Blood miRNAs Are Linked to Frequent Asthma Exacerbations in Childhood Asthma and Adult COPD
Abstract: Rationale
1. Introduction
2. Methods
2.1. Study Population
2.2. Generalization Population
2.3. Primary Outcome
2.4. Sample Sequencing and Quality Control
2.5. Identification of Differentially Expressed miRNAs
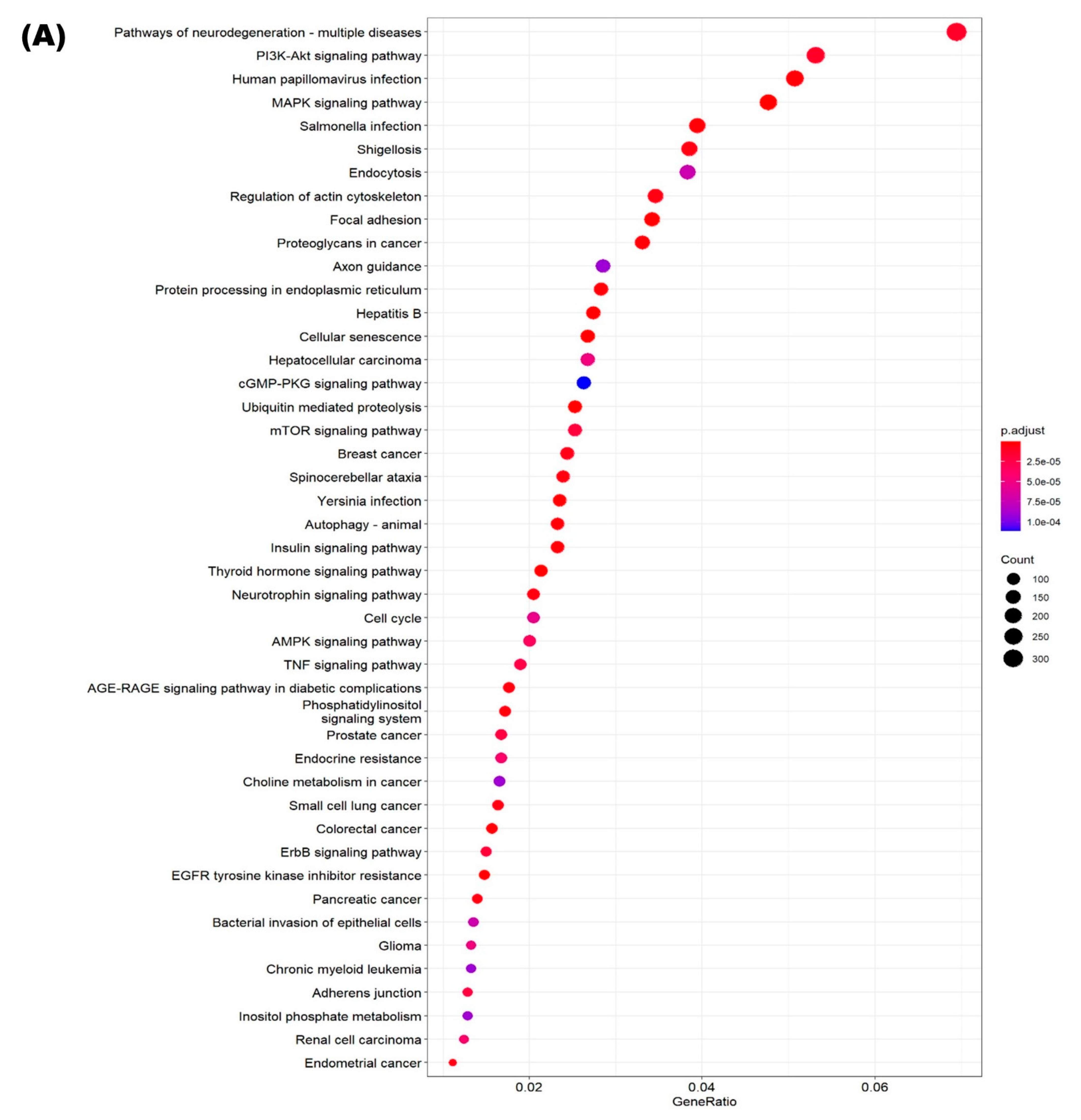
2.6. Functional Annotation of Differentially Expressed miRNAs
3. Results
3.1. Cohort Characteristics
3.2. miRNA Sequencing
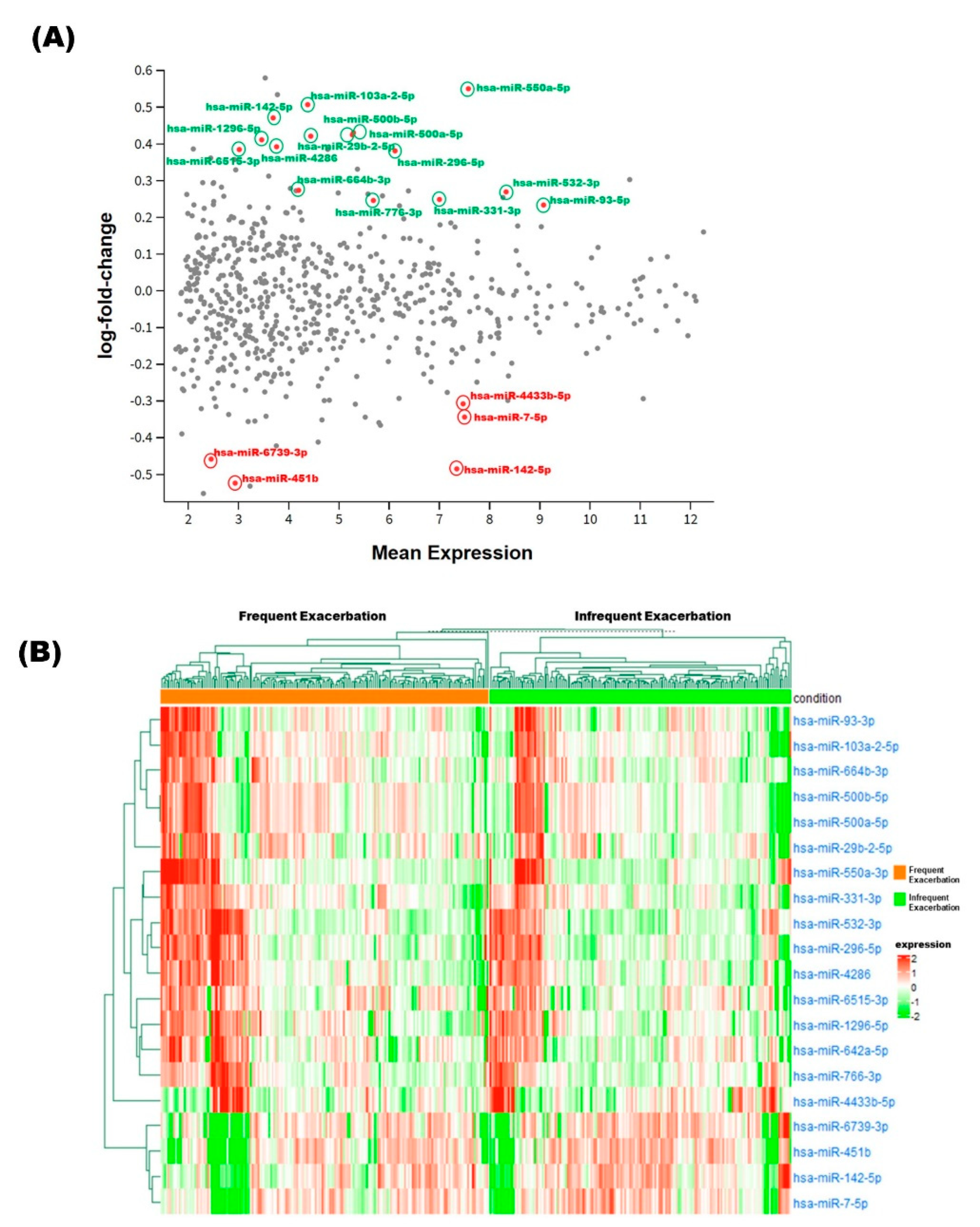
3.3. Identification of Differentially Expressed miRNAs
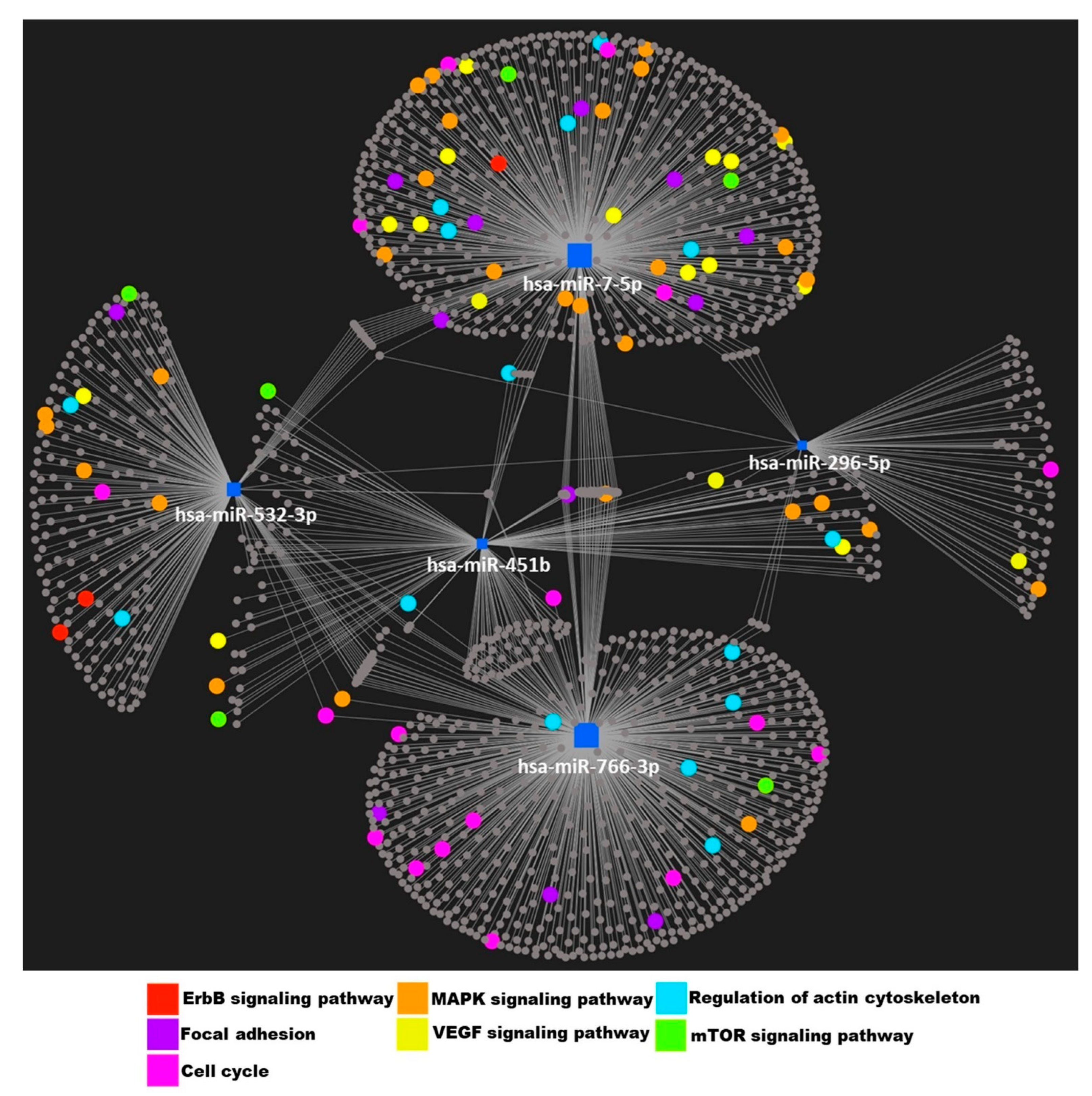
3.4. Identification of Putative Targets and Functional Assessment of Differentially Expressed miRNAs
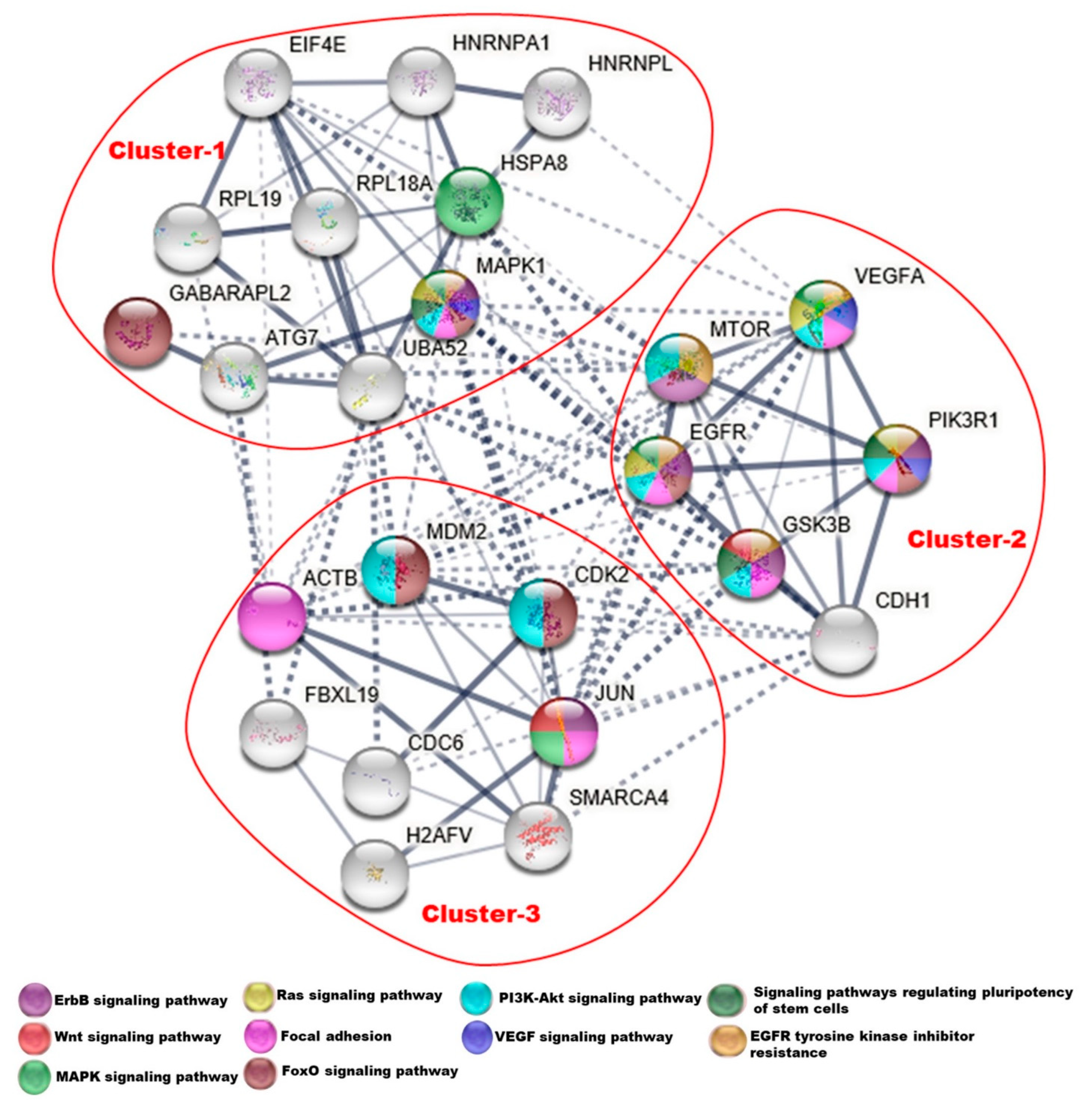
3.5. Protein–Protein Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- May, S.M.; Li, J.T. Burden of chronic obstructive pulmonary disease: Healthcare costs and beyond. Allergy Asthma Proc. 2015, 36, 4–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donaldson, G.C.; Seemungal, T.A.; Bhowmik, A.; Wedzicha, J.A. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax 2002, 57, 847–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postma, D.S.; Weiss, S.T.; van den Berge, M.; Kerstjens, H.A.; Koppelman, G.H. Revisiting the Dutch hypothesis. J. Allergy Clin. Immunol. 2015, 136, 521–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, A.; Li, J.; Kho, A.T.; Sun, M.; Lu, Q.; Weiss, S.T.; Tantisira, K.G.; McGeachie, M.J. COPD-associated miR-145-5p is downregulated in early-decline FEV1 trajectories in childhood asthma. J. Allergy Clin. Immunol. 2021, 147, 2181–2190. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, E.G.; Mestdagh, P.; Vandesompele, J.; Brusselle, G.G.; Bracke, K.R. Non-coding RNAs in the pathogenesis of COPD. Thorax 2015, 70, 782–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandan, K.; Gupta, M.; Sarwat, M. Role of Host and Pathogen-Derived MicroRNAs in Immune Regulation During Infectious and Inflammatory Diseases. Front. Immunol. 2019, 10, 3081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupani, H.; Sanchez-Elsner, T.; Howarth, P. MicroRNAs and respiratory diseases. Eur. Respir. J. 2013, 41, 695–705. [Google Scholar] [CrossRef]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [Green Version]
- Kho, A.T.; McGeachie, M.J.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Tantisira, K.G. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir. Res. 2018, 19, 128. [Google Scholar] [CrossRef] [Green Version]
- Gomez, J.L.; Chen, A.; Diaz, M.P.; Zirn, N.; Gupta, A.; Britto, C.; Sauler, M.; Yan, X.; Stewart, E.; Santerian, K.; et al. A Network of Sputum MicroRNAs Is Associated with Neutrophilic Airway Inflammation in Asthma. Am. J. Respir. Crit Care Med. 2020, 202, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, U.S.; Holgate, S.T.; Reddy, P.S.; Hill, A.A.; McKee, C.M.; Csimma, C.I.; Weaver, A.A.; Legault, H.M.; Small, C.G.; Ramsey, R.C.; et al. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PLoS ONE 2011, 6, e21902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsalik, E.L.; Henao, R.; Nichols, M.; Burke, T.; Ko, E.R.; McClain, M.T.; Hudson, L.L.; Mazur, A.; Freeman, D.H.; Veldman, T.; et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 2016, 8, 322ra311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lydon, E.C.; Bullard, C.; Aydin, M.; Better, O.M.; Mazur, A.; Nicholson, B.P.; Ko, E.R.; McClain, M.T.; Ginsburg, G.S.; Woods, C.W.; et al. A host gene expression approach for identifying triggers of asthma exacerbations. PLoS ONE 2019, 14, e0214871. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.L.; Diaz, M.P.; Nino, G.; Britto, C.J. Impaired type I interferon regulation in the blood transcriptome of recurrent asthma exacerbations. BMC Med. Genom. 2018, 11, 21. [Google Scholar] [CrossRef]
- Croteau-Chonka, D.C.; Qiu, W.; Martinez, F.D.; Strunk, R.C.; Lemanske, R.F., Jr.; Liu, A.H.; Gilliland, F.D.; Millstein, J.; Gauderman, W.J.; Ober, C.; et al. Gene Expression Profiling in Blood Provides Reproducible Molecular Insights into Asthma Control. Am. J. Respir. Crit. Care Med. 2017, 195, 179–188. [Google Scholar] [CrossRef]
- Barnes, P.J. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008, 8, 183–192. [Google Scholar] [CrossRef]
- Morrow, J.D.; Qiu, W.; Chhabra, D.; Rennard, S.I.; Belloni, P.; Belousov, A.; Pillai, S.G.; Hersh, C.P. Identifying a gene expression signature of frequent COPD exacerbations in peripheral blood using network methods. BMC Med. Genom. 2015, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Kho, A.T.; Sordillo, J.; Wu, A.C.; Cho, M.H.; Sharma, S.; Tiwari, A.; Lasky-Su, J.; Weiss, S.T.; Tantisira, K.G.; McGeachie, M.J. CASTER: Cross-Sectional Asthma STEroid Response Measurement. J. Pers. Med. 2020, 10, 95. [Google Scholar] [CrossRef]
- Hunninghake, G.M.; Soto-Quiros, M.E.; Avila, L.; Ly, N.P.; Liang, C.; Sylvia, J.S.; Klanderman, B.J.; Silverman, E.K.; Celedon, J.C. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J. Allergy Clin. Immunol. 2007, 119, 654–661. [Google Scholar] [CrossRef]
- Blumenthal, M.N.; Banks-Schlegel, S.; Bleecker, E.R.; Marsh, D.G.; Ober, C. Collaborative studies on the genetics of asthma--National Heart, Lung and Blood Institute. Clin. Exp. Allergy 1995, 25 (Suppl. S2), 29–32. [Google Scholar] [CrossRef] [PubMed]
- Regan, E.A.; Hokanson, J.E.; Murphy, J.R.; Make, B.; Lynch, D.A.; Beaty, T.H.; Curran-Everett, D.; Silverman, E.K.; Crapo, J.D. Genetic epidemiology of COPD (COPDGene) study design. COPD J. Chronic Obstr. Pulm. Dis. 2010, 7, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Han, M.K.; Kazerooni, E.A.; Lynch, D.A.; Liu, L.X.; Murray, S.; Curtis, J.L.; Criner, G.J.; Kim, V.; Bowler, R.P.; Hanania, N.A.; et al. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: Associated radiologic phenotypes. Radiology 2011, 261, 274–282. [Google Scholar] [CrossRef] [PubMed]
- LaBelle, J.; Bowser, M.; Brown, A.; Farnam, L.; Kho, A.; Li, J.; McGeachie, M.; Chase, R.; Piehl, S.; Allen, K.; et al. Commercially Available Blocking Oligonucleotides Effectively Suppress Unwanted Hemolysis Related miRNAs in a Large Whole Blood RNA Cohort. J. Mol. Diagn. 2021, 23, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kho, A.T.; Chase, R.P.; Pantano, L.; Farnam, L.; Amr, S.S.; Tantisira, K.G. COMPSRA: A COMprehensive Platform for Small RNA-Seq data Analysis. Sci. Rep. 2020, 10, 4552. [Google Scholar] [CrossRef] [PubMed]
- Reese, S.E.; Archer, K.J.; Therneau, T.M.; Atkinson, E.J.; Vachon, C.M.; de Andrade, M.; Kocher, J.P.; Eckel-Passow, J.E. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics 2013, 29, 2877–2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Davis, J.S.; Sun, M.; Kho, A.T.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating microRNAs and association with methacholine PC20 in the Childhood Asthma Management Program (CAMP) cohort. PLoS ONE 2017, 12, e0180329. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.; Yang, Y.; Zhang, C.; Sun, Y.F.; Zhu, W.; Ma, C.L.; Zhou, X.Y. [Prediction of microRNA-296-5p target genes and its application in lung development]. Zhongguo Dang Dai Er Ke Za Zhi 2016, 18, 1302–1307. [Google Scholar]
- Skronska-Wasek, W.; Mutze, K.; Baarsma, H.A.; Bracke, K.R.; Alsafadi, H.N.; Lehmann, M.; Costa, R.; Stornaiuolo, M.; Novellino, E.; Brusselle, G.G.; et al. Reduced Frizzled Receptor 4 Expression Prevents WNT/beta-Catenin-driven Alveolar Lung Repair in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2017, 196, 172–185. [Google Scholar] [CrossRef]
- Verhamme, F.M.; Bracke, K.R.; Joos, G.F.; Brusselle, G.G. Transforming growth factor-beta superfamily in obstructive lung diseases. more suspects than TGF-beta alone. Am. J. Respir. Cell Mol. Biol. 2015, 52, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Panganiban, R.; Kho, A.T.; McGeachie, M.J.; Farnam, L.; Chase, R.P.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating MicroRNAs and Treatment Response in Childhood Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Kawasaki, M.; Hirai, T.; Yoshida, Y.; Tsushima, H.; Fujishiro, M.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. MicroRNA-766-3p Contributes to Anti-Inflammatory Responses through the Indirect Inhibition of NF-kappaB Signaling. Int. J. Mol. Sci. 2019, 20, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, W.C.; Kwan, C.K.; Yau, S.; So, P.P.; Poon, P.C.; Au, J.S. The role of inflammation in the pathogenesis of lung cancer. Expert Opin. Ther. Targets 2011, 15, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Peng, R.; Peng, H.; Yao, L.; Sun, Y.; Wen, L.; Wu, T.; Zhou, J.; Zhang, Z. MiR-451 suppresses cell proliferation and metastasis in A549 lung cancer cells. Mol. Biotechnol. 2015, 57, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pelaia, C.; Vatrella, A.; Gallelli, L.; Lombardo, N.; Sciacqua, A.; Savino, R.; Pelaia, G. Role of p38 Mitogen-Activated Protein Kinase in Asthma and COPD: Pathogenic Aspects and Potential Targeted Therapies. Drug Des. Devel Ther. 2021, 15, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, B.N.; Hammad, H.; Fahy, J.V. The Cytokines of Asthma. Immunity 2019, 50, 975–991. [Google Scholar] [CrossRef]
- Pelaia, C.; Vatrella, A.; Sciacqua, A.; Terracciano, R.; Pelaia, G. Role of p38-mitogen-activated protein kinase in COPD: Pathobiological implications and therapeutic perspectives. Expert Rev. Respir. Med. 2020, 14, 485–491. [Google Scholar] [CrossRef]
- DiMango, E.; Rogers, L.; Reibman, J.; Gerald, L.B.; Brown, M.; Sugar, E.A.; Henderson, R.; Holbrook, J.T. Risk Factors for Asthma Exacerbation and Treatment Failure in Adults and Adolescents with Well-controlled Asthma during Continuation and Step-Down Therapy. Ann. Am. Thorac. Soc. 2018, 15, 955–961. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, Z.; Zeng, J.; Zheng, S.; Sun, L.; Zhu, L.; Liao, W. Th17/Treg imbalance is associated with reduced indoleamine 2,3 dioxygenase activity in childhood allergic asthma. Allergy Asthma Clin. Immunol. 2020, 16, 61. [Google Scholar] [CrossRef]
- Athari, S.S. Targeting cell signaling in allergic asthma. Signal Transduct. Target. Ther. 2019, 4, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Miao, Y.; Gao, X.; Wang, Y.Y.; Wang, H.; Zheng, Y.W.; Zhao, Z.Y. MicroRNA-200a Affects the Proliferation of Airway Smooth Muscle Cells and Airway Remodeling by Targeting FOXC1 via the PI3K/AKT Signaling Pathway in Ovalbumin-Induced Asthmatic Mice. Cell Physiol. Biochem. 2018, 50, 2365–2389. [Google Scholar] [CrossRef] [PubMed]
- Bozinovski, S.; Vlahos, R.; Hansen, M.; Liu, K.; Anderson, G.P. Akt in the pathogenesis of COPD. Int. J. Chronic Obs. Pulmon Dis. 2006, 1, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Uliczka, K.; Bossen, J.; Niu, X.; Fink, C.; Thiedmann, M.; Knop, M.; Vock, C.; Abdelsadik, A.; Zissler, U.M.; et al. Constitutive immune activity promotes JNK- and FoxO-dependent remodeling of Drosophila airways. Cell Rep. 2021, 35, 108956. [Google Scholar] [CrossRef] [PubMed]
- Zaslona, Z.; Peters-Golden, M. Prostanoids in Asthma and COPD: Actions, Dysregulation, and Therapeutic Opportunities. Chest 2015, 148, 1300–1306. [Google Scholar] [CrossRef] [Green Version]
- Bartel, S.; Schulz, N.; Alessandrini, F.; Schamberger, A.C.; Pagel, P.; Theis, F.J.; Milger, K.; Noessner, E.; Stick, S.M.; Kicic, A.; et al. Pulmonary microRNA profiles identify involvement of Creb1 and Sec14l3 in bronchial epithelial changes in allergic asthma. Sci. Rep. 2017, 7, 46026. [Google Scholar] [CrossRef] [Green Version]
- Alashkar Alhamwe, B.; Miethe, S.; Pogge von Strandmann, E.; Potaczek, D.P.; Garn, H. Epigenetic Regulation of Airway Epithelium Immune Functions in Asthma. Front. Immunol. 2020, 11, 1747. [Google Scholar] [CrossRef]
- Papi, A.; Bellettato, C.M.; Braccioni, F.; Romagnoli, M.; Casolari, P.; Caramori, G.; Fabbri, L.M.; Johnston, S.L. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am. J. Respir. Crit. Care Med. 2006, 173, 1114–1121. [Google Scholar] [CrossRef]
- Roberts, B.S.; Hardigan, A.A.; Kirby, M.K.; Fitz-Gerald, M.B.; Wilcox, C.M.; Kimberly, R.P.; Myers, R.M. Blocking of targeted microRNAs from next-generation sequencing libraries. Nucleic Acids Res. 2015, 43, e145. [Google Scholar] [CrossRef] [Green Version]

|
Non-Exacerbation (N = 168) |
Exacerbation (N = 183) | p-Value | |
|---|---|---|---|
| Gender | |||
| Male | 73 (43.5%) | 69 (37.7%) | 0.324 |
| Female | 95 (56.5%) | 114 (62.3%) | |
| Age (years) | |||
| Mean (SD) | 9.40 (1.83) | 8.89 (1.88) | 0.0109 |
| Median [Min, Max] | 9.29 [4.50, 13.3] | 8.65 [0.271, 13.0] | |
| Height (cm) | |||
| Mean (SD) | 133 (15.7) | 131 (11.7) | 0.161 |
| Median [Min, Max] | 133 [0, 163] | 129 [108, 167] | |
| Weight (kg) | |||
| Mean (SD) | 34.2 (12.2) | 31.4 (11.1) | 0.0245 |
| Median [Min, Max] | 30.9 [15.0, 71.7] | 27.9 [17.0, 81.6] | |
| BMI | |||
| Mean (SD) | 18.6 (4.35) | 17.8 (3.56) | 0.0634 |
| Median [Min, Max] | 17.2 [11.3, 41.4] | 16.8 [12.7, 34.0] | |
| % Predicted Pre-BD FEV1 | |||
| Mean (SD) | 98.6 (15.1) | 100 (17.1) | 0.423 |
| Median [Min, Max] | 97.5 [53.1, 144] | 99.6 [46.2, 180] | |
| % Predicted Pre-BD FVC | |||
| Mean (SD) | 101 (15.3) | 104 (16.0) | 0.0662 |
| Median [Min, Max] | 99.0 [52.5, 151] | 102 [54.6, 174] | |
| FEV1/FVC pre-bronchodilator | |||
| Mean (SD) | 87.5 (7.60) | 85.6 (7.99) | 0.0283 |
| Median [Min, Max] | 87.5 [65.3,100] | 86.0 [61.8,99.9] | |
| FEV1/FVC post-bronchodilator | |||
| Mean (SD) | 89.9 (6.02) | 88.2 (7.07) | 0.0179 |
| Median [Min, Max] | 89.4 [67.0,100] | 89.2 [66.4,100] | |
| Bronchodilator Response as % of baseline FEV1 | |||
| Mean (SD) | 4.38 (8.47) | 6.37 (9.57) | 0.0405 |
| Median [Min, Max] | 2.88 [−15.3,48.6] | 5.10 [−16.3,47.2] | |
| Inhaled Steroids | |||
| No | 88 (52.4%) | 70 (38.3%) | 0.0108 |
| Yes | 80 (47.6%) | 113 (61.7%) | |
| Total IgE | |||
| Mean (SD) | 1.79 (0.408) | 1.81 (0.391) | 0.596 |
| Median [Min, Max] | 2.00 [1.00,2.00] | 2.00 [1.00,2.00] | |
| Eosinophil Count | |||
| Mean (SD) | 1.40 (0.491) | 1.51 (0.501) | 0.0444 |
| Median [Min, Max] | 1.00 [1.00,2.00] | 2.00 [1.00,2.00] |
| No Exacerbations (N = 122) | Severe Exacerbations (N = 24) | p-Value | |
|---|---|---|---|
| Gender | |||
| Male | 54 (44.3%) | 10 (42.7%) | 0.99 |
| Female | 68 (55.7%) | 14 (58.3%) | |
| Age (years) | |||
| Mean (SD) | 68.6 (8.8) | 69.5 (9.16) | 0.64 |
| Race (% African American) | 27 (22.1) | 7 (29.2) | 0.63 |
| Current Smoking (%) | 46 (37.7) | 8 (33.3) | 0.86 |
| Pack Years Smoking | |||
| Mean (SD) | 51.7 (26.6) | 51.9 (28.8) | 0.97 |
| % predicted FEV1 | |||
| Mean (SD) | 53.9 (16.5) | 53.2 (17.6) | 0.85 |
| FEV1/FVC | |||
| Mean (SD) | 0.52 (0.12) | 0.52 (0.11) | 0.95 |
| Asthma Diagnosed before Age 40 (%) | 15 (12.3) | 5 (20.8) | 0.26 |
| Base Mean | log2FC | Beta | Odds Ratio | p-Value | FDR | |
|---|---|---|---|---|---|---|
| hsa-miR-451b | 18.29 | −0.524 | −0.065 | 0.57 | 1.88 × 10−4 | 2.65 × 10−2 |
| hsa-miR-142-5p | 1550.70 | −0.485 | −0.057 | 0.62 | 5.76× 10−4 | 4.15× 10−2 |
| hsa-miR-6739-3p | 11.14 | −0.458 | −0.063 | 0.64 | 8.29× 10−4 | 4.50× 10−2 |
| hsa-miR-7-5p | 1811.60 | −0.344 | −0.041 | 0.80 | 3.02× 10−3 | 9.80× 10−2 |
| hsa-miR-4433b-5p | 1759.78 | −0.308 | −0.059 | 0.80 | 2.36× 10−3 | 9.00× 10−2 |
| hsa-miR-93-3p | 8737.45 | 0.234 | 0.105 | 1.22 | 2.32×10−3 | 9.00× 10−2 |
| hsa-miR-766-3p | 294.18 | 0.246 | 0.065 | 1.31 | 2.80× 10−3 | 9.56× 10−2 |
| hsa-miR-331-3p | 1098.62 | 0.249 | 0.052 | 1.19 | 2.64× 10−3 | 9.53× 10−2 |
| hsa-miR-532-3p | 4142.15 | 0.27 | 0.102 | 1.25 | 9.33× 10−4 | 4.50× 10−2 |
| hsa-miR-664b-3p | 66.08 | 0.274 | 0.074 | 1.27 | 1.04× 10−3 | 4.50× 10−2 |
| hsa-miR-296-5p | 452.86 | 0.381 | 0.063 | 1.35 | 9.60× 10−4 | 4.50× 10−2 |
| hsa-miR-6515-3p | 19.91 | 0.385 | 0.064 | 1.43 | 4.79× 10−4 | 4.15× 10−2 |
| hsa-miR-4286 | 42.35 | 0.392 | 0.049 | 1.37 | 9.82× 10−4 | 4.50× 10−2 |
| hsa-miR-1296-5p | 31.37 | 0.411 | 0.038 | 1.31 | 9.00× 10−4 | 4.50× 10−2 |
| hsa-miR-29b-2-5p | 84.16 | 0.421 | 0.081 | 1.48 | 5.79× 10−5 | 2.65× 10−2 |
| hsa-miR-500b-5p | 193.56 | 0.425 | 0.072 | 1.59 | 1.48× 10−4 | 2.65× 10−2 |
| hsa-miR-500a-5p | 197.54 | 0.431 | 0.072 | 1.59 | 1.20× 10−4 | 2.65× 10−2 |
| hsa-miR-642a-5p | 39.54 | 0.471 | 0.045 | 1.44 | 5.20× 10−4 | 4.15× 10−2 |
| hsa-miR-103a-2-5p | 79.21 | 0.507 | 0.043 | 1.50 | 2.04× 10−4 | 2.65× 10−2 |
| hsa-miR-550a-3p | 1953.34 | 0.55 | 0.045 | 1.40 | 3.17× 10−4 | 3.43× 10−2 |
| log2FC | p-Value | |
|---|---|---|
| hsa-miR-451b | −0.636 | 0.054 |
| hsa-miR-7-5p | −0.524 | 0.064 |
| hsa-miR-532-3p | 0.311 | 0.02 |
| hsa-miR-296-5p | 0.391 | 0.021 |
| hsa-miR-766-3p | 0.289 | 0.036 |
| p-Value | Beta | OR | 2.50% | 97.50% | |
|---|---|---|---|---|---|
| hsa-miR-7-5p | 0.094 | 0.031 | 0.031 | −0.005 | 0.068 |
| hsa-miR-451b | 0.023 | 0.033 | 0.033 | 0.004 | 0.062 |
| hsa-miR-296-5p | 0.909 | 0.002 | 0.002 | −0.036 | 0.041 |
| hsa-miR-532-3p | 0.039 | −0.060 | −0.060 | −0.118 | −0.003 |
| hsa-miR-766-3p | 0.962 | 0.001 | 0.001 | −0.041 | 0.043 |
| p-Value | Beta | OR | 2.50% | 97.50% | |
|---|---|---|---|---|---|
| hsa-miR-7-5p | 0.325 | 0.023 | 0.023 | −0.023 | 0.071 |
| hsa-miR-451b | 0.904 | 0.002 | 0.002 | −0.034 | 0.039 |
| hsa-miR-296-5p | 0.516 | 0.016 | 0.016 | −0.033 | 0.066 |
| hsa-miR-532-3p | 0.449 | −0.028 | −0.028 | −0.103 | 0.045 |
| hsa-miR-766-3p | 0.183 | 0.036 | 0.036 | −0.017 | 0.090 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, A.; Hobbs, B.D.; Li, J.; Kho, A.T.; Amr, S.; Celedón, J.C.; Weiss, S.T.; Hersh, C.P.; Tantisira, K.G.; McGeachie, M.J. Blood miRNAs Are Linked to Frequent Asthma Exacerbations in Childhood Asthma and Adult COPD. Non-Coding RNA 2022, 8, 27. https://doi.org/10.3390/ncrna8020027
Tiwari A, Hobbs BD, Li J, Kho AT, Amr S, Celedón JC, Weiss ST, Hersh CP, Tantisira KG, McGeachie MJ. Blood miRNAs Are Linked to Frequent Asthma Exacerbations in Childhood Asthma and Adult COPD. Non-Coding RNA. 2022; 8(2):27. https://doi.org/10.3390/ncrna8020027
Chicago/Turabian StyleTiwari, Anshul, Brian D. Hobbs, Jiang Li, Alvin T. Kho, Samir Amr, Juan C. Celedón, Scott T. Weiss, Craig P. Hersh, Kelan G. Tantisira, and Michael J. McGeachie. 2022. "Blood miRNAs Are Linked to Frequent Asthma Exacerbations in Childhood Asthma and Adult COPD" Non-Coding RNA 8, no. 2: 27. https://doi.org/10.3390/ncrna8020027
APA StyleTiwari, A., Hobbs, B. D., Li, J., Kho, A. T., Amr, S., Celedón, J. C., Weiss, S. T., Hersh, C. P., Tantisira, K. G., & McGeachie, M. J. (2022). Blood miRNAs Are Linked to Frequent Asthma Exacerbations in Childhood Asthma and Adult COPD. Non-Coding RNA, 8(2), 27. https://doi.org/10.3390/ncrna8020027







