CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders
Abstract
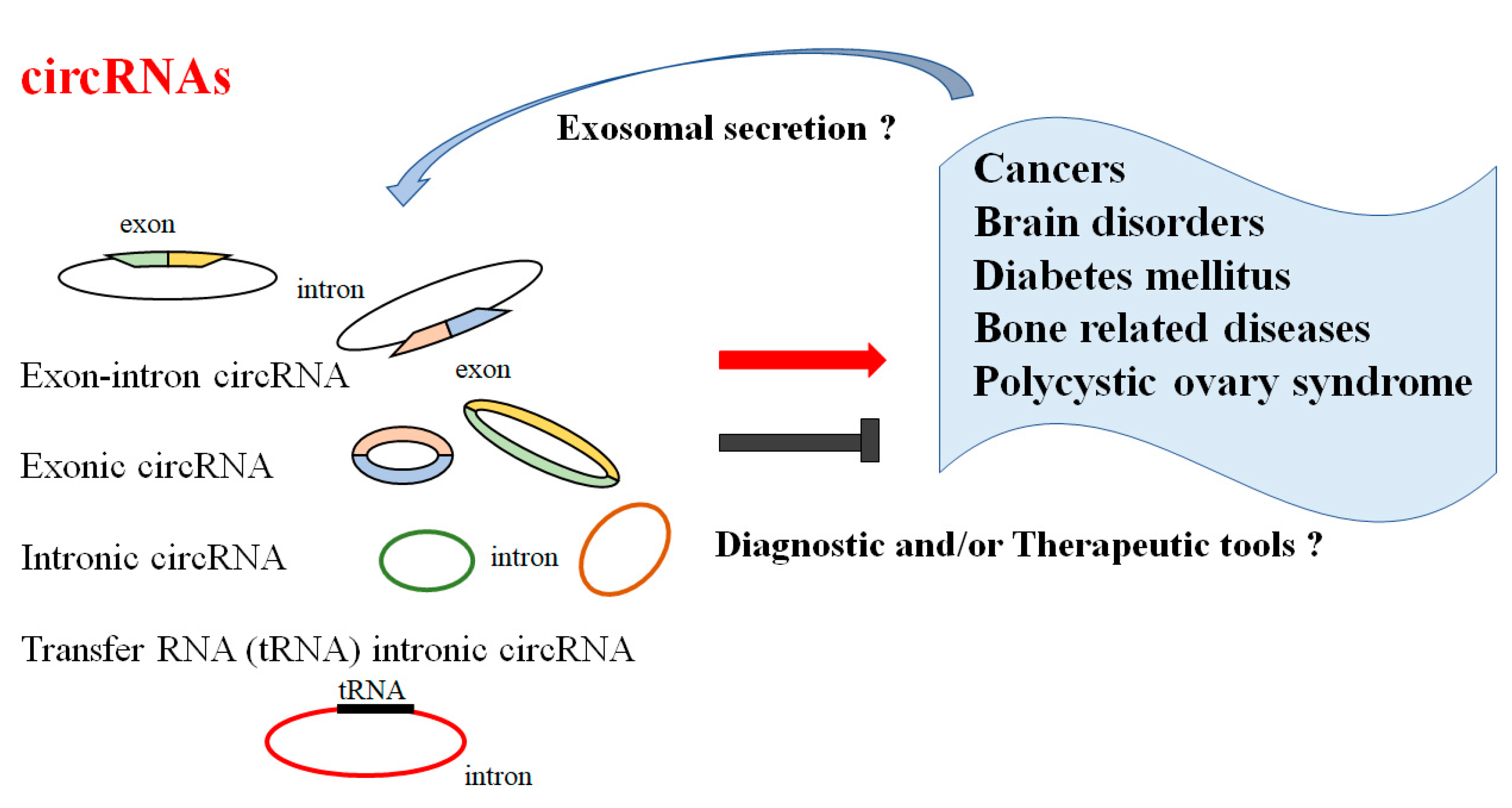
1. Introduction
2. Roles of CircRNAs in Pathophysiology
3. CircRNAs and Several Diseases
3.1. CircRNAs and Cancers
3.2. CircRNAs and CNS Disorders
3.3. CircRNAs and Diabetes Mellitus
3.4. CircRNAs and Bone Related Diseases
3.5. Polycystic Ovary Syndrome
4. Mechanism of circRNAs’ Action with PABP and APROs
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AGO | Argonaute |
| ALS | amyotrophic lateral sclerosis |
| APRO | antiproliferative |
| BBB | blood–brain barrier |
| BMSC | bone marrow derived mesenchymal stem cell |
| BMP6 | bone morphogenetic protein-6 |
| BTG2 | B-cell translocation gene 2 |
| BTG3 | B-cell translocation gene 3 |
| CNS | central nervous system |
| circRNA | circular RNA |
| miRNA | microRNA |
| miRISC | miRNA-loaded RNA-induced silencing complex |
| MSC | mesenchymal stem cell |
| ncRNA | non-coding RNA |
| PABP | poly(A)-binding protein |
| PAM2 | PABP-interacting motif 2 |
| PCOS | Polycystic ovary syndrome |
| RBP | RNA-binding protein |
| T1DM | type 1 diabetes mellitus |
| T2DM | type 2 diabetes mellitus |
| VDR | vitamin D receptor |
References
- ElMonier, A.A.; El-Boghdady, N.A.; Fahim, S.A.; Sabry, D.; Elsetohy, K.A.; Shaheen, A.A. LncRNA NEAT1 and MALAT1 are involved in polycystic ovary syndrome pathogenesis by functioning as competing endogenous RNAs to control the expression of PCOS-related target genes. Non-Coding RNA Res. 2023, 8, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, Y.; Han, B.; Yang, L.; Chen, X.; Huang, R.; Wu, F.; Chao, J.; Liu, P.; Hu, G.; et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood–Brain Barrier Integrity. J. Neurosci. 2017, 38, 32–50. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Zhang, P.; Chao, Z.; Zhang, R.; Ding, R.; Wang, Y.; Wu, W.; Han, Q.; Li, C.; Xu, H.; Wang, L.; et al. Circular RNA Regulation of Myogenesis. Cells 2019, 8, 885. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, Y.; Jiang, Z. CircRNAs: A new perspective of biomarkers in the nervous system. Biomed. Pharmacother. 2020, 128, 110251. [Google Scholar] [CrossRef]
- Tang, X.; Ren, H.; Guo, M.; Qian, J.; Yang, Y.; Gu, C. Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 2021, 19, 910–928. [Google Scholar] [CrossRef]
- Dong, W.; Dai, Z.H.; Liu, F.C.; Guo, X.G.; Ge, C.M.; Ding, J.; Liu, H.; Yang, F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 2019, 45, 155–167. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sharp, P.A. A Circuitous Route to Noncoding RNA. Science 2013, 340, 440–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Zaphiropoulos, P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: Correlation with exon skipping. Proc. Natl. Acad. Sci. USA 1996, 93, 6536–6541. [Google Scholar] [CrossRef]
- Dong, W.; Bi, J.; Liu, H.; Yan, D.; He, Q.; Zhou, Q.; Wang, Q.; Xie, R.; Su, Y.; Yang, M.; et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol. Cancer 2019, 18, 95. [Google Scholar] [CrossRef]
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018, 32, 639–644. [Google Scholar] [CrossRef]
- Aufiero, S.; Reckman, Y.J.; Pinto, Y.M.; Creemers, E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019, 16, 503–514. [Google Scholar] [CrossRef]
- Yang, S.; Gao, S.; Liu, T.; Liu, J.; Zheng, X.; Li, Z. Circular RNA SMARCA5 functions as an anti-tumor candidate in colon cancer by sponging microRNA-552. Cell Cycle 2021, 20, 689–701. [Google Scholar] [CrossRef]
- Videira, R.F.; Martins, P.A.D.C. Non-coding RNAs in Cardiac Intercellular Communication. Front. Physiol. 2020, 11, 738. [Google Scholar] [CrossRef]
- Das, D.; Das, A.; Panda, A.C. Antisense Oligo Pulldown of Circular RNA for Downstream Analysis. Bio-Protocol 2021, 11, e4088. [Google Scholar] [CrossRef] [PubMed]
- Guria, A.; Sharma, P.; Natesan, S.; Pandi, G. Circular RNAs-The Road Less Traveled. Front. Mol. Biosci. 2020, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Kakumani, P.K. AGO-RBP crosstalk on target mRNAs: Implications in miRNA-guided gene silencing and cancer. Transl. Oncol. 2022, 21, 101434. [Google Scholar] [CrossRef]
- Wakiyama, M.; Ogami, K.; Iwaoka, R.; Aoki, K.; Hoshino, S.I. MicroRNP-mediated translational activation of nonadenylated mRNAs in a mammalian cell-free system. Genes Cells 2018, 23, 332–344. [Google Scholar] [CrossRef]
- Ezzeddine, N.; Chang, T.-C.; Zhu, W.; Yamashita, A.; Chen, C.-Y.A.; Zhong, Z.; Yamashita, Y.; Zheng, D.; Shyu, A.-B. Human TOB, an Antiproliferative Transcription Factor, Is a Poly(A)-Binding Protein-Dependent Positive Regulator of Cytoplasmic mRNA Deadenylation. Mol. Cell. Biol. 2007, 27, 7791–7801. [Google Scholar] [CrossRef]
- Chen, C.-Y.A.; Strouz, K.; Huang, K.-L.; Shyu, A.-B. Tob2 phosphorylation regulates global mRNA turnover to reshape transcriptome and impact cell proliferation. RNA 2020, 26, 1143–1159. [Google Scholar] [CrossRef]
- Matsuda, S.; Rouault, J.; Magaud, J.; Berthet, C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001, 497, 67–72. [Google Scholar] [CrossRef]
- Yuniati, L.; Scheijen, B.; van der Meer, L.T.; van Leeuwen, F.N. Tumor suppressors BTG1 and BTG2: Beyond growth control. J. Cell Physiol. 2019, 234, 5379–5389. [Google Scholar] [CrossRef]
- Hentze, M.W.; Preiss, T. Circular RNAs: Splicing’s enigma variations. EMBO J. 2013, 32, 923–925. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [PubMed]
- Larsson, O.; Perlman, D.M.; Fan, D.; Reilly, C.S.; Peterson, M.; Dahlgren, C.; Liang, Z.; Li, S.; Polunovsky, V.A.; Wahlestedt, C.; et al. Apoptosis resistance downstream of eIF4E: Posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 2006, 34, 4375–4386. [Google Scholar] [CrossRef]
- Wu, N.; Yuan, Z.; Du, K.Y.; Fang, L.; Lyu, J.; Zhang, C.; He, A.; Eshaghi, E.; Zeng, K.; Ma, J.; et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019, 26, 2758–2773. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Liu, Y.; Su, H.; Zhang, J.; Liu, Y.; Feng, C.; Han, F. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol. 2020, 18, e3000582. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Liu, C.; Liu, B.-H.; Chen, X.; Dong, R.; Liu, X.; Zhang, Y.-Y.; Liu, B.; Zhang, S.-J.; Wang, J.-J.; et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017, 136, 1629–1642. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef]
- Vasei, M.; Moch, H.; Mousavi, A.; Kajbafzadeh, A.M.; Sauter, G. Immunohistochemical profiling of Wilms tumor: A tissue microarray study. Appl. Immunohistochem. Mol. Morphol. 2008, 16, 128–134. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, K.; Shang, J.; Wang, B.; Zhong, J.; Wu, B.; Li, H.; Xu, X.; Lu, H. Comprehensive expression profiles of CircRNAs, LncRNAs, and mRNAs in PBMCs from patients with the ossification of the posterior longitudinal ligament. Mol. Omics 2021, 17, 607–619. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Yang, R.; Xu, B.; Yang, B.; Fu, J.; Chen, H.; Wang, X. Non-coding RNAs: The extensive and interactive regulators of the blood-brain barrier permeability. RNA Biol. 2021, 18, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Ruffo, P.; Strafella, C.; Cascella, R.; Caputo, V.; Conforti, F.L.; Andò, S.; Giardina, E. Deregulation of ncRNA in Neurodegenerative Disease: Focus on circRNA, lncRNA and miRNA in Amyotrophic Lateral Sclerosis. Front. Genet. 2021, 12, 784996. [Google Scholar] [CrossRef] [PubMed]
- Reinoso-Sánchez, J.; Baroli, G.; Duranti, G.; Scaricamazza, S.; Sabatini, S.; Valle, C.; Morlando, M.; Casero, R.; Bozzoni, I.; Mariottini, P.; et al. Emerging Role for Linear and Circular Spermine Oxidase RNAs in Skeletal Muscle Physiopathology. Int. J. Mol. Sci. 2020, 21, 8227. [Google Scholar] [CrossRef] [PubMed]
- Bachmayr-Heyda, A.; Reiner, A.T.; Auer, K.; Sukhbaatar, N.; Aust, S.; Bachleitner-Hofmann, T.; Mesteri, I.; Grunt, T.W.; Zeillinger, R.; Pils, D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015, 5, 8057. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Song, H.; Yin, H.; Jiang, T.; Xu, Y.; Liu, L.; Wang, H.; Gao, H.; Wang, R.; et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol. Cancer 2021, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Prats, A.C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the Key for Translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, X.; Zhan, X.; Kang, S.; Liu, H.; Luo, Y.; Lin, L. Advance in circular RNA modulation effects of heart failure. Gene 2020, 763, 100036. [Google Scholar] [CrossRef]
- Yan, F.; Xie, X.; Huo, Q.; Zhang, W.; Wu, T.; Daniyaer, D.; Shi, L. circ-CCND1 regulates the CCND1/P53/P21 pathway through sponging miR-138-5p in valve interstitial cells to aggravate aortic valve calcification. J. Physiol. Biochem. 2022, 78, 845–854. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Y.; Zhao, M.; Zou, J.; Zhu, Y.; Yuan, X.; Liu, Q.; Cai, H.; Chu, C.-Q.; Liu, Y. Differential Profile of Plasma Circular RNAs in Type 1 Diabetes Mellitus. Diabetes Metab. J. 2020, 44, 854–865. [Google Scholar] [CrossRef]
- Su, Y.; Lv, X.; Yin, W.; Zhou, L.; Hu, Y.; Zhou, A.; Qi, F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging 2019, 11, 8183–8203. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, C.; Liu, Y.; Hu, Y.; Wu, H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging 2019, 11, 3362–3375. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Liu, Z.; Liang, M.; Pan, J.; Lin, M.; Lin, H.; Luo, Y.; Zhou, X.; Yao, W. Identification of circRNA-miRNA-mRNA networks contributes to explore underlying pathogenesis and therapy strategy of gastric cancer. J. Transl. Med. 2021, 19, 226. [Google Scholar] [CrossRef]
- Wang, S.; Hu, Y.; Lv, X.; Li, B.; Gu, D.; Li, Y.; Sun, Y.; Su, Y. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin. Sci. 2019, 133, 1935–1953. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Cao, Z.; Du, J.; Liu, T.; Wang, T. Circular RNA circPITX1 knockdown inhibits glycolysis to enhance radiosensitivity of glioma cells by miR-329-3p/NEK2 axis. Cancer Cell Int. 2020, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Liu, M.; Tian, Q.; Liu, X. Microarray analysis of circRNAs expression profile in gliomas reveals that circ_0037655 could promote glioma progression by regulating miR-214/PI3K signaling. Life Sci. 2020, 245, 117363. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Tan, C.; Liu, X. Circular RNAs: A new frontier in the study of human diseases. J. Med. Genet. 2016, 53, 359–365. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, Y.; Gao, P.; Cui, Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem. Biophys. Res. Commun. 2018, 509, 138–142. [Google Scholar] [CrossRef]
- Chen, L.; Nan, A.; Zhang, N.; Jia, Y.; Li, X.; Ling, Y.; Dai, J.; Zhang, S.; Yang, Q.; Yi, Y.; et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer 2019, 18, 13. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, G.; Li, S.; Xiong, H.; Chen, R.; Zuo, L.; Liu, H. CircSMARCA5: A key circular RNA in various human diseases. Front. Genet. 2022, 13, 921306. [Google Scholar] [CrossRef]
- Huang, L.; Han, J.; Yu, H.; Liu, J.; Gui, L.; Wu, Z.; Zhao, X.; Su, S.; Fu, G.; Li, F. CircRNA_000864 Upregulates B-cell Translocation Gene 2 Expression and Represses Migration and Invasion in Pancreatic Cancer Cells by Binding to miR-361-3p. Front. Oncol. 2020, 10, 547942. [Google Scholar] [CrossRef]
- Shi, L.; Cao, Y.; Yuan, W.; Guo, J.; Sun, G. Exosomal circRNA BTG2 derived from RBP-J overexpressed-macrophages inhibits glioma progression via miR-25-3p/PTEN. Cell Death Dis. 2022, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Xu, Y.; Chen, Y.; Yan, F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed. Pharmacother. 2017, 88, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhao, G.; Ma, X.; Dong, Q.; Zhang, H.; Wang, Y.; Cui, J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem. Biophys. Res. Commun. 2018, 503, 2429–2435. [Google Scholar] [CrossRef]
- Enkhnaran, B.; Zhang, G.C.; Zhang, N.P.; Liu, H.N.; Wu, H.; Xuan, S.; Yu, X.N.; Song, G.Q.; Shen, X.Z.; Zhu, J.M.; et al. microRNA-106b-5p Promotes Cell Growth and Sensitizes Chemosensitivity to Sorafenib by Targeting the BTG3/Bcl-xL/p27 Signaling Pathway in Hepatocellular Carcinoma. J. Oncol. 2022, 2022, 1971559. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, S.; Chen, S.; Wu, H.; Jiang, M.; Liu, A. Exosomal miR-552-5p promotes tumorigenesis and disease progression via the PTEN/TOB1 axis in gastric cancer. J. Cancer 2022, 13, 890–905. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Li, Y.; Fan, H.; Sun, J.; Ni, M.; Zhang, L.; Chen, C.; Hong, X.; Fang, F.; Zhang, W.; Ma, P. Circular RNA expression profile of Alzheimer’s disease and its clinical significance as biomarkers for the disease risk and progression. Int. J. Biochem. Cell Biol. 2020, 123, 105747. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.L.; Dempsey, R.J.; Vemuganti, R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020, 186, 101746. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Ju, K.; Chen, A.; Cao, H. Circulating CircRNAs Panel Acts as a Biomarker for the Early Diagnosis and Severity of Parkinson’s Disease. Front. Aging Neurosci. 2021, 13, 684289. [Google Scholar] [CrossRef] [PubMed]
- Dube, U.; Del-Aguila, J.L.; Li, Z.; Budde, J.P.; Jiang, S.; Hsu, S.; Ibanez, L.; Fernandez, M.V.; Farias, F.; Norton, J.; et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019, 22, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, E.; Cairns, M.J. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci. Rep. 2019, 9, 2564. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, J.; Wu, Y.; Luo, H.; Ke, Y. Decrease of circARID1A retards glioblastoma invasion by modulating miR-370-3p/TGFBR2 pathway. Int. J. Biol. Sci. 2022, 18, 5123–5135. [Google Scholar] [CrossRef]
- Khoutorsky, A.; Yanagiya, A.; Gkogkas, C.G.; Fabian, M.R.; Prager-Khoutorsky, M.; Cao, R.; Gamache, K.; Bouthiette, F.; Parsyan, A.; Sorge, R.E.; et al. Control of Synaptic Plasticity and Memory via Suppression of Poly(A)-Binding Protein. Neuron 2013, 78, 298–311. [Google Scholar] [CrossRef]
- Beilerli, A.; Gareev, I.; Beylerli, O.; Yang, G.; Pavlov, V.; Aliev, G.; Ahmad, A. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin. Cancer Biol. 2022, 83, 242–252. [Google Scholar] [CrossRef]
- Okochi, K.; Suzuki, T.; Inoue, J.-I.; Matsuda, S.; Yamamoto, T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: Implication of Tob in translational control. Genes Cells 2005, 10, 151–163. [Google Scholar] [CrossRef]
- Mao, L.; Zeng, Q.; Su, W.; Song, M.; Li, J.; Xie, M. Elevation of miR-146a Inhibits BTG2/BAX Expression to Ameliorate Postoperative Cognitive Dysfunction Following Probiotics (VSL#3) Treatment. Mol. Neurobiol. 2021, 58, 3457–3470. [Google Scholar]
- Yoshida, Y.; Matsuda, S.; Ikematsu, N.; Kawamura-Tsuzuku, J.; Inazawa, J.; Umemori, H.; Yamamoto, T. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene 1998, 16, 2687–2693. [Google Scholar] [CrossRef]
- Pang, H.; Fan, W.; Shi, X.; Luo, S.; Wang, Y.; Lin, J.; Xiao, Y.; Li, X.; Huang, G.; Xie, Z.; et al. Differential Expression and Bioinformatics Analysis of Plasma-Derived Exosomal circRNA in Type 1 Diabetes Mellitus. J. Immunol. Res. 2022, 2022, 3625052. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Kruit, J.K.; Rome, S.; Menoud, V.; Mulder, N.L.; Jurdzinski, A.; Mancarella, F.; Sebastiani, G.; Donda, A.; Gonzalez, B.J.; et al. Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic β Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab. 2019, 29, 348–361.e6. [Google Scholar] [CrossRef]
- Cianciaruso, C.; Phelps, E.A.; Pasquier, M.; Hamelin, R.; Demurtas, D.; Alibashe Ahmed, M.; Piemonti, L.; Hirosue, S.; Swartz, M.A.; De Palma, M.; et al. Primary Human and Rat β-Cells Release the Intracellular Autoantigens GAD65, IA-2, and Proinsulin in Exosomes Together With Cytokine-Induced Enhancers of Immunity. Diabetes 2017, 66, 460–473. [Google Scholar] [CrossRef]
- Krishnan, P.; Syed, F.; Jiyun Kang, N.; Mirmira, R.G.; Evans-Molina, C. Profiling of RNAs from Human Islet-Derived Exosomes in a Model of Type 1 Diabetes. Int. J. Mol. Sci. 2019, 20, 5903. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Pang, H.; Shi, X.; Li, J.; Wang, Y.; Luo, S.; Lin, J.; Yu, H.; Xiao, Y.; Li, X.; et al. Plasma-derived exosomal mRNA profiles associated with type 1 diabetes mellitus. Front. Immunol. 2022, 13, 995610. [Google Scholar] [CrossRef]
- Pang, H.; Fan, W.; Shi, X.; Li, J.; Wang, Y.; Luo, S.; Lin, J.; Huang, G.; Li, X.; Xie, Z.; et al. Characterization of lncRNA Profiles of Plasma-Derived Exosomes From Type 1 Diabetes Mellitus. Front. Endocrinol. 2022, 13, 822221. [Google Scholar] [CrossRef]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef]
- Nojehdehi, S.; Soudi, S.; Hesampour, A.; Rasouli, S.; Soleimani, M.; Hashemi, S.M. Immunomodulatory effects of mesenchymal stem cell–derived exosomes on experimental type-1 autoimmune diabetes. J. Cell. Biochem. 2018, 119, 9433–9443. [Google Scholar] [CrossRef]
- Chen, X.; Yin, J.; Zhang, F.; Xiao, T.; Zhao, M. has_circ_CCNB1 and has_circ_0009024 function as potential biomarkers for the diagnosis of type 2 diabetes mellitus. J. Clin. Lab. Anal. 2020, 34, e23439. [Google Scholar] [CrossRef]
- Wu, L.; Xiong, L.; Li, J.; Peng, Z.; Zhang, L.; Shi, P.; Gong, Y.; Xiao, H. Circ-Tulp4 promotes β-cell adaptation to lipotoxicity by regulating soat1 expression. J. Mol. Endocrinol. 2020, 65, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wu, S.; Zhu, Y.; Ye, M.; Shen, J.; Liu, Y.; Zhang, Y.; Bu, S. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin. Epigenet. 2019, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Rezaeinejad, F.; Mirzaei, A.; Khalvati, B.; Sabz, G.; Alipoor, B. Circulating expression levels of CircHIPK3 and CDR1as circular-RNAs in type 2 diabetes patients. Mol. Biol. Rep. 2021, 49, 131–138. [Google Scholar] [CrossRef]
- Khan, S.; Jha, A.; Panda, A.C.; Dixit, A. Cancer-Associated circRNA-miRNA-mRNA Regulatory Networks: A Meta-Analysis. Front. Mol. Biosci. 2021, 8, 671309. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Muralidharan, B.; Panda, A.C.; Bakthavachalu, B.; Vindu, A.; Seshadri, V. Glucose-stimulated Translation Regulation of Insulin by the 5′ UTR-binding Proteins. J. Biol. Chem. 2011, 286, 14146–14156. [Google Scholar] [CrossRef]
- Dou, C.; Cao, Z.; Yang, B.; Ding, N.; Hou, T.; Luo, F.; Kang, F.; Li, J.; Yang, X.; Jiang, H.; et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 2016, 6, 21499. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Liu, Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem. Biophys. Res. Commun. 2019, 516, 546–550. [Google Scholar] [CrossRef]
- Zhai, N.; Lu, Y.; Wang, Y.; Ren, X.; Han, J. Circular RNAs and hereditary bone diseases. Intractable Rare Dis. Res. 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, X.; Jian, D.; Hao, P.; Rao, L.; Li, M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2016, 54, 237–245. [Google Scholar] [CrossRef]
- Miao, F.; Yin, B.H.; Zhang, X.; Xue, D.D.; Ma, C. CircRNA_009934 induces osteoclast bone resorption via silencing miR-5107. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7580–7588. [Google Scholar]
- Guan, J.; Gan, L.; Jin, D.; Wu, X.; Cheng, L.; Liu, M.; Fan, Y.; Zhou, J.; Zhang, H.; Zhang, Y.; et al. Overexpression of circ_0021739 in Peripheral Blood Mononuclear Cells in Women with Postmenopausal Osteoporosis Is Associated with Reduced Expression of microRNA-194-5p in Osteoclasts. Experiment 2021, 27, e929170. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, D.; Huang, S.; Zhuang, J.; Zheng, X.; Chang, Y.; Yin, D. Circular RNA YAP1 attenuates osteoporosis through up-regulation of YAP1 and activation of Wnt/β-catenin pathway. Biomed. Pharmacother. 2020, 129, 110365. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, H.; Zou, G.; Cang, D.; Shen, Y. Circular RNA_0062582 promotes osteogenic differentiation of human bone marrow mesenchymal stem cells via regulation of microRNA-145/CBFB axis. Bioengineered 2021, 12, 1952–1963. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Cui, X.; Yang, Y.; Zhou, X. CircRNA hsa_circ_0006215 promotes osteogenic differentiation of BMSCs and enhances osteogenesis–angiogenesis coupling by competitively binding to miR-942-5p and regulating RUNX2 and VEGF. Aging 2021, 13, 10275–10288. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Wang, J.; Fu, Q.; Gu, S.; Rui, Y. CircRUNX2 through has-miR-203 regulates RUNX2 to prevent osteoporosis. J. Cell Mol. Med. 2018, 22, 6112–6121. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, J.; Hou, Y.; Chen, C.; Long, W.; Jiang, H. Alteration of circular RNA expression in rat dental follicle cells during osteogenic differentiation. J. Cell. Biochem. 2019, 120, 13289–13301. [Google Scholar] [CrossRef]
- Guo, Z.; Zhao, L.; Ji, S.; Long, T.; Huang, Y.; Ju, R.; Tang, W.; Tian, W.; Long, J. CircRNA-23525 regulates osteogenic differentiation of adipose-derived mesenchymal stem cells via miR-30a-3p. Cell Tissue Res. 2020, 383, 795–807. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Tan, B.; Wang, D.; Yuan, Z.; Wang, F. Circular RNA circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression. J. Cell. Biochem. 2020, 121, 4819–4826. [Google Scholar] [CrossRef]
- Li, Y.; Fan, L.; Hu, J.; Zhang, L.; Liao, L.; Liu, S.; Wu, D.; Yang, P.; Shen, L.; Chen, J.; et al. MiR-26a Rescues Bone Regeneration Deficiency of Mesenchymal Stem Cells Derived From Osteoporotic Mice. Mol. Ther. 2015, 23, 1349–1357. [Google Scholar] [CrossRef]
- Ajima, R.; Akiyama, T.; Usui, M.; Yoneda, M.; Yoshida, Y.; Nakamura, T.; Minowa, O.; Noda, M.; Tanaka, S.; Noda, T.; et al. Osteoporotic bone formation in mice lacking tob2; involvement of Tob2 in RANK ligand expression and osteoclasts differentiation. FEBS Lett. 2008, 582, 1313–1318. [Google Scholar] [CrossRef]
- Yang, Q.; Jin, L.; Ding, Q.; Hu, W.; Zou, H.; Xiao, M.; Chen, K.; Yu, Y.; Shang, J.; Huang, X.; et al. Novel Therapeutic Mechanism of Adipose-Derived Mesenchymal Stem Cells in Osteoarthritis via Upregulation of BTG2. Oxidative Med. Cell. Longev. 2022, 2022, 9252319. [Google Scholar] [CrossRef]
- Cai, Y.; Lei, X.; Chen, Z.; Mo, Z. The roles of cirRNA in the development of germ cells. Acta Histochem. 2020, 122, 151506. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Morreale, H.F. Polycystic ovary syndrome: Definition, aetiology, diagnosis and treatment. Nat. Rev. Endocrinol. 2018, 14, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Givens, J.R. Familial polycystic ovarian disease. Endocrinol. Metab. Clin. N. Am. 1988, 17, 771–783. [Google Scholar] [CrossRef]
- Escobar-Morreale, H.F.; Samino, S.; Insenser, M.; Vinaixa, M.; Luque-Ramírez, M.; Lasunción, M.A.; Correig, X. Metabolic heterogeneity in polycystic ovary syndrome is determined by obesity: Plasma metabolomic approach using GC-MS. Clin. Chem. 2012, 58, 999–1009. [Google Scholar] [CrossRef]
- Azziz, R.; Woods, K.S.; Reyna, R.; Key, T.J.; Knochenhauer, E.S.; Yildiz, B.O. The Prevalence and Features of the Polycystic Ovary Syndrome in an Unselected Population. J. Clin. Endocrinol. Metab. 2004, 89, 2745–2749. [Google Scholar] [CrossRef]
- Azziz, R.; Carmina, E.; Dewailly, D.; Diamanti-Kandarakis, E.; Escobar-Morreale, H.F.; Futterweit, W.; Janssen, O.E.; Legro, R.S.; Norman, R.J.; Taylor, A.E.; et al. Positions statement: Criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: An Androgen Excess Society guideline. J. Clin. Endocrinol. Metab. 2006, 91, 4237–4245. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017, 66, 22–37e9. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, S.; Cai, B.; Wang, Z.; Chen, M.; Zheng, Z.; Zhou, C. Involvement of ferroptosis in the granulosa cells proliferation of PCOS through the circRHBG/miR-515/SLC7A11 axis. Ann. Transl. Med. 2021, 9, 1348. [Google Scholar] [CrossRef]
- Deng, L.; Chen, Q.; Xie, J.; Wei, W.; Hui, H. circPUM1 promotes polycystic ovary syndrome progression by sponging to miR-760. Gene 2020, 754, 144903. [Google Scholar] [CrossRef]
- Weng, W.; Liu, N.; Toiyama, Y.; Kusunoki, M.; Nagasaka, T.; Fujiwara, T.; Wei, Q.; Qin, H.; Lin, H.; Ma, Y.; et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer 2018, 17, 16. [Google Scholar] [CrossRef]
- Huang, H.; Yu, X.; Han, X.; Hao, J.; Zhao, J.; Bebek, G.; Bao, S.; Prayson, R.A.; Khalil, A.M.; Jankowsky, E.; et al. Piwil1 Regulates Glioma Stem Cell Maintenance and Glioblastoma Progression. Cell Rep. 2021, 34, 108522. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Qi, M.; Xu, L.; Huang, P.; Wang, X.; Sun, J.; Shi, J.; Hu, Y. Differential host circRNA expression profiles in human lung epithelial cells infected with SARS-CoV-2. Infect. Genet. Evol. 2021, 93, 104923. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wang, X.; Sun, H.; Xu, B.; Song, R.; Tian, Y.; Zhao, L.; Xu, Y.; Zhao, Y.; Yang, F.; et al. Oxidant stress-sensitive circRNA Mdc1 controls cardiomyocyte chromosome stability and cell cycle re-entry during heart regeneration. Pharmacol. Res. 2022, 184, 106422. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m6A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef]
- Winkler, G.S. The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 2010, 222, 66–72. [Google Scholar] [CrossRef]
- Doidge, R.; Mittal, S.; Aslam, A.; Winkler, G.S. The Anti-Proliferative Activity of BTG/TOB Proteins Is Mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) Deadenylase Subunits of the Ccr4-Not Complex. PLoS ONE 2012, 7, e51331. [Google Scholar] [CrossRef]
- Tirone, F. The gene PC3TIS21/BTG2, prototype member of the PC3/BTG/TOB family: Regulator in control of cell growth, differentiation, and DNA repair? J. Cell. Physiol. 2001, 187, 155–165. [Google Scholar] [CrossRef]
- Lim, N.S.; Kozlov, G.; Chang, T.-C.; Groover, O.; Siddiqui, N.; Volpon, L.; De Crescenzo, G.; Shyu, A.-B.; Gehring, K. Comparative Peptide Binding Studies of the PABC Domains from the Ubiquitin-protein Isopeptide Ligase HYD and Poly(A)-binding Protein. J. Biol. Chem. 2006, 281, 14376–14382. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, M.; Takeuchi, K.; Noda, N.; Muroya, N.; Suzuki, T.; Nakamura, T.; Kawamura-Tsuzuku, J.; Takahasi, K.; Yamamoto, T.; Inagaki, F. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 2009, 284, 13244–13255. [Google Scholar] [CrossRef]
- Ezzeddine, N.; Chen, C.-Y.A.; Shyu, A.-B. Evidence Providing New Insights into TOB-Promoted Deadenylation and Supporting a Link between TOB’s Deadenylation-Enhancing and Antiproliferative Activities. Mol. Cell. Biol. 2012, 32, 1089–1098. [Google Scholar] [CrossRef]
- Stupfler, B.; Birck, C.; Séraphin, B.; Mauxion, F. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat. Commun. 2016, 7, 10811. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.S.; Lim, J.; Yu, Z.; Kong, P.; Sefik, E.; Xu, H.; Harman, C.C.D.; Kim, L.K.; Lee, G.R.; Li, H.-B.; et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 2020, 367, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Flamand, M.N.; Wu, E.; Vashisht, A.; Jannot, G.; Keiper, B.D.; Simard, M.J.; Wohlschlegel, J.; Duchaine, T.F. Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Res. 2016, 44, 5924–5935. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Mathonnet, G.; Sundermeier, T.; Mathys, H.; Zipprich, J.T.; Svitkin, Y.V.; Rivas, F.; Jinek, M.; Wohlschlegel, J.; Doudna, J.A.; et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 2009, 35, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Cao, Z.; Huang, L.; Liu, S.; Shen, Z.; Wang, Y.; Wang, H.; Zhang, H.; Li, D.; Song, F. CCR4-Not Complex Subunit Not2 Plays Critical Roles in Vegetative Growth, Conidiation and Virulence in Watermelon Fusarium Wilt Pathogen Fusarium oxysporum f. sp. niveum. Front. Microbiol. 2016, 7, 1449. [Google Scholar] [CrossRef]
- Cheng, Q.; Li, Q.; Xu, L.; Jiang, H. Exosomal microRNA-301a-3p promotes the proliferation and invasion of nasopharyngeal carcinoma cells by targeting BTG1 mRNA. Mol. Med. Rep. 2021, 23, 328. [Google Scholar] [CrossRef]
- Wu, X.B.; Wu, Y.T.; Guo, X.X.; Xiang, C.; Chen, P.S.; Qin, W.; Shi, Z.S. Circular RNA hsa_circ_0007990 as a blood biomarker for unruptured intracranial aneurysm with aneurysm wall enhancement. Front. Immunol. 2022, 13, 1061592. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, Y.; Morikawa, S.; Nakashima, M.; Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Suga, N.; Tsuji, A.; Matsuda, S. CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders. Non-Coding RNA 2023, 9, 23. https://doi.org/10.3390/ncrna9020023
Ikeda Y, Morikawa S, Nakashima M, Yoshikawa S, Taniguchi K, Sawamura H, Suga N, Tsuji A, Matsuda S. CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders. Non-Coding RNA. 2023; 9(2):23. https://doi.org/10.3390/ncrna9020023
Chicago/Turabian StyleIkeda, Yuka, Sae Morikawa, Moeka Nakashima, Sayuri Yoshikawa, Kurumi Taniguchi, Haruka Sawamura, Naoko Suga, Ai Tsuji, and Satoru Matsuda. 2023. "CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders" Non-Coding RNA 9, no. 2: 23. https://doi.org/10.3390/ncrna9020023
APA StyleIkeda, Y., Morikawa, S., Nakashima, M., Yoshikawa, S., Taniguchi, K., Sawamura, H., Suga, N., Tsuji, A., & Matsuda, S. (2023). CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders. Non-Coding RNA, 9(2), 23. https://doi.org/10.3390/ncrna9020023






