Abstract
Plasmids are commonly used tools in microbiology and molecular biology and have important and wide-ranging applications in the study of gene function, protein expression, and compound synthesis. The complex relationship between necessary antibiotic addition, compatibility between multiple plasmids, and the growth burden of host bacteria has plagued the wider use of compatibility plasmids. In this study, we constructed the terminal polymerization pathway of PSA by exogenously expressing the neuA, neuD, and neuS genes after the knockdown of Eschesrichia coli BL21 (DE3). Duet series vectors were utilized to regulate the expression level of neuA, neuD, and neuS genes to study the gene expression level, plasmid copy number growth burden, pressure of antibiotic addition, stability of compatible plasmids, and the level of expression stability of exogenous genes, as well as the effect on the biological reaction process. The results showed that the three genes, neuA, neuD, and neuS, were enhanced in the recombinant strain E. coli NA-05, with low copy, medium copy, and high copy, respectively. The effect of PSA synthesis under standard antibiotic pressure was remarkable. The results of this thesis suggest the use of a Duet series of compatible expression vectors to achieve the stable existence and co-expression of multiple genes in recombinant bacteria, which is a good reason for further research.
1. Introduction
Polysialic acid (PSA) is a linear polyanionic polysaccharide with a degree of polymerization of 8–400, composed of N-acetylneuraminic acid (Neu5Ac), a monomer with α-2, 8 and/or α-2, 9 [1] glycosidic bonds [2], and a chain-proteinase-resistant macromolecule [3]; moreover, a glycoprotein component exists in some mammalian cells, which plays an important role in biological processes such as embryogenesis, neuronal cell growth, differentiation, intercellular regulation, and membrane transport [4,5]. Extensive research into PSA synthesis is currently based on long traditional monosaccharide metabolic pathways, and the inherent resistance to metabolic flow hinders efficient PSA synthesis [6]. There is a relative lack of research on the polymerization of sialic acid monomers into PSA, as well as a lack of in-depth understanding of its polymerization-associated genes and their intensity-regulating mechanisms, which has severely limited the development of PSA functions and the popularization of its applications. In this study, we used Escherichia coli BL21 (DE3) as the starting strain and first used CRISPR Cas9 to target and edit the host’s bacterial genome, synthesize key genes, and construct the PSA polymerization pathway. The level of the expression of key genes was regulated and correlated with polysialic acid production using expression vectors of different copy numbers, and the stability of the plasmids was characterized by measuring the intensity of the expression of resistance genes (Amp, Kan, and Chl) in vectors using quantitative fluorescence PCR.
Polysialic acid is found in higher animals and a few bacteria and is considered an ideal drug control and scaffold material due to its poor immunogenicity and biodegradability, and its good biocompatibility. However, to date there are no reports on the extraction of polysialic acid from animal body tissues. Currently, microbial fermentation with carbon-source substances, such as glucose, is the main production method for polysialic acids [6,7].
The biosynthetic pathway of PSA in E. coli has been intensively studied, including the synthesis of sialic acid monomers, the polymerization of PSA from sialic acid, and the transport of PSA to the cell surface, as shown in Figure 1. [8] In E. coli, the KPS gene cluster is involved in the biosynthesis, modification, and transport of the bacterial PSA chain. Neu5Ac is the precursor of PSA, and the KPS cluster includes three conserved regions [9,10,11]. The central region of KPS (region II) contains six key genes, namely, neuD, neuB, neuA, neuC, neuE, and neuS, which direct the biosynthesis, activation, and polymerization of Neu5Ac [12]. Neu5Ac synthase (neuB) [9,13] catalyzes the energetic reaction of ManNAc and phosphoenolpyruvate (PEP) to form Neu5Ac [14], and Neu5Ac aldolase (nanA) catalyzes the reversible cleavage of Neu5Ac to ManNAc and pyruvate [15,16,17]. N-acylneuraminate cytidylyltransferase (neuA), a bifunctional enzyme with cytidine 5-monophosphate-sialic acid synthase (CMP-Neu5Ac) [18,19], catalyzes the generation of free sialic acid to CMP-Neu5Ac, thus generating a candidate sialic acid donor for all known sialic acid transferases; neuD is a Neu5Ac 7-O- (or 9-O)-acetyltransferase that acetylates residues in the seven or nine positions of Neu5Ac [20]; and neuS encodes an α-2,8 sialyltransferase, responsible for polymerizing the homopolymer of Neu5Ac in an α-2,8 glycosidic bond to the end of adjacent Neu5Ac to form PSA. neuA, neuD, and neuS are key enzymes in the polysialic acid biosynthesis pathway [8,18,20].
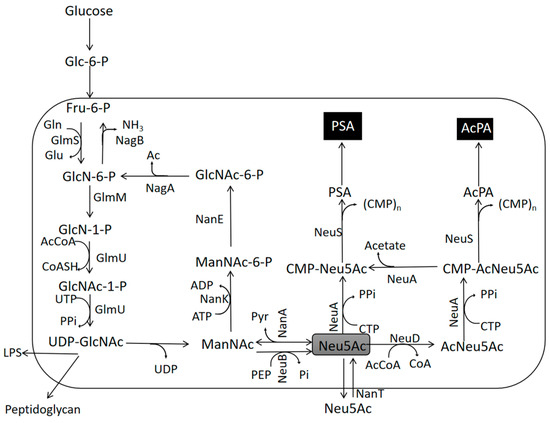
Figure 1.
Anabolic pathway of PSA synthesis using glucose as a substrate. NanA Neu5Ac aldolase, NanT sialic acid transporter, NanK ManNAc kinase, NanE ManNAc-6-P epimerase, GlmS L-glutamine:D-fructose-6-phosphate aminotransferase, GlmM phosphoglucosamine mutase, GlmU bifunctional UDP-N-acetylglucosamine pyrophosphorylase/glucosamine-1-phosphate N-acetyltransferase, NagA N-acetylglucosamine-6-phosphate deacetylase, NagB glucosamine-6-phosphate deaminase, NeuA CMP-Neu5Ac synthetase, NeuS α-Neu5Ac α-2,8-sialyltransferase, NeuB Neu5Ac synthase, NeuD Neu5Ac 7-O (or 9-O)-acetyltransferase. Fru-6-P fructose-6-phosphate, GlcN-6-P D-glucosamine-6-phosphate, GlcN-1-P aminoglucose, GlcNAc-1-P N-acetyl-D-aminoglucose1-phosphate, UDP-GlcNAc Nucleotide sugar, ManNAc N-acetyl-D-mannosamine, ManNAc-6-P ManNAc-6-phosphate, GlcNAc-6-P N-acetyl-D-Glucosamine phosphate, Neu5Ac N-Acetylneuraminic acid, AcNeu5Ac acetylated N-Acetylneuraminic acid, CMP-Neu5Ac sialic acid glycine monophosphate, CMP-AcNeu5Ac acetylated sialylglycerol monophosphate, PSA polysialic acid.
Existing studies have been successful in the efficient synthesis of Neu5Ac; the principle is based on a nanA-catalyzed reversible reaction. However, the main flow of the nanA-catalyzed reaction is the cleavage of ManNAc and pyruvate, with Neu5Ac as the substrate. Excess pyruvate, which is more readily available in extracellular reaction systems than PEP, drives the balance of the nanA-catalyzed reaction towards Neu5Ac synthesis. Therefore, this method has been widely used for the extracellular enzyme-catalyzed synthesis of Neu5Ac monomers, as a titer of 74.2 g/L or even more Neu5Ac can be synthesized in 12 h [17].
Based on the above research progress, in this study we designed the polysialic acid terminal synthesis pathway, using the Neu5Ac monomer as a starting point, and with the help of the co-expression of three enzymes, neuA, neuD, and neuS, we achieved the efficient intracellular polymerization of polysialic acid by regulating the expression level of three key genes. At the same time, in the case of multiple plasmids co-existing, although there are now widely used compatible plasmids, it is still worth discussing the stability of the transmission. In this study, we explored the stability of the compatibility plasmid using the example of polysialic acid end polymerization and investigated its effect on polysialic acid synthesis.
2. Materials and Methods
2.1. Plasmids and Bacterial Strains
The bacterial strains used in this study are listed in Table 1. E. coli DH5α was used as the host for the manipulation of recombinant DNA and the construction of the plasmid. These six strains of bacteria (E. coli NA-01~E. coli NA-06) have undergone multiple passages and cultures, and they have overcome the randomness in the transformation process.

Table 1.
Strains.
2.2. Media, and Culture Conditions
E. coli BL21 (DE3) and derived strains were used for PSA production. All strains were grown in LuriaBertani (LB) for seed preparation and PSA biosynthesis. The LB medium contained 10 g/L peptone, 5 g/L yeast extract, and 10 g/L NaCl. Yeast powder and peptone were provided by Angel Yeast Co., Ltd. (Yichang, China). The corresponding solid LB medium was supplemented with 2% agar powder. The sterilization conditions were 121 °C for 20 min. When necessary, the medium was supplemented with 50 mg/L kanamycin (Kan), 50 mg/L ampicillin (Amp), 25 mg/L chloramphenicol (Chl), and 30 mg/L spectinomycin (Spe) prior to inoculation.
For the terminal polymerization of PSA, Neu5Ac was added through a 0.22 μm membrane to LB medium prior to inoculation, so that the starting concentration of the solution was 10 g/L. Microorganisms were inoculated in 250 mL flasks containing 50 mL of LB medium and incubated overnight at 200 rpm in a shaker at 37 °C [21].
2.3. Deletion of Chromosomal Genes Using CRISPR/Cas9 System
The gene nanA was inactivated using the CRISPR/Cas9 system. The two-plasmid system containing the plasmids pCas and pTargetF was used, in which pCas provides the Cas9 gene and pTargetF harbors sgRNA with a certain N20 sequence, targeting the gene for editing [22]. E. coli cells containing pCas were prepared for electroporation, and arabinose was added to the culture at a final concentration of 10 mM for λ-red induction. After electroporating the mixture of the pTarget and the donor DNA fragment, the E. coli cells were recovered at 37 °C for 1 h and then spread onto an LB agar medium containing Kan and Spc for overnight growth. The identified colonies were cured for pTargetF via IPTG induction and cured for pCas9 via incubation at 42 °C. The plasmids and primers for gene deletions are listed in Tables S1 and S2, respectively.
2.4. Terminal Polymerization Pathway Construction and Assembly
All constructed plasmids and all primers for plasmid construction are listed in Tables S1 and S2, respectively. neuA, neuD, and neuS optimized for codons were synthesized by Sangon Biotech (Shanghai, China) and inserted into three vectors: pRSFDuet-1, pETDuet-1, and pACYCDuet-1, respectively. Subsequently, a series of plasmids were constructed by varying the copy number based on Duet vectors (as shown in Table 1). All successfully constructed plasmids were transformed into E. coli BL21 (DE3) or the derived strains to generate the recombinant strains.
2.5. Quantitative Measurement of Polymerized PSA
2.5.1. Measurement of PSA Based on Resorcinol Colorimetric Assay
In this experiment, the exogenous addition of Neu5Ac was used as a substrate for the synthesis of PSA. The method for determining PSA using the resorcinol method actually hydrolyzes PSA into monomers using a strong acid, and then determines the amounts of all sialic acid monomers [23]. Both exogenously added sialic acid monomers and monomers from the breakdown of the product PSA were included. The amount of Neu5Ac monomer in the fermentation broth was then determined using the HPLC method under neutral conditions. The amount of PSA produced via fermentation was then calculated by subtracting the HPLC score from the resorcinol score.
2.5.2. Measurement of Monomer Neu5Ac Based on HPLC
Neu5Ac was determined using high-performance liquid chromatography (Shimadzu, LC-20A, Kyoto, Japan). The chromatographic column used in the experiment was a Carbomix Ca-NP column from Sai Fen Technology Co. (Oakington, UK). The column temperature was set at 75 °C, the mobile phase was ultrapure water with an injection volume of 5 μL, the flow rate of the mobile phase was 0.6 mL/min, and the detector was an ultraviolet detector with a detection wavelength of 196 nm.
2.5.3. Calculation of Polymerization to Produce Polysialic Acid
The resorcinol colorimetric method measures the content of all sialic acid residues, and the high-performance liquid chromatography method measures the content of sialic acid monomers. In this work, we take the value measured using resorcinol colorimetry minus the value measured using the liquid phase as the yield of polysialic acid produced via polymerization.
2.6. Fluorescence Quantitative PCR Methods
The cells were collected and the total RNA was extracted, and then reverse transcription cDNA was synthesized. 16S was selected as the internal reference gene for this study, and the primers were designed using Oligo7, as in Table S3, and synthesized by Shanghai Bioengineering Co. (Shanghai, China). The cDNA obtained from reverse transcription was used as template for a fluorescence quantitative PCR reaction. The data were processed and that of 12 h in this experiment was set as 1. The relative expression level at different time periods was analyzed and ploted.
2.7. Fermentation Culture Conditions
All strains were cultured overnight in LB medium for activation, and the activated strains were transferred to the fermentation medium at a 4% inoculation rate. The fermentation medium contains 40 g/L sorbitol, 5 g/L (NH4)2SO4, 20 g/L K2HPO4, 1.5 g/L tryptone, 0.9 g/L MgSO4 (sterilized separately), pH was adjusted to 7.8, sterilization conditions were 121 °C for 20 min. Neu5Ac was sterilized via a 0.22 μm membrane filtration, and was added into the culture medium to a final concentration of 10 g/L before inoculation. Corresponding concentrations of antibiotics were added to the fermentation medium, inoculated and fermented for 3 h in 37 °C, with IPTG added to a final concentration of 0.6 mM for induction. The fermentation cycle was 48 h.
3. Results
3.1. Phenotypic Verification of nanA Gene Knockout
In the process of producing PSA using Escherichia coli, a small amount of Neu5Ac enters the PSA synthesis branch, and most of it is decomposed by the Escherichia coli Neu5Ac decomposition pathway [24]. To explore the effect of knocking out the nanA gene on blocking the Neu5Ac decomposition pathway in Escherichia coli, a certain amount of Neu5Ac was added externally before sterilizing the liquid LB medium, so that the initial concentration of solution was 0.5 g/L; the E. coli BL21 (DE3)ΔnanA and E. coli BL21 (DE3) were inoculated separately. During the cultivation process, samples were taken at 0 h, 6 h, 12 h, 18 h, and 24 h to compare changes in Neu5Ac concentration in the culture medium before and after knocking out the nanA gene, and to analyze the degradation of Neu5Ac after knocking out the nanA gene. A gradual decrease was observed in the concentration of Neu5Ac in the E. coli BL21 (DE3) medium, as shown in Figure 2; in contrast, Neu5Ac concentrations in the medium of E. coli BL21 (DE3)ΔnanA were almost stable for 24 h. It was demonstrated that the successful knockout of the nanA gene deprived E. coli BL21 (DE3) of its ability to catabolize Neu5Ac.
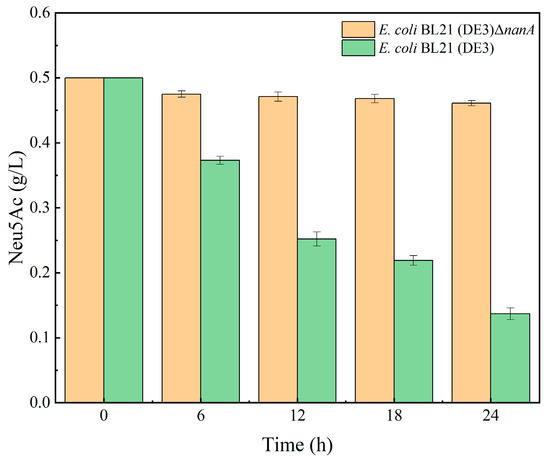
Figure 2.
Effect of nanA knockout on Neu5Ac decomposition.
3.2. Construction and Validation of Recombinant Bacteria
Nine recombinant plasmids were constructed and transformed into E. coli BL21 (DE3) with nanA knocked out. The three Duet series compatible expression vectors have different replicon properties and different copy numbers, and can therefore be classified into three categories: high copy, pRSFDuet-1 (copy number > 100); medium copy, pETDuet-1 (copy number 20~40); and low copy, pACYCDuet-1 (copy number 10~12) [25].
The recombinant plasmids of different copy numbers were then transferred into the same E. coli for expression, and a total of six recombinant-engineered bacteria were constructed, which were named E. coli NA-01–E. coli NA-06 (shown in Table 1). After plasmid transformation, all were screened by adding the appropriate resistance and validated using colony PCR.
In the preliminary validation experiment of the LB culture medium, the fermentation results for the six strains are shown in Figure 3, with E. coli NA-05 strains producing more than the remaining five. This result indicates that the constructed expression vector was successfully expressed in the host bacterium and catalyzed the polymerization of the sialic acid monomer, leading to the production of polysialic acid. To further investigate the relationship between the neuD gene driven by the high copy vector and high PSA yield, the correlation between copy number, expression, and PSA yield was investigated using quantitative fluorescence PCR.
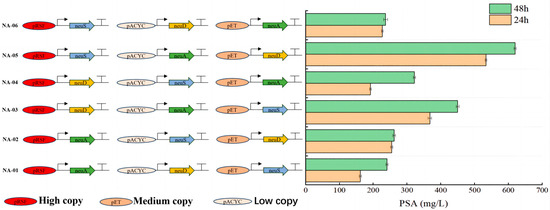
Figure 3.
Comparison of the PSA fermentation yield over different time periods.
3.3. Plasmid Stability at Different Resistance Concentrations
The stability of recombinantly engineered bacteria is essential for high levels of efficient fermentation. The stable and efficient nature of the plasmid in the engineered strain also results in a significant increase in the expression products of the genes on the plasmid. Duet vectors carry compatible replicons and antibiotic resistance markers for the efficient propagation and maintenance of multiple plasmids in a single cell. pETDuet-1 carries the ColE1 replicon and ampicillin (Amp) resistance, pACYCDuet-1 carries the P15A replicon and chloramphenicol (Chl) resistance, and pRSFDuet-1 carries the RSF1030 replicon and kanamycin (Kan) resistance. Therefore, theoretically, Duet series vectors can be stably inherited within the host bacteria without loss. Next, we tested the stability of the recombinant vectors by fermenting and culturing the six constructed recombinant-engineered bacteria at different antibiotic concentrations. The stability of the plasmid and the intensity of expression of the resistance genes were examined in standard antibiotic concentrations, weak antibiotic concentrations, and in the absence of antibiotics. The stability of the vector was characterized using fluorescence quantitative PCR to detect the expression level of resistance genes on the vector. We performed a fluorescence quantitative PCR reaction using cDNA as a template, and analyzed the products of the fluorescence quantitative PCR. We characterized the plasmid stability by measuring the relative expression changes in the resistance genes on Duet vectors.
3.3.1. Plasmid Stability at Standard Antibiotic Concentrations
As can be seen from Figure 4,the stability of the vectors behind the changes in the expression strength of the resistance genes, considering the error of quantitative fluorescence PCR, in the first 36 h, gradually increased across all resistance genes, indicating that the exogenous genes in the vectors did not show toxicity towards the host bacteria and that the stability of the plasmids pRSFDuet-US, pACYCDuet-UA, and pETDuet-UD was very good; meanwhile, the expression level of the neuS gene decreased between 36 and 48 h, probably due to the deterioration of the shake flask culture environment and antibiotic inactivation leading to the loss of the plasmids pETDuet-US and pACYCDuet-US.
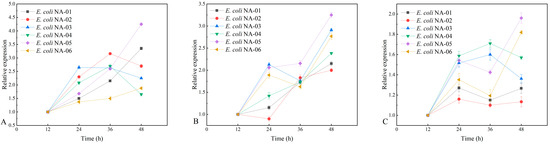
Figure 4.
(A) Relative expression level of Amp at standard antibiotic concentration. (B) Relative expression level of Kan at standard antibiotic concentration. (C) Relative expression level of Chl at standard antibiotic concentration.
3.3.2. Plasmid Stability at 50% Antibiotic Concentration
In contrast to the situation with standard antibiotic concentrations, the expression level of the three resistance genes was consistently reduced in the half-dose antibiotic environment, suggesting a persistent loss of the compatible expression vectors in which they reside. The results are shown in Figure 5: at 50% antibiotic concentration, the relative expression of Kan, Amp, and Chl decreased compared to 12 h, and there was a loss of plasmids for different recombinant strains for different genes after 48 h of fermentation.
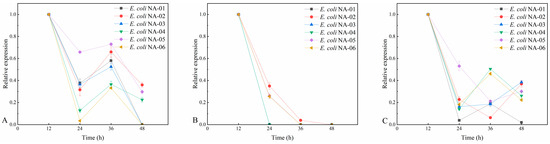
Figure 5.
(A) Relative expression level of Amp at 50% antibiotic concentration. (B) Relative expression level of Kan at 50% antibiotic concentration. (C) Relative expression level of Chl at 50% antibiotic concentration.
3.3.3. Plasmid Stability at Antibiotic-Free Concentrations
As can be seen from Figure 6, under antibiotic-free conditions, the loss of different-resistance plasmids was more frequent in some recombinant strains than in others. This indicates that antibiotic concentration affects gene expression and that resistant plasmids are most likely to be lost under resistance-free conditions.
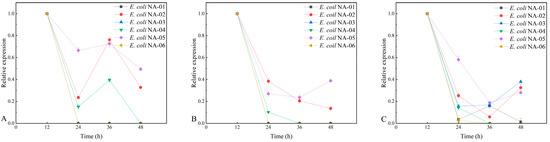
Figure 6.
(A) Relative expression level of Amp at 0% antibiotic concentration. (B) Relative expression level of Kan at 0% antibiotic concentration. (C) Relative expression level of Chl at 0% antibiotic concentration.
Although all six recombinant-engineered bacteria contain compatible plasmids, standard antibiotic concentrations are still required to maintain plasmid stability. Six fermentations were carried out at standard antibiotic concentrations, with normal expression of Amp, Kan, and Chl genes; Kan and Amp started to show zero expression at 50% antibiotic concentration; and under antibiotic-free conditions, only the fermentates of E. coli NA-02 and E. coli NA-05 showed a normal expression of Amp, Kan, and Chl genes at 48 h. Therefore, this experiment demonstrates that standard antibiotic concentrations should be used in the following fermentation experiments. The comparison of the fluorescence quantitative PCR experimental results showed the expression of exogenous genes in E. coli NA-05 was more stable than those in other strains. The PSA yield of E. coli NA-05 was higher than the other five strains in Figure 3, and E. coli NA-05 could be used as the strain for subsequent fermentation optimization experiments.
3.4. Association between Expression of Key Genes and Yield
The six engineered strains differed considerably in their ability to synthesize polysialic acid, and it is clear that differences in the expression of their genes under the influence of different copy number vectors are a potentially important reason for the differences in their ability to polymerize sialic acid. High-copy-number vector-expressed neuS drives the synthesis of high yield polysialic acid, whereas low-copy neuD constitutes a bottleneck in the polymerization of sialic acid. In the plasmid stability experiment, the level of plasmid loss was higher with fermentation. Therefore, we continue to determine the stability of exogenous gene expression in six strains of engineering bacteria using quantitative fluorescence PCR to analyze the relationship between it and the yield of polysialic acid, and to identify engineering bacteria with optimal performance. The fermentation broth of six recombinant-engineered bacteria at different fermentation times (12 h, 24 h, 36 h, and 48 h) was subjected to quantitative fluorescent PCR analysis at standard antibiotic concentrations to determine the relative expression changes of the three genes (neuA, neuD, and neuS), and the results are shown in Figure 7.
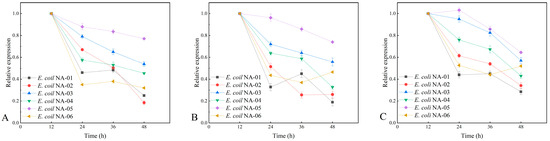
Figure 7.
(A) Expression level of neuA gene in different strains. (B) Expression level of neuD gene in different strains. (C) Expression level of neuS gene in different strains.
For the neuA, neuD, and neuS genes, the highest expression in the E. coli NA-05 fermentation broth was observed at 48 h of fermentation. Comparing the PSA yield of the recombinant bacteria with the expression level of the key genes, it can be hypothesized that the magnitude of the expression of the key genes neuA, neuD, and neuS correlates with the PSA yield. The stability of the three plasmids in the engineered bacterium E. coli NA-05 was better and the expression levels of the three exogenous genes it introduced were more stable compared to the other engineered bacteria, which also contributed to its higher capacity for polysialic acid synthesis.
3.5. PSA Polymerization in Fermenter Level
After optimizing the culture conditions and fermentation with the sialic acid monomer as the precursor, the recombinant strain E. coli NA-05 was fermented and cultured in a small fermenter for 60 h. 6.15 g/L of polysialic acid and 14.52 g/L of wet cell weight were obtained with a significant effect on the PSA synthesis (Figure 8). The yield of PSA from the sialic acid monomer reached more than 61.5%.
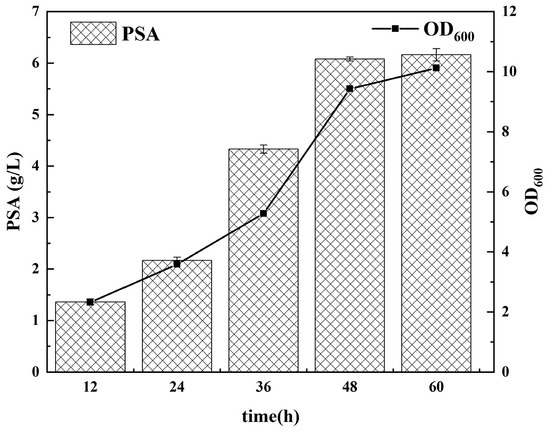
Figure 8.
Fermentation of PSA with E. coli NA-05.
4. Discussion
4.1. The Potential of Neu5Ac-Based PSA Synthesis and the Way Forward
It can be seen that the limiting link in the polysialic acid polymerization process is the activity of α-2,8 sialyltransferase, which brings about a higher product generation when expressed in larger amounts. Compared to E. coli NA-01 and E. coli NA-06, neuD was expressed by a low-copy plasmid (pACYCDuet-1), thus demonstrating that the low activity of neuD caused a bottleneck in polysialic acid synthesis.
The main methods for the production of sialic acid are chemical synthesis, enzymatic synthesis, and natural product extraction, while polysialic acid can only be obtained through microbial fermentation. The enzymatic synthesis method, which has the advantage of good product efficiency and being highly established, uses N-acetylglucosamine to synthesize N-acetylneuraminic acid [26]. The synthesis of PSA eliminates the need for the traditional metabolic pathway starts with monose (glucose), and, thereby, the synthesis of polysialic acid based on the enhanced polymerization of sialic acid monomers becomes economically viable. For biotechnological production, E. coli has been subjected to process modification and metabolic modification for the efficient production of PSA [6]. In the present study, we confirm and improve the biosynthetic process to obtain PSA, with the polymerization of Neu5Ac as a precursor, demonstrating the good feasibility of the process and the potential for further improvement. During the fermentation of E. coli, its culture conditions can be optimized for optimal salt, organic matter, and parameter conditions to further increase PSA production.
4.2. Compatibility of Expression Vectors in Synthetic Biology
Compatible expression vectors are widely used in synthetic biology, and are of great importance for the efficient co-expression of multiple genes. Synthetic biology is a discipline that integrates the application of science, technology, and engineering to facilitate and accelerate the design, modification, and alteration of genetic material in living organisms. Synthetic biology breaks away from the limits of natural evolution and focuses on “artificial design and genome editing” to produce specific target products. In this study, the Duet family of expression vectors was genetically edited using biosynthetic techniques, and plasmids were constructed, designed, and modified. The Duet family of expression vector replicons is mutually compatible and contains different antibiotic markers, allowing the stable co-expression of multiple genes in E. coli [25,27]. The synthesis, assembly, and editing of genetic material such as DNA are of great importance to biosynthetics. Using gene-editing techniques, recombinant plasmids are edited by ligating the target gene to a compatible expression vector. The recombinant plasmids of different copy numbers were then transferred to the same E. coli BL21 (DE3)ΔnanA to achieve the co-expression of multiple genes. The change from “discovery” to “creation” opens up new ideas for genetic modification. Many biological functions are not controlled by a single gene, but require the coordinated action of multiple related genes. The use of genetic engineering techniques to co-express multiple genes in the same host bacterium is an effective tool for developmental regulation studies and metabolic pathway modification in E. coli. The target gene is integrated into a compatible expression vector using gene assembly technology. The target gene is cleaved with the same fast cutting enzyme as the vector, and the two are then ligated to create the desired expression vector. Synthetic biology has a wide range of applications in health care. PSA plays an important role in numerous biogenic reactions in living organisms, and is also an ideal material for controlled drug release and scaffolding. The multiple biological effects of PSA make it promising for a wide range of applications in the biomedical field.
5. Conclusions
E. coli BL21 (DE3) was used as the starting strain and a salivary acid monomer was used as the precursor substance to construct the shortest efficient PSA synthesis pathway using end-enhanced polymerization technology. The successful knockdown of the nanA gene using CRISPR Cas9 caused E. coli BL21 (DE3) to lose its ability to catabolize Neu5Ac, which promoted the efficiency of PSA synthesis and increased the positive utilization of the substrate. Compatible expression vectors (pRSFDuet-1, pETDuet-1, and pACYCDuet-1) were selected, and key gene fragments (neuA, neuD, and neuS) were inserted downstream of the T7 promoter and imported into the recombinant host bacterium; the gradient expression matrix of key genes was established, and nine recombinant plasmids were successfully constructed. Expression vectors with different copy numbers were introduced into the same recombinant host bacteria for expression. By regulating the transcription level of target genes through vector copy number, additional gene copy support enzyme levels are provided, thereby affecting the synthesis of PSA. It is known that a high level of transcription of the neuD gene is necessary for efficient PSA synthesis [28]. Although Duet series vectors are mutually compatible vectors, their stable existence in the host still requires antibiotic pressure. Among the six engineered strains constructed in this study, E. coli NA-05 showed the highest PSA production. The highest stability of the expression level of key genes was detected in this strain, which, together with the optimized combination of the expression level of key genes, was found to be the intrinsic reason for its efficient polymerization for PSA production.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation10010064/s1: Figure S1: PCR products amplified upstream and downstream of nanA gene; Figure S2: Upstream and downstream fragments of nanA gene linked by fusion PCR; Figure S3: Agarose gel electrophoresis for pTarget-nanA plasmid digestion; Figure S4: Agarose gel electrophoresis of pCas transformed into BL21 (DE3) for plasmid extraction and single digestion verification; Figure S5: PCR verification of nanA gene knockout; Figure S6: Plasmid elimination plate; Figure S7: Agarose gel electrophoresis for plasmid elimination validation; Figure S8: Dual digestion agarose gel electrophoresis of the gene and vector; Figure S9: Single digestion validation of recombinant plasmid; Table S1: Plasmid; Table S2: Primer; Table S3: qPCR primer.
Author Contributions
Conceptualization, D.Z. and C.W.; methodology, C.W., H.C., X.L. (Xiaomeng Liu), H.Z., J.G., S.P. and W.W.; software, C.W. and H.C.; validation, C.W., H.C. and D.Z.; formal analysis, D.Z. and C.W.; investigation, D.Z., C.W., H.C., X.L. (Xiaomeng Liu), H.Z., J.G., S.P. and W.W.; resources, D.Z.; data curation, D.Z. and C.W.; writing-original draft preparation, C.W., X.L. (Xiaomeng Liu) and D.Z.; writing-review and editing, D.Z. and X.L. (Xinli Liu); visualization, D.Z.; supervision, D.Z. and X.L. (Xinli Liu); project administration, D.Z.; funding acquisition, D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Pilot Project for the Integration of Science, Education and Industry, Qilu University of Technology, Shandong Academy of Sciences (2023PY007), the Shandong Province Science and Technology SMES Innovation Ability Improvement Project (2023TSGC0525), and the Dezhou Major Science and Technology Innovation Project and Rizhao Key Research and Development Plan (2023ZDYF010140).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are contained within the article.
Conflicts of Interest
The authors declare that they have no competing financial interests.
References
- McGowen, M.M.; Vionnet, J.; Vann, W.F. Elongation of alternating α2,8/2,9 polysialic acid by the Escherichia coli K92 polysialyltransferase. Glycobiology 2001, 11, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Azurmendi, H.F.; Vionnet, J.; Wrightson, L.; Trinh, L.B.; Shiloach, J.; Freedberg, D.I. Extracellular structure of polysialic acid explored by on cell solution NMR. Proc. Natl. Acad. Sci. USA 2007, 104, 11557–11561. [Google Scholar] [CrossRef]
- Brusés, J.L.; Rutishauser, U. Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie 2001, 83, 635–643. [Google Scholar] [CrossRef] [PubMed]
- El Maarouf, A.; Petridis, A.K.; Rutishauser, U. Use of polysialic acid in repair of the central nervous system. Proc. Natl. Acad. Sci. USA 2006, 103, 16989–16994. [Google Scholar] [CrossRef]
- Mühlenhoff, M.; Eckhardt, M.; Gerardy-Schahn, R. Polysialic acid: Three-dimensional structure, biosynthesis and function. Curr. Opin. Struct. Biol. 1998, 8, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.-X.; Qiao, Y.; Shi, B.; Tao, Y. Polysialic acid biosynthesis and production in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 100, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.Y.; Wang, S.Z.; Li, G.S.; Zhan, X.B.; Lin, C.C.; Wu, J.R.; Zhu, L. A new polysialic acid production process based on dual-stage pH control and fed-batch fermentation for higher yield and resulting high molecular weight product. Appl. Microbiol. Biotechnol. 2013, 97, 2405–2412. [Google Scholar] [CrossRef]
- Ferrero, M.A.; Aparicio, L.R. Biosynthesis and production of polysialic acids in bacteria. Appl. Microbiol. Biotechnol. 2010, 86, 1621–1635. [Google Scholar] [CrossRef]
- Ishikawa, M.; Koizumi, S. Microbial production of N-acetylneuraminic acid by genetically engineered Escherichia coli. Carbohydr. Res. 2010, 345, 2605–2609. [Google Scholar] [CrossRef]
- Roberts, I.S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996, 50, 285–315. [Google Scholar] [CrossRef]
- Whitfield, C.; Roberts, I.S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999, 31, 1307–1319. [Google Scholar] [CrossRef]
- Daines, D.A.; Silver, R.P. Evidence for Multimerization of Neu Proteins Involved in Polysialic Acid Synthesis in Escherichia coli K1 Using Improved LexA-Based Vectors. J. Bacteriol. 2000, 182, 5267–5270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tian, R.Z.; Shen, Q.Y.; Liu, Y.F.; Liu, L.; Li, J.H.; Du, G.C. Pathway Engineering of Bacillus subtilis for Enhanced N-Acetylneuraminic Acid Production via Whole-Cell Biocatalysis. Biotechnol. J. 2019, 14, 8. [Google Scholar] [CrossRef]
- Lundgren, B.R.; Boddy, C.N. Sialic acid and N-acyl sialic acid analog production by fermentation of metabolically and genetically engineered Escherichia coli. Org. Biomol. Chem. 2007, 5, 1903–1909. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Gu, P.F.; Wang, Y.; Li, Y.K.; Yang, F.; Wang, Q.; Qi, Q.S. Engineering of an N-acetylneuraminic acid synthetic pathway in Escherichia coli. Metab. Eng. 2012, 14, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Yu, H.; Cao, H.Z.; Lau, K.; Muthana, S.; Tiwari, V.K.; Son, B.; Chen, X. Pasteurella multocida sialic acid aldolase: A promising biocatalyst. Appl. Microbiol. Biotechnol. 2008, 79, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.X.; Zhang, Z.J.; Liu, W.F.; Dong, Z.Y.; Tao, Y. Enhanced production of N-acetyl-D-neuraminic acid by multi-approach whole-cell biocatalyst. Appl. Microbiol. Biotechnol. 2013, 97, 4775–4784. [Google Scholar] [CrossRef] [PubMed]
- Song, L.L.; Zhou, H.; Cai, X.H.; Li, C.Y.; Liang, J.N.; Jin, C. NeuA O-acetylesterase activity is specific for CMP-activated O-acetyl sialic acid in Streptococcus suis serotype 2. Biochem. Biophys. Res. Commun. 2011, 410, 212–217. [Google Scholar] [CrossRef]
- Zapata, G.; Vann, W.F.; Aaronson, W.; Lewis, M.S.; Moos, M. Sequence of the Cloned Escherichia coli K1 CMP-N-acetylneuraminic Acid Synthetase Gene. J. Biol. Chem. 1989, 264, 14769–14774. [Google Scholar] [CrossRef]
- Steenbergen, S.M.; Lee, Y.C.; Vann, W.F.; Vionnet, J.; Wright, L.F.; Vimr, E.R. Separate pathways for O acetylation of polymeric and monomeric sialic acidsand identification of sialyl O-acetyl esterase in Escherichia coli K1. J. Bacteriol. 2006, 188, 6195–6206. [Google Scholar] [CrossRef]
- Zhu, Y.Y.; Wan, L.; Meng, J.W.; Luo, G.C.; Chen, G.; Wu, H.; Zhang, W.L.; Mu, W.M. Metabolic Engineering of Escherichia coli for Lacto-N-triose II Production with High Productivity. J. Agric. Food Chem. 2021, 69, 3702–3711. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.L.; Sun, B.B.; Yang, J.J.; Yang, S. Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9 System. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L. Quantitive estimation of sialic acids: II. A colorimetric resorcinol-hydrochloric acid method. Biochim. Et Biophys. Acta 1957, 24, 604–611. [Google Scholar] [CrossRef]
- Vimr, E.R.; Troy, F.A. Regulation of sialic acid metabolism in Escherichia coli: Role of N-acylneuraminate pyruvate-lyase. J. Bacteriol. 1985, 164, 854–860. [Google Scholar] [CrossRef]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.J.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid Pathway Optimization for Taxol Precursor Overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.S.; Bednarski, M.D.; Whitesides, G.M. Synthesis of CMP-NeuAc from N-acetylglucosamine—Generation of CTP from CMP Using Adenylate Kinase. J. Am. Chem. Soc. 1988, 110, 7159–7163. [Google Scholar] [CrossRef]
- Wu, J.J.; Du, G.C.; Zhou, J.W.; Chen, J. Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab. Eng. 2013, 16, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tao, Y.; Jin, C.; Xu, Y.; Lin, B.X. Enhanced production of polysialic acid by metabolic engineering of Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).