other vacuolar hydrolases, which lead to the degradation of LegH [37,38,39].
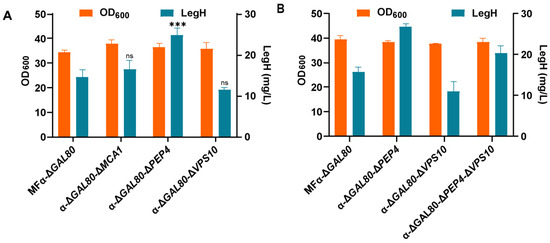
Figure 6.
Knockout of genes related to the degradation of heterologous proteins: (A) the effect of protease knockout on LegH secretion; (B) the effects of PEP4 and VPS10 knockout on LegH secretion. “***” p < 0.001 vs. compared group; “ns” stands for “not significant”.
3.6. The Secretory Expression of LegH at the Fermenter Level
The optimal signal peptide (pre-FLO10 with the single mutation L63S of the pro-α) and the optimal modified strain (ΔGAL80-ΔPEP4) were used to obtain the final secretory expression strain FLO10αL63S-ΔGAL80-ΔPEP4 (FL63S), which had a titer of LegH that was 1.78-fold higher (28.3 mg/L) than that of the MFα-ΔGAL80 strain (Figure 7A). Then, batch and fed-batch fermentations were carried out for high-cell-density culture in accordance with the optimal fermentation conditions. In batch fermentation, LegH was secreted from 12 h until it reached the highest titer of 47.7 mg/L at 48 h (Figure 7B). At 12 h, almost no protein was present in the fermentation supernatant, and after glucose was depleted at 24 h, the promoter PGAL1 was activated and LegH began to be gradually secreted (Figure 7C). Then, the concentration of total proteins still increased, but the secreted level of LegH began to decrease. Compared with the original MFα-ΔGAL80 strain, the secretory expression of LegH increased 3.26-fold.
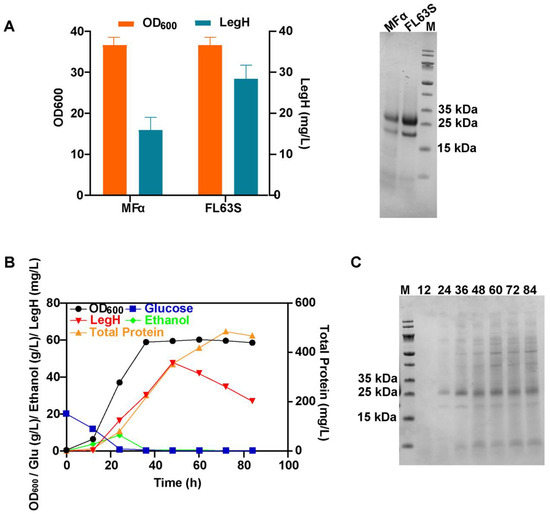
Figure 7.
The secreted LegH obtained in FL63S strain via batch fermentation at the 5-L fermenter level: (A) the LegH secreted by the FL63S and original MF-α strains at the shaking-flask level; (B) the results of batch fermentation at the 5-L fermenter level; (C) the results of SDS-PAGE for batch fermentations.
Finally, the two-stage fed-batch fermentation was performed for the best secretory strain (FL63S). It was observed that the titer of secreted LegH significantly increased from 12 to 60 h, and the highest titer reached 88.5 mg/L at 60 h (Figure 8A,B). The concentration of total supernatant proteins continued to rise in the late fermentation period, which may have been caused by autolysis [40].
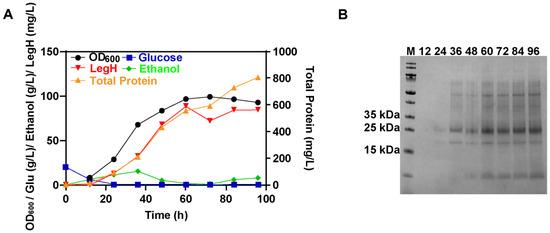
Figure 8.
The LegH obtained in FL63S strain via fed-batch fermentation at the 5-L fermenter level: (A) the results of fed-batch fermentation at the 5-L fermenter level; (B) the results of SDS-PAGE for fed-batch fermentations.
4. Conclusions
In this study, the efficient secretory production of LegH was achieved in S. cerevisiae. At first, by knocking out GAL80, the activity of PGAL1 was enhanced, resulting in an increasing level of LegH expression. Subsequently, a hybrid signal peptide, FLO10αL63S, was engineered to optimize the secretion of LegH by comparing various engineering strategies for signal peptides. To inhibit the degradation of LegH during fermentation, the protease PEP4 was knocked out. Finally, by employing a two-stage fed-batch strategy, the secretory expression of LegH reached 88.5 mg/L, which was the highest secretion of LegH in S. cerevisiae (Table 1). The results laid the foundation for the large-scale industrial production of LegH.

Table 1.
The latest studies of microbial synthetic leghemoglobin.
Apart from S. cerevisiae, K. phaffii is also regarded as one of the most promising current hosts for the large-scale synthesis of LegH. Compared to S. cerevisiae, which boasts a well-established safety record and extensive research on its impact on human metabolism, K. phaffii is considered to be a suitable host for LegH production due to its high capability for protein expression and rapid growth rate. The yield of LegH in K. phaffii has reached 3.5 g/L (including degraded bands), which is promising for future scaling up. Additionally, with the advancements in synthetic biology, other species of yeast or non-yeast microorganisms may be developed, offering new possibilities for the large-scale production of LegH.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation10030146/s1, Figure S1: The DO-STAT strategy used for fed-batch fermentation at 5-L fermenter level (S4 strain); Figure S2: The pH-STAT strategy used for fed-batch fermentation at 5-L fermenter level (S4 strain); Figure S3: The exponential feeding strategy used for fed-batch fermentation at 5-L fermenter level (S4 strain); Table S1: Strains and plasmids used in this study; Table S2: Primers used in this study; Table S3: Elements of the promoters used in this study; Table S4: Hybrid signal peptides used in this study.
Author Contributions
J.C., J.L., G.D., J.Z. and X.Z. conceived the idea and design for this work. Y.H. carried out the experiments and data analyses. X.Z. and Y.H. drafted the paper. X.Z. revised the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2021YFC2101400) and the National First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-08).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hargrove, M.S.; Barry, J.K.; Brucker, E.A.; Berry, M.B.; Phillips, G.N.; Olson, J.S.; Arredondo-Peter, R.; Dean, J.M.; Klucas, R.V.; Sarath, G. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J. Mol. Biol. 1997, 266, 1032–1042. [Google Scholar] [CrossRef]
- Fraser, R.Z.; Shitut, M.; Agrawal, P.; Mendes, O.; Klapholz, S. Safety evaluation of soy leghemoglobin protein preparation derived from Pichia pastoris, intended for use as a flavor catalyst in plant-based meat. Int. J. Toxicol. 2018, 37, 241–262. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, J.; Du, G.; Chen, J. Recent advances in the microbial synthesis of hemoglobin. Trends Biotechnol. 2021, 39, 286–297. [Google Scholar] [CrossRef]
- Simsa, R.; Yuen, J.; Stout, A.; Rubio, N.; Fogelstrand, P.; Kaplan, D.L. Extracellular heme proteins influence bovine myosatellite cell proliferation and the color of cell-based meat. Foods 2019, 8, 521. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, Y. Progress in the study of leghemoglobin. Acta Bot. Sin. 2000, 20, 684–689. [Google Scholar]
- Linjie, C.; Changlu, X.; Yue, S.; Gongnian, X.; Zhinan, X.; Peilian, W. Optimization of leghemoglobin expression conditions in Pichia pastoris. Microbiology 2022, 49, 2050–2061. [Google Scholar]
- Anwised, P.; Kabbua, T.; Temsiripong, T.; Dhiravisit, A.; Jitrapakdee, S.; Araki, T.; Yoneda, K.; Thammasirirak, S. Molecular cloning and expression of α-globin and β-globin genes from crocodile (Crocodylus siamensis). Protein J. 2013, 32, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Kabbua, T.; Anwised, P.; Boonmee, A.; Subedi, B.P.; Pierce, B.S.; Thammasirirak, S. Autoinduction, purification, and characterization of soluble α-globin chains of crocodile (Crocodylus siamensis) hemoglobin in Escherichia coli. Protein. Expr. Purif. 2014, 103, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Wng, M.; Shi, Z.; Gao, N.; Zhou, Y.; Ni, X.; Chen, J.; Liu, J.; Zhou, W.; Guo, X.; Xin, B.; et al. Sustainable and high-level microbial production of plant hemoglobin in Corynebacterium glutamicum. Biotechnol. Biofuels Bioprod. 2023, 16, 80. [Google Scholar]
- Shao, Y.; Xue, C.; Liu, W.; Zuo, S.; Wei, P.; Huang, L.; Lian, J.; Xu, Z. High-level secretory production of leghemoglobin in Pichia pastoris through enhanced globin expression and heme biosynthesis. Bioresour. Technol. 2022, 363, 127884. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yue, C.; Zhang, Y.; Li, Y.; Da, X.; Zhou, X.; Ye, L. Alleviation of the byproducts formation enables highly efficient biosynthesis of rosmarinic acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 2022, 70, 5077–5087. [Google Scholar] [CrossRef]
- Martínez, J.L.; Liu, L.; Petranovic, D.; Nielsen, J. Pharmaceutical protein production by yeast: Towards production of human blood proteins by microbial fermentation. Curr. Opin. Biotechnol. 2012, 23, 965–971. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, J.; Li, J.; Du, G.; Chen, J.; Wang, M.; Zhao, X. Systematic engineering of Saccharomyces cerevisiae for efficient synthesis of hemoglobins and myoglobins. Bioresour. Technol. 2023, 370, 128556. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, H.; Liang, Q.; Zhou, J.; Li, J.; Du, G.; Chen, J. Stepwise optimization of inducible expression system for the functional secretion of horseradish peroxidase in Saccharomyces cerevisiae. J. Agric. Food Chem. 2023, 71, 4059–4068. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wu, Y.; Deng, J.; Chen, N.; Zheng, Z.; Wei, Y.; Luo, X.; Keasling, J.D. Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 2020, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ma, L.; Fu, W.; Su, R.; Zhao, Y.; Deng, Y. Programmable synthetic upstream activating sequence library for fine-tuning gene expression levels in Saccharomyces cerevisiae. ACS Synth. Biol. 2022, 11, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Pannala, V.R.; Bhat, P.J.; Bhartiya, S.; Venkatesh, K.V. Systems biology of GAL regulon in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wu, Y.; Zheng, Z.; Chen, N.; Luo, X.; Tang, H.; Keasling, J.D. A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae. Microb. Cell Fact. 2021, 20, 202. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Liu, X.; Pan, Y.; Xiao, C.; Feng, Y.; Zheng, L.; Zhao, M.; Huang, M. Comprehensive analysis of signal peptides in Sccharomyces cerevisiae reveals features for efficient secretion. Adv. Sci. 2023, 10, e2203433. [Google Scholar] [CrossRef] [PubMed]
- Ishchuk, O.P.; Frost, A.T.; Muñiz-Paredes, F.; Matsumoto, S.; Laforge, N.; Eriksson, N.L.; Martínez, J.L.; Petranovic, D. Improved production of human hemoglobin in yeast by engineering hemoglobin degradation. Metab. Eng. 2021, 66, 259–267. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Laughery, M.F.; Hunter, T.; Brown, A.; Hoopes, J.; Ostbye, T.; Shumaker, T.; Wyrick, J.J. New vectors for simple and streamlined CRISPR–Cas9 genome editing in Saccharomyces cerevisiae. Yeast 2015, 32, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhou, P.; Chen, M.; Yu, H.; Ye, L. Spatiotemporal regulation of astaxanthin synthesis in S. cerevisiae. ACS Synth. Biol. 2022, 11, 2636–2649. [Google Scholar] [CrossRef] [PubMed]
- Whang, J.; Ahn, J.; Chun, C.S.; Son, Y.J.; Lee, H.; Choi, E.S. Efficient, galactose-free production of Candida antarctica lipase B by GAL10 promoter in Δgal80 mutant of Saccharomyces cerevisiae. Process Biochem. 2009, 44, 1190–1192. [Google Scholar] [CrossRef]
- van Hoek, P.; de Hulster, E.; van Dijken, J.P.; Pronk, J.T. Fermentative capacity in high-cell-density fed-batch cultures of baker’s yeast. Biotechnol. Bioeng. 2000, 68, 517–523. [Google Scholar] [CrossRef]
- Blazeck, J.; Garg, R.; Reed, B.; Alper, H.S. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 2012, 109, 2884–2895. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Xie, W.; Yao, Z.; Zhu, Y.; Ye, L.; Yu, H. Development of a temperature-responsive yeast cell factory using engineered Gal4 as a protein switch. Biotechnol. Bioeng. 2018, 115, 1321–1330. [Google Scholar] [CrossRef]
- Xiong, Z.Q.; Guo, M.J.; Guo, Y.X.; Chu, J.; Zhuang, Y.P.; Zhang, S.L. Real-time viable-cell mass monitoring in high-cell-density fed-batch glutathione fermentation by Saccharomyces cerevisiae T65 in industrial complex medium. J. Biosci. Bioeng. 2008, 105, 409–413. [Google Scholar] [CrossRef]
- Delic, M.; Valli, M.; Graf, A.B.; Pfeffer, M.; Mattanovich, D.; Gasser, B. The secretory pathway: Exploring yeast diversity. FEMS Microbiol. Rev. 2013, 37, 872–914. [Google Scholar] [CrossRef]
- Son, S.H.; Kim, J.E.; Park, G.; Ko, Y.J.; Sung, B.H.; Seo, J.; Oh, S.S.; Lee, J.Y. Metabolite trafficking enables membrane-impermeable-terpene secretion by yeast. Nat. Commun. 2022, 13, 2605. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Chen, X.; Nielsen, J.; Petranovic, D.; Siewers, V. Expression of antibody fragments in Saccharomyces cerevisiae strains evolved for enhanced protein secretion. Microb. Cell Fact. 2021, 20, 134. [Google Scholar] [CrossRef]
- Ito, Y.; Ishigami, M.; Hashiba, N.; Nakamura, Y.; Terai, G.; Hasunuma, T.; Ishii, J.; Kondo, A. Avoiding entry into intracellular protein degradation pathways by signal mutations increases protein secretion in Pichia pastoris. Microb. Biotechnol. 2022, 15, 2364–2378. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhao, X.; Zhou, J.; Lu, W.; Li, J.; Chen, J.; Du, G. Biosynthesis of high-active hemoproteins by the efficient heme-supply Pichia Pastoris chassis. Adv. Sci. 2023, 10, e2302826. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, I.; Glick, B.S. Secretion of a foreign protein from budding yeasts is enhanced by cotranslational translocation and by suppression of vacuolar targeting. Microb. Cell Fact. 2014, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.E.C.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.A.; Stevens, T.H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 1996, 133, 529–541. [Google Scholar] [CrossRef]
- Westphal, V.; Marcusson, E.G.; Winther, J.R.; Emr, S.D.; van den Hazel, H.B. Multiple pathways for vacuolar sorting of yeast proteinase A. J. Biol. Chem. 1996, 271, 11865–11870. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, M.U.; Emr, S.D.; Winther, J.R. Ligand recognition and domain structure of Vps10p, a vacuolar protein sorting receptor in Saccharomyces cerevisiae. Eur. J. Biochem. 1999, 260, 461–469. [Google Scholar] [CrossRef]
- Wang, J.; Li, M.; Zheng, F.; Niu, C.; Liu, C.; Li, Q.; Sun, J. Cell wall polysaccharides: Before and after autolysis of brewer’s yeast. World J. Microbiol. Biotechnol. 2018, 34, 137. [Google Scholar] [CrossRef]
- Tian, T.; Wu, X.; Wu, P.; Lu, X.; Wang, Q.; Lin, Y.; Liu, C.; Zhou, J.; Yu, Y.; Lu, H. High-level expression of leghemoglobin in Kluyveromyces marxianus by remodeling the heme metabolism pathway. Front. Bioeng. Biotechnol. 2024, 11, 1329016. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).