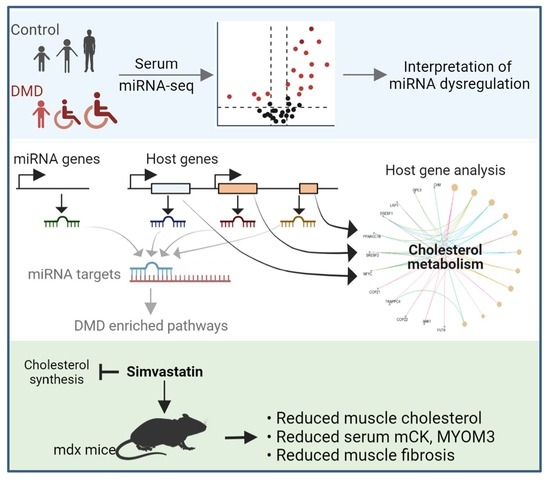
Deciphering the Molecular Mechanism of Incurable Muscle Disease by a Novel Method for the Interpretation of miRNA Dysregulation
Abstract
:1. MicroRNA Dysregulation in Human Diseases
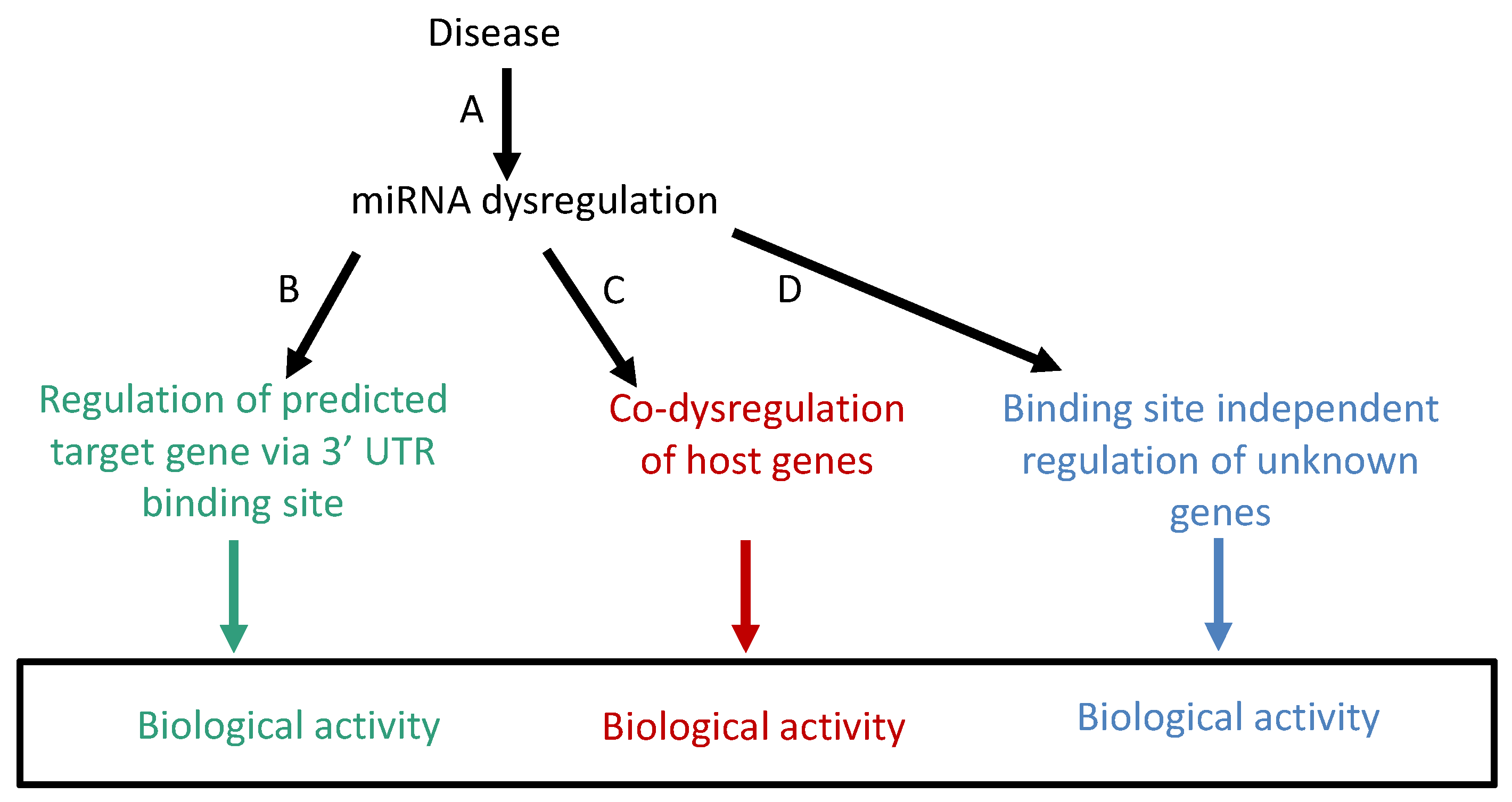
2. Problems and Weaknesses of Biological Interpretations of miRNA Dysregulation Using the Target Gene Method
3. Development of a Complementary Method for the Interpretation of miRNA Dysregulation
4. Interpretation of miRNA Dysregulation in DMD Using the Target Gene Method
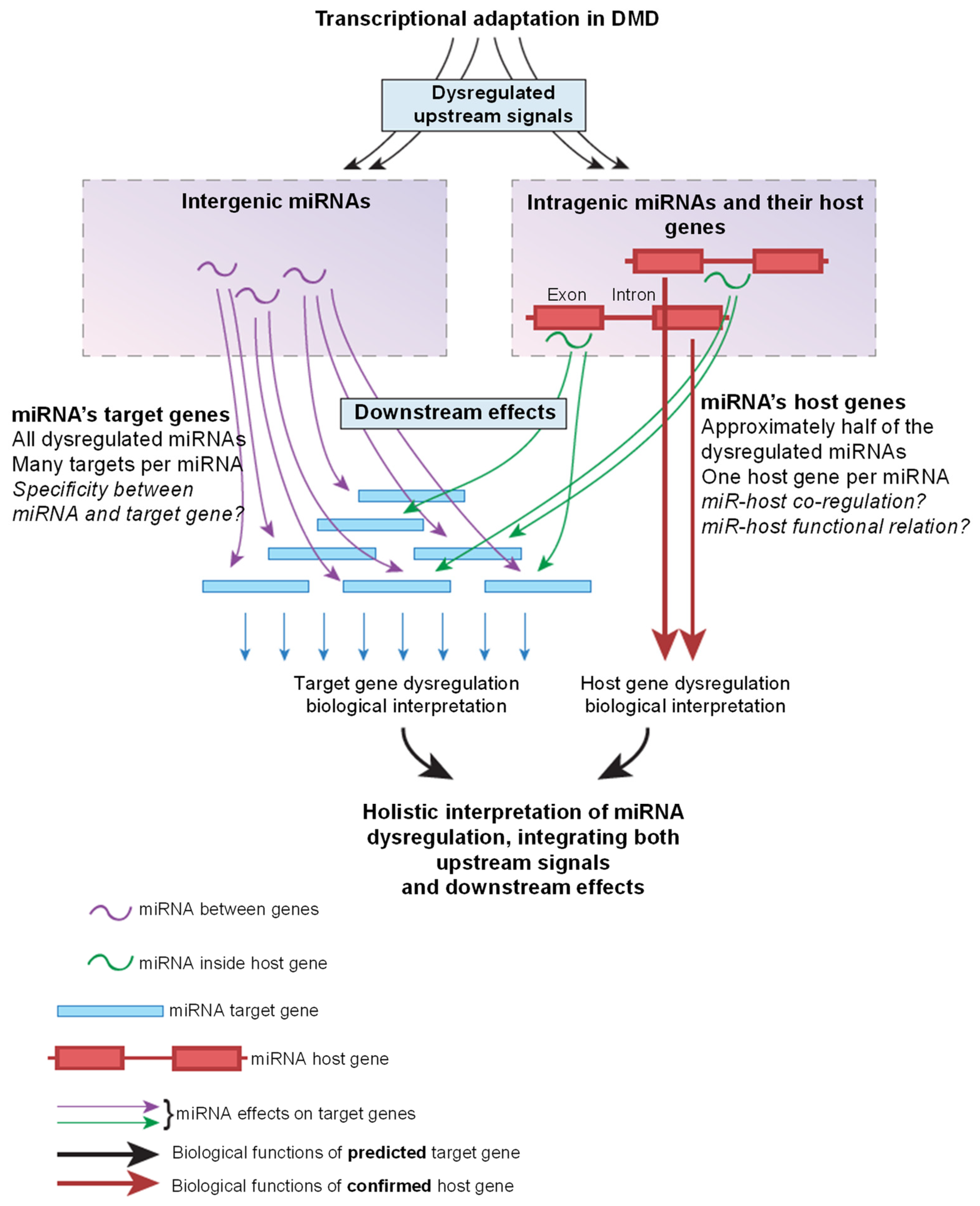
5. Interrelations between miRNAs and Their Host Genes
6. Co-Dysregulation of miRNA and Host Genes in DMD
7. miRNAs and Their Host Genes May Coordinately Regulate Disease State in DMD
8. Interpretation of miRNA Dysregulation in DMD Using the Host Gene Method
9. Target Genes versus Host Genes for the Interpretation of miRNA Dysregulation
10. Experimental Validation in DMD
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Miska, E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Munker, R.; Calin, G.A. MicroRNA profiling in cancer. Clin. Sci. 2011, 121, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. Causes and Consequences of microRNA Dysregulation. Cancer J. 2012, 18, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Giza, D.E.; Vasilescu, C.; Calin, G.A. Key principles of miRNA involvement in human diseases. Discoveries 2014, 2, e34. [Google Scholar] [CrossRef]
- Juźwik, C.A.; Drake, S.S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.S.; Fournier, A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 2003, 113, 673–676. [Google Scholar] [CrossRef] [Green Version]
- Bleazard, T.; Lamb, J.A.; Griffiths-Jones, S. Bias in microRNA functional enrichment analysis. Bioinformatics 2015, 31, 1592–1598. [Google Scholar] [CrossRef] [Green Version]
- Godard, P.; van Eyll, J. Pathway analysis from lists of microRNAs: Common pitfalls and alternative strategy. Nucleic Acids Res. 2015, 43, 3490–3497. [Google Scholar] [CrossRef]
- Pinzón, N.; Li, B.; Martinez, L.; Sergeeva, A.; Presumey, J.; Apparailly, F.; Seitz, H. microRNA target prediction programs predict many false positives. Genome Res. 2017, 27, 234–245. [Google Scholar] [CrossRef] [Green Version]
- Fridrich, A.; Hazan, Y.; Moran, Y. Too Many False Targets for MicroRNAs: Challenges and Pitfalls in Prediction of miRNA Targets and Their Gene Ontology in Model and Non-model Organisms. Bioessays 2019, 41, 1800169. [Google Scholar] [CrossRef] [Green Version]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [Green Version]
- Odame, E.; Chen, Y.; Zheng, S.; Dai, D.; Kyei, B.; Zhan, S.; Cao, J.; Guo, J.; Zhong, T.; Wang, L.; et al. Enhancer RNAs: Transcriptional regulators and workmates of NamiRNAs in myogenesis. Cell. Mol. Biol. Lett. 2021, 26, 4. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 2022, 38, 379–394. [Google Scholar] [CrossRef]
- Amor, F.; Vu Hong, A.; Corre, G.; Sanson, M.; Suel, L.; Blaie, S.; Servais, L.; Voit, T.; Richard, I.; Israeli, D.; et al. Cholesterol metabolism is a potential therapeutic target in Duchenne muscular dystrophy. J. Cachexia. Sarcopenia Muscle 2021, 12, 677–693. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Hinske, L.C.; França, G.S.; Torres, H.A.M.; Ohara, D.T.; Lopes-Ramos, C.M.; Heyn, J.; Reis, L.F.L.; Ohno-Machado, L.; Kreth, S.; Galante, P.A.F. miRIAD-integrating microRNA inter- and intragenic data. Database 2014, 2014, bau099. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [Green Version]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Lu, M.; Miao, J.; Li, T.; Wang, E.; Cui, Q. Cepred: Predicting the Co-Expression Patterns of the Human Intronic microRNAs with Their Host Genes. PLoS ONE 2009, 4, e4421. [Google Scholar] [CrossRef] [PubMed]
- Godnic, I.; Zorc, M.; Jevsinek Skok, D.; Calin, G.A.; Horvat, S.; Dovc, P.; Kovac, M.; Kunej, T. Genome-wide and species-wide in silico screening for intragenic MicroRNAs in human, mouse and chicken. PLoS ONE 2013, 8, e65165. [Google Scholar] [CrossRef] [Green Version]
- Hinske, L.; Galante, P.A.; Kuo, W.P.; Ohno-Machado, L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genom. 2010, 11, 533. [Google Scholar] [CrossRef] [Green Version]
- Lutter, D.; Marr, C.; Krumsiek, J.; Lang, E.W.; Theis, F.J. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genom. 2010, 11, 224. [Google Scholar] [CrossRef] [Green Version]
- Boivin, V.; Deschamps-Francoeur, G.; Scott, M.S. Protein coding genes as hosts for noncoding RNA expression. Semin. Cell Dev. Biol. 2018, 75, 3–12. [Google Scholar] [CrossRef]
- Zeidler, M.; Hüttenhofer, A.; Kress, M.; Kummer, K.K. Intragenic MicroRNAs Autoregulate Their Host Genes in Both Direct and Indirect Ways-A Cross-Species Analysis. Cells 2020, 9, 232. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Xie, J.; Sun, H. Three miRNAs cooperate with host genes involved in human cardiovascular disease. Hum. Genom. 2019, 13, 40. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Shyr, Y.; Cai, J.; Liu, Q. Interplay between miRNAs and host genes and their role in cancer. Brief. Funct. Genom. 2018, 18, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Biasiolo, M.; Sales, G.; Lionetti, M.; Agnelli, L.; Todoerti, K.; Bisognin, A.; Coppe, A.; Romualdi, C.; Neri, A.; Bortoluzzi, S. Impact of Host Genes and Strand Selection on miRNA and miRNA* Expression. PLoS ONE 2011, 6, e23854. [Google Scholar] [CrossRef]
- Steiman-Shimony, A.; Shtrikman, O.; Margalit, H. Assessing the functional association of intronic miRNAs with their host genes. RNA 2018, 24, 991–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilfred, B.R.; Wang, W.X.; Nelson, P.T. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007, 91, 209–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dansie, L.E.; Reeves, S.; Miller, K.; Zano, S.P.; Frank, M.; Pate, C.; Wang, J.; Jackowski, S. Physiological roles of the pantothenate kinases. Biochem. Soc. Trans. 2014, 42, 1033–1036. [Google Scholar] [CrossRef] [Green Version]
- Czumaj, A.; Szrok-Jurga, S.; Hebanowska, A.; Turyn, J.; Swierczynski, J.; Sledzinski, T.; Stelmanska, E. The Pathophysiological Role of CoA. Int. J. Mol. Sci. 2020, 21, 9057. [Google Scholar] [CrossRef]
- Pegoraro, E.; Hoffman, E.P.; Piva, L.; Gavassini, B.F.; Cagnin, S.; Ermani, M.; Bello, L.; Soraru, G.; Pacchioni, B.; Bonifati, M.D.; et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology 2011, 76, 219–226. [Google Scholar] [CrossRef] [Green Version]
- Ohsakaya, S.; Fujikawa, M.; Hisabori, T.; Yoshida, M. Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J. Biol. Chem. 2011, 286, 20292–20296. [Google Scholar] [CrossRef] [Green Version]
- Sanson, M.; Hog Vu, A.; Massourides, E.; Bourg, N.; Suel, L.; Amor, F.; Corre, G.; Bénit, P.; Barthelemy, I.; Blot, S.; et al. miR-379 links glucocorticoid treatment with mitochondrial response in Duchenne muscular dystrophy. Sci. Rep. 2020, 10, 9139. [Google Scholar] [CrossRef] [PubMed]
- Vu Hong, A.; Sanson, M.; Richard, I.; Israeli, D. A revised model for mitochondrial dysfunction in Duchenne muscular dystrophy. Eur. J. Transl. Myol. 2021, 31, 2021. [Google Scholar] [CrossRef]
- Xie, S.; Jiang, X.; Qin, R.; Song, S.; Lu, Y.; Wang, L.; Chen, Y.; Lu, D. miR-1307 promotes hepatocarcinogenesis by CALR-OSTC-endoplasmic reticulum protein folding pathway. iScience 2021, 24, 103271. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, M.; Wu, J.; Jie, Z.; Chang, S.; Shuang, T. The clinicopathological significance of miR-1307 in chemotherapy resistant epithelial ovarian cancer. J. Ovarian Res. 2015, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Bible, K.L.; Regnier, M.; Adams, M.E.; Froehner, S.C.; Whitehead, N.P. Simvastatin provides long-term improvement of left ventricular function and prevents cardiac fibrosis in muscular dystrophy. Physiol. Rep. 2019, 7, e14018. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, N.P.; Kim, M.J.; Bible, K.L.; Adams, M.E.; Froehner, S.C. A new therapeutic effect of simvastatin revealed by functional improvement in muscular dystrophy. Proc. Natl. Acad. Sci. USA 2015, 112, 12864–12869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, Z.; Theret, M.; Milad, N.; Tung, L.W.; Chen, W.W.; Sirois, M.G.; Rossi, F.; Bernatchez, P. Cholesterol absorption blocker ezetimibe prevents muscle wasting in severe dysferlin-deficient and mdx mice. J. Cachexia. Sarcopenia Muscle 2021, 13, 544–560. [Google Scholar] [CrossRef] [PubMed]
- Bourg, N.; Hong, A.V.; Lostal, W.; Jaber, A.; Guerchet, N.; Tanniou, G.; Bordier, F.; Bertil-Froidevaux, E.; Georger, C.; Daniele, N.; et al. Co-Administration of Simvastatin Does Not Potentiate the Benefit of Gene Therapy in the mdx Mouse Model for Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2022, 23, 2016. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Israeli, D.; Vu Hong, A.; Corre, G.; Miagoux, Q.; Richard, I. Deciphering the Molecular Mechanism of Incurable Muscle Disease by a Novel Method for the Interpretation of miRNA Dysregulation. Non-Coding RNA 2022, 8, 48. https://doi.org/10.3390/ncrna8040048
Israeli D, Vu Hong A, Corre G, Miagoux Q, Richard I. Deciphering the Molecular Mechanism of Incurable Muscle Disease by a Novel Method for the Interpretation of miRNA Dysregulation. Non-Coding RNA. 2022; 8(4):48. https://doi.org/10.3390/ncrna8040048
Chicago/Turabian StyleIsraeli, David, Ai Vu Hong, Guillaume Corre, Quentin Miagoux, and Isabelle Richard. 2022. "Deciphering the Molecular Mechanism of Incurable Muscle Disease by a Novel Method for the Interpretation of miRNA Dysregulation" Non-Coding RNA 8, no. 4: 48. https://doi.org/10.3390/ncrna8040048
APA StyleIsraeli, D., Vu Hong, A., Corre, G., Miagoux, Q., & Richard, I. (2022). Deciphering the Molecular Mechanism of Incurable Muscle Disease by a Novel Method for the Interpretation of miRNA Dysregulation. Non-Coding RNA, 8(4), 48. https://doi.org/10.3390/ncrna8040048