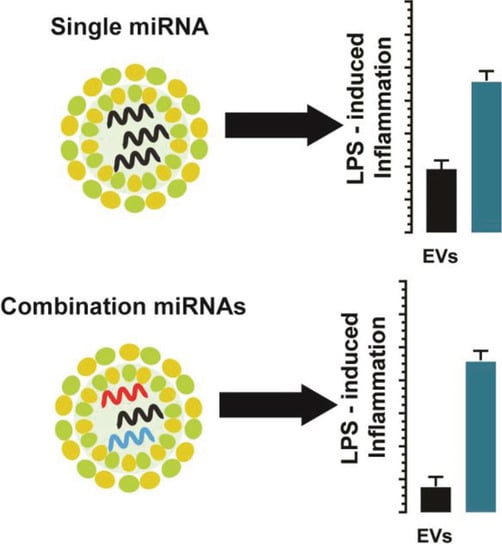
Combinatorial microRNA Loading into Extracellular Vesicles for Increased Anti-Inflammatory Efficacy
Abstract
1. Introduction
2. Results
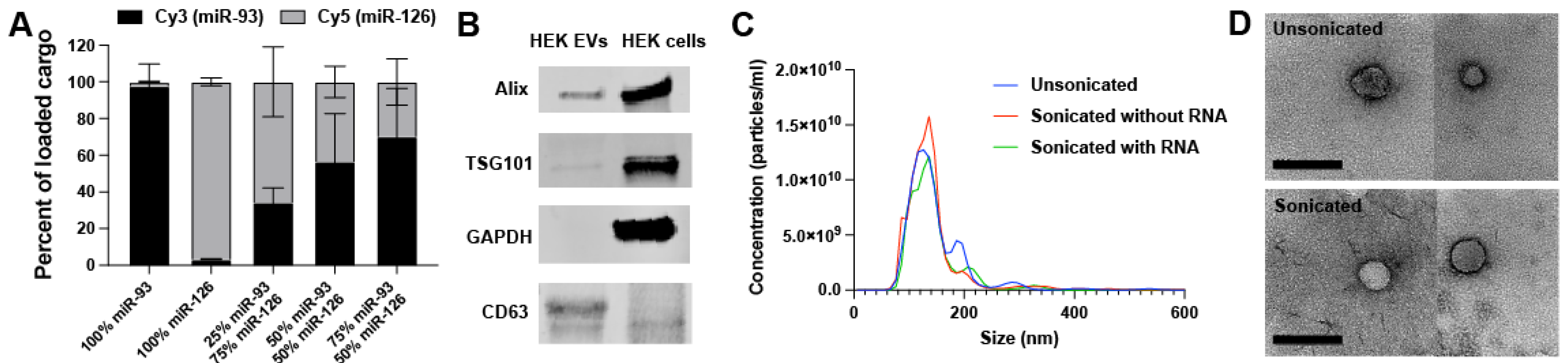
2.1. EV Loading and Characterization
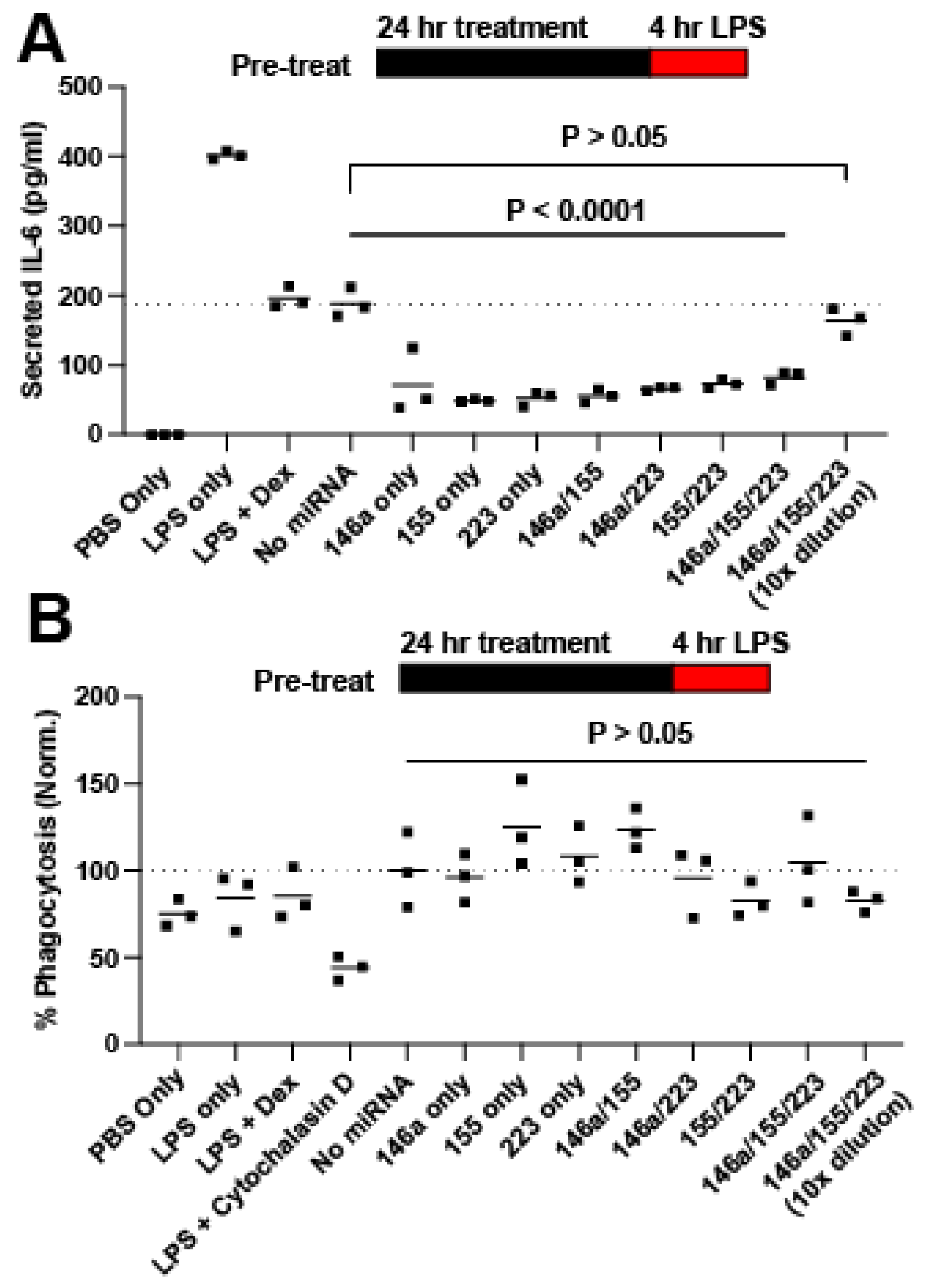
2.2. Screening for Anti-Inflammatory miRNA
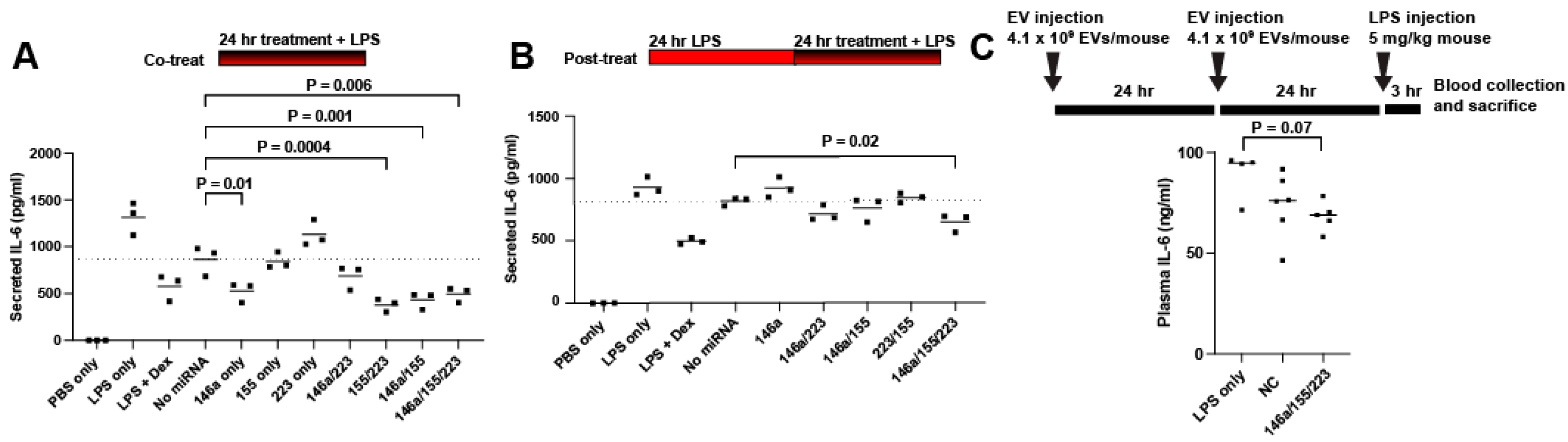
2.3. Delivery of miRNAs 146a/155/223 Has Variable Anti-Inflammatory Effects Aside from Reducing IL-6 Secretion
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Extracellular Vesicle Isolation
4.3. Extracellular Vesicle Loading
4.4. Transmission Electron Microscopy (TEM)
4.5. Fluorescent-Labeled RNA Co-Loading Test
4.6. In Vitro RAW264.7 Inflammatory Assay
4.7. ELISA
4.8. Proteome Array
4.9. In Vivo Endotoxemia Study
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic Inflammation in the Etiology of Disease across the Life Span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Feng, Y.; Jeyaram, A.; Jay, S.M.; Zou, L.; Chao, W. Circulating Plasma Extracellular Vesicles from Septic Mice Induce Inflammation via MicroRNA- and TLR7-Dependent Mechanisms. J. Immunol. 2018, 201, 3392–3400. [Google Scholar] [CrossRef]
- Szilágyi, B.; Fejes, Z.; Pócsi, M.; Kappelmayer, J.; Nagy, B., Jr. Role of Sepsis Modulated Circulating MicroRNAs. EJIFCC 2019, 30, 128–145. [Google Scholar] [PubMed]
- Bai, X.; Zhang, J.; Cao, M.; Han, S.; Liu, Y.; Wang, K.; Han, F.; Li, X.; Jia, Y.; Wang, X.; et al. MicroRNA-146a Protects against LPS-Induced Organ Damage by Inhibiting Notch1 in Macrophage. Int. Immunopharmacol. 2018, 63, 220–226. [Google Scholar] [CrossRef]
- Wang, X.; Gu, H.; Qin, D.; Yang, L.; Huang, W.; Essandoh, K.; Wang, Y.; Caldwell, C.C.; Peng, T.; Zingarelli, B.; et al. Exosomal MIR-223 Contributes to Mesenchymal Stem Cell-Elicited Cardioprotection in Polymicrobial Sepsis. Sci. Rep. 2015, 5, 13721. [Google Scholar] [CrossRef]
- Essandoh, K.; Fan, G.C. Role of Extracellular and Intracellular MicroRNAs in Sepsis. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Feng, Y.; Xu, G.; Jian, W.; Chao, W. Splenic RNA and MicroRNA Mimics Promote Complement Factor B Production and Alternative Pathway Activation via Innate Immune Signaling. J. Immunol. 2016, 196, 2788–2798. [Google Scholar] [CrossRef]
- Shimada, B.K.; Yang, Y.; Zhu, J.; Wang, S.; Suen, A.; Kronstadt, S.M.; Jeyaram, A.; Jay, S.M.; Zou, L.; Chao, W. Extracellular MiR-146a-5p Induces Cardiac Innate Immune Response and Cardiomyocyte Dysfunction. ImmunoHorizons 2020, 4, 561–572. [Google Scholar] [CrossRef]
- Hashemian, S.M.R.; Pourhanifeh, M.H.; Fadaei, S.; Velayati, A.A.; Mirzaei, H.; Hamblin, M.R. Non-Coding RNAs and Exosomes: Their Role in the Pathogenesis of Sepsis. Mol. Ther.-Nucleic Acids 2020, 21, 51–74. [Google Scholar] [CrossRef]
- Feng, Y.; Zou, L.; Yan, D.; Chen, H.; Xu, G.; Jian, W.; Cui, P.; Chao, W. Extracellular MicroRNAs Induce Potent Innate Immune Responses via TLR7/MyD88-Dependent Mechanisms. J. Immunol. 2017, 199, 2106–2117. [Google Scholar] [CrossRef] [PubMed]
- Gurien, S.D.; Aziz, M.; Jin, H.; Wang, H.; He, M.; Al-Abed, Y.; Nicastro, J.M.; Coppa, G.F.; Wang, P. Extracellular Micro RNA 130b-3p Inhibits ECIRP -induced Inflammation. EMBO Rep. 2020, 21, e48075. [Google Scholar] [CrossRef] [PubMed]
- Dumache, R.; Rogobete, A.F.; Bedreag, O.H.; Sarandan, M.; Cradigati, A.C.; Papurica, M.; Dumbuleu, C.M.; Nartita, R.; Sandesc, D. Use of MiRNAs as Biomarkers in Sepsis. Anal. Cell. Pathol. 2015, 2015, 186716. [Google Scholar] [CrossRef]
- Song, Y.; Dou, H.; Li, X.; Zhao, X.; Li, Y.; Liu, D.; Ji, J.; Liu, F.; Ding, L.; Ni, Y.; et al. Exosomal MiR-146a Contributes to the Enhanced Therapeutic Efficacy of Interleukin-1β-Primed Mesenchymal Stem Cells Against Sepsis. Stem Cells 2017, 35, 1208–1221. [Google Scholar] [CrossRef]
- Zhang, L.; He, S.; Wang, Y.; Zhu, X.; Shao, W.; Xu, Q.; Cui, Z. MiRNA-20a Suppressed Lipopolysaccharide-induced HK-2 Cells Injury via NFκB and ERK1/2 Signaling by Targeting CXCL12. Mol. Immunol. 2020, 118, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Piffoux, M.; Volatron, J.; Silva, A.K.A.; Gazeau, F. Thinking Quantitatively of RNA-Based Information Transfer via Extracellular Vesicles: Lessons to Learn for the Design of RNA-Loaded EVs. Pharmaceutics 2021, 13, 1931. [Google Scholar] [CrossRef]
- Albanese, M.; Chen, Y.F.A.; Hüls, C.; Gärtner, K.; Tagawa, T.; Mejias-Perez, E.; Keppler, O.T.; Göbel, C.; Zeidler, R.; Shein, M.; et al. MicroRNAs Are Minor Constituents of Extracellular Vesicles That Are Rarely Delivered to Target Cells. PLoS Genet. 2021, 17, e1009951. [Google Scholar] [CrossRef]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and Stoichiometric Analysis of the MicroRNA Content of Exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef]
- Didiot, M.C.; Hall, L.M.; Coles, A.H.; Haraszti, R.A.; Godinho, B.M.D.C.; Chase, K.; Sapp, E.; Ly, S.; Alterman, J.F.; Hassler, M.R.; et al. Exosome-Mediated Delivery of Hydrophobically Modified SiRNA for Huntingtin MRNA Silencing. Mol. Ther. 2016, 24, 1836–1847. [Google Scholar] [CrossRef]
- Kamerkar, S.; Lebleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Perets, N.; Betzer, O.; Ben-Shaul, S.; Sheinin, A.; Michaelevski, I.; Popovtzer, R.; Offen, D.; Levenberg, S. Intranasal Delivery of Mesenchymal Stem Cell Derived Exosomes Loaded with Phosphatase and Tensin Homolog SiRNA Repairs Complete Spinal Cord Injury. ACS Nano 2019, 13, 10015–10028. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Querejeta, I.; Carregal-Romero, S.; Ayerdi-Izquierdo, A.; Mäger, I.; Nash, L.A.; Wood, M.; Egimendia, A.; Betanzos, M.; Alberro, A.; Iparraguirre, L.; et al. MiR-219a-5p Enriched Extracellular Vesicles Induce OPC Differentiation and EAE Improvement More Efficiently than Liposomes and Polymeric Nanoparticles. Pharmaceutics 2020, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Wolfram, J. Extracellular Vesicles versus Synthetic Nanoparticles for Drug Delivery. Nat. Rev. Mater. 2021, 6, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.E.; de Jong, O.G.; Evers, M.J.W.; Nurazizah, M.; Schiffelers, R.M.; Vader, P. Natural or Synthetic RNA Delivery: A Stoichiometric Comparison of Extracellular Vesicles and Synthetic Nanoparticles. Nano Lett. 2021, 21, 1888–1895. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Jeyaram, A.; Patel, D.B.; Parajuli, B.; Livingston, N.K.; Arumugasaamy, N.; Schardt, J.S.; Jay, S.M. Oncogene Knockdown via Active Loading of Small RNAs into Extracellular Vesicles by Sonication. Cell. Mol. Bioeng. 2016, 9, 315–324. [Google Scholar] [CrossRef]
- Pacienza, N.; Lee, R.H.; Bae, E.H.; Kim, D.K.; Liu, Q.; Prockop, D.J.; Yannarelli, G. In Vitro Macrophage Assay Predicts the In Vivo Anti-Inflammatory Potential of Exosomes from Human Mesenchymal Stromal Cells. Mol. Ther.-Methods Clin. Dev. 2018, 13, 612–632. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Lamichhane, T.N.; Raiker, R.S.; Jay, S.M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation Is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharm. 2015, 12, 3650–3657. [Google Scholar] [CrossRef]
- Arenas-Padilla, M.; Mata-Haro, V. Regulation of TLR Signaling Pathways by MicroRNAs: Implications in Inflammatory Diseases. Cent. Eur. J. Immunol. 2018, 43, 482–489. [Google Scholar] [CrossRef]
- Yan, Y.; Lu, K.; Ye, T.; Zhang, Z. MicroRNA-223 Attenuates LPS-Induced Inflammation in an Acute Lung Injury Model via the NLRP3 Inflammasome and TLR4/NF-ΚB Signaling Pathway via RHOB. Int. J. Mol. Med. 2019, 43, 1467–1477. [Google Scholar] [CrossRef] [PubMed]
- Raza Naqvi, A.; Zhong, S. Expression Profiling of LPS Responsive MiRNA in Primary Human Macrophages. J. Microb. Biochem. Technol. 2016, 08, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, H.; Liu, Y.; Song, Y.; Lai, L.; Han, Q.; Cao, X.; Wang, Q. Inducible MicroRNA-223 down-Regulation Promotes TLR-Triggered IL-6 and IL-1β Production in Macrophages by Targeting STAT3. PLoS ONE 2012, 7, e42971. [Google Scholar] [CrossRef]
- Chen, M.; Wang, F.; Xia, H.; Yao, S. MicroRNA-155: Regulation of Immune Cells in Sepsis. Mediat. Inflamm. 2021, 2021, 8874854. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA MiR-223 as Regulator of Innate Immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi-Jahromi, S.S.; Aslani, M.; Mirshafiey, A. A Comprehensive Review on MiR-146a Molecular Mechanisms in a Wide Spectrum of Immune and Non-Immune Inflammatory Diseases. Immunol. Lett. 2020, 227, 8–27. [Google Scholar] [CrossRef]
- Huang, R.; Hu, G.; Lin, B.; Lin, Z.; Sun, C. MicroRNA-155 Silencing Enhances Inflammatory Response and Lipid Uptake in Oxidized Low-Density Lipoprotein-Stimulated Human THP-1 Macrophages. J. Investig. Med. 2010, 58, 961–967. [Google Scholar] [CrossRef]
- Tang, B.; Xiao, B.; Liu, Z.; Li, N.; Zhu, E.-D.; Li, B.-S.; Xie, Q.-H.; Zhuang, Y.; Zou, Q.-M.; Mao, X.-H. Identification of MyD88 as a Novel Target of MiR-155, Involved in Negative Regulation of Helicobacter Pylori-Induced Inflammation. FEBS Lett. 2010, 584, 1481–1486. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Y.; Suen, A.; Zhu, J.; Williams, B.; Hu, J.; Chen, F.; Kozar, R.; Shen, S.; Li, Z.; et al. Role of Extracellular MicroRNA-146a-5p in Host Innate Immunity and Bacterial Sepsis. iScience 2021, 24, 103441. [Google Scholar] [CrossRef]
- Huang, H.; Zhu, J.; Gu, L.; Hu, J.; Feng, X.; Huang, W.; Wang, S.; Yang, Y.; Cui, P.; Lin, S.-H.; et al. TLR7 Mediates Acute Respiratory Distress Syndrome in Sepsis by Sensing Extracellular MiR-146a. Am. J. Respir. Cell Mol. Biol. 2022, 67, 375–388. [Google Scholar] [CrossRef]
- Choi, H.; Kim, Y.; Mirzaaghasi, A.; Heo, J.; Kim, Y.N.; Shin, J.H.; Kim, S.; Kim, N.H.; Cho, E.S.; Yook, J.I.; et al. Exosome-Based Delivery of Super-Repressor IκBα Relieves Sepsis-Associated Organ Damage and Mortality. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, V.; Nowicki, M.O.; Idriss, M.; Jimenez, M.A.; Lugli, G.; Hayes, J.L.; Mahmoud, A.B.; Zane, R.E.; Passaro, C.; Ligon, K.L.; et al. The Functional Synergism of MicroRNA Clustering Provides Therapeutically Relevant Epigenetic Interference in Glioblastoma. Nat. Commun. 2019, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Marik, P.E.; Khangoora, V.; Rivera, R.; Hooper, M.H.; Catravas, J. Hydrocortisone, Vitamin C, and Thiamine for the Treatment of Severe Sepsis and Septic Shock: A Retrospective Before-After Study. Chest 2017, 151, 1229–1238. [Google Scholar] [CrossRef]
- Marik, P.E. Hydrocortisone, Ascorbic Acid and Thiamine (HAT Therapy) for the Treatment of Sepsis. Focus on Ascorbic Acid. Nutrients 2018, 10, 1762. [Google Scholar] [CrossRef]
- Bayraktar, R.; Bertilaccio, M.T.S.; Calin, G.A. The Interaction between Two Worlds: MicroRNAs and Toll-like Receptors. Front. Immunol. 2019, 10, 1053. [Google Scholar] [CrossRef]
- Schulte, L.N.; Westermann, A.J.; Vogel, J. Differential Activation and Functional Specialization of MiR-146 and MiR-155 in Innate Immune Sensing. Nucleic Acids Res. 2013, 41, 542–553. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol Phosphatase SHIP1 Is a Primary Target of MiR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.-J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of MiR-155 and MiR-125b Levels Following Lipopolysaccharide/TNF-α Stimulation and Their Possible Roles in Regulating the Response to Endotoxin Shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-Delivered MicroRNAs Modulate the Inflammatory Response to Endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef]
- Chvatchko, Y.; Hoogewerf, A.J.; Meyer, A.; Alouani, S.; Juillard, P.; Buser, R.; Conquet, F.; Proudfoot, A.E.I.; Wells, T.N.C.; Power, C.A. A Key Role for CC Chemokine Receptor 4 in Lipopolysaccharide-Induced Endotoxic Shock. J. Exp. Med. 2000, 191, 1755–1763. [Google Scholar] [CrossRef]
- Molinaro, R.; Pecli, C.; Guilherme, R.F.; Alves-Filho, J.C.; Cunha, F.Q.; Canetti, C.; Kunkel, S.L.; Bozza, M.T.; Benjamim, C.F. CCR4 Controls the Suppressive Effects of Regulatory T Cells on Early and Late Events during Severe Sepsis. PLoS ONE 2015, 10, e0133227. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wiklander, O.P.B.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular Vesicle in Vivo Biodistribution Is Determined by Cell Source, Route of Administration and Targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef]
- Mirzaaghasi, A.; Han, Y.; Ahn, S.H.; Choi, C.; Park, J.H. Biodistribution and Pharmacokinectics of Liposomes and Exosomes in a Mouse Model of Sepsis. Pharmaceutics 2021, 13, 427. [Google Scholar] [CrossRef] [PubMed]
- Born, L.J.; Chang, K.H.; Shoureshi, P.; Lay, F.; Bengali, S.; Hsu, A.T.W.; Abadchi, S.N.; Harmon, J.W.; Jay, S.M. HOTAIR-Loaded Mesenchymal Stem/Stromal Cell Extracellular Vesicles Enhance Angiogenesis and Wound Healing. Adv. Healthc. Mater. 2022, 11, 2002070. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pottash, A.E.; Levy, D.; Jeyaram, A.; Kuo, L.; Kronstadt, S.M.; Chao, W.; Jay, S.M. Combinatorial microRNA Loading into Extracellular Vesicles for Increased Anti-Inflammatory Efficacy. Non-Coding RNA 2022, 8, 71. https://doi.org/10.3390/ncrna8050071
Pottash AE, Levy D, Jeyaram A, Kuo L, Kronstadt SM, Chao W, Jay SM. Combinatorial microRNA Loading into Extracellular Vesicles for Increased Anti-Inflammatory Efficacy. Non-Coding RNA. 2022; 8(5):71. https://doi.org/10.3390/ncrna8050071
Chicago/Turabian StylePottash, Alex Eli, Daniel Levy, Anjana Jeyaram, Leo Kuo, Stephanie M. Kronstadt, Wei Chao, and Steven M. Jay. 2022. "Combinatorial microRNA Loading into Extracellular Vesicles for Increased Anti-Inflammatory Efficacy" Non-Coding RNA 8, no. 5: 71. https://doi.org/10.3390/ncrna8050071
APA StylePottash, A. E., Levy, D., Jeyaram, A., Kuo, L., Kronstadt, S. M., Chao, W., & Jay, S. M. (2022). Combinatorial microRNA Loading into Extracellular Vesicles for Increased Anti-Inflammatory Efficacy. Non-Coding RNA, 8(5), 71. https://doi.org/10.3390/ncrna8050071