Exosomes in Epilepsy of Tuberous Sclerosis Complex: Carriers of Pro-Inflammatory MicroRNAs
Abstract
1. Introduction
2. Results
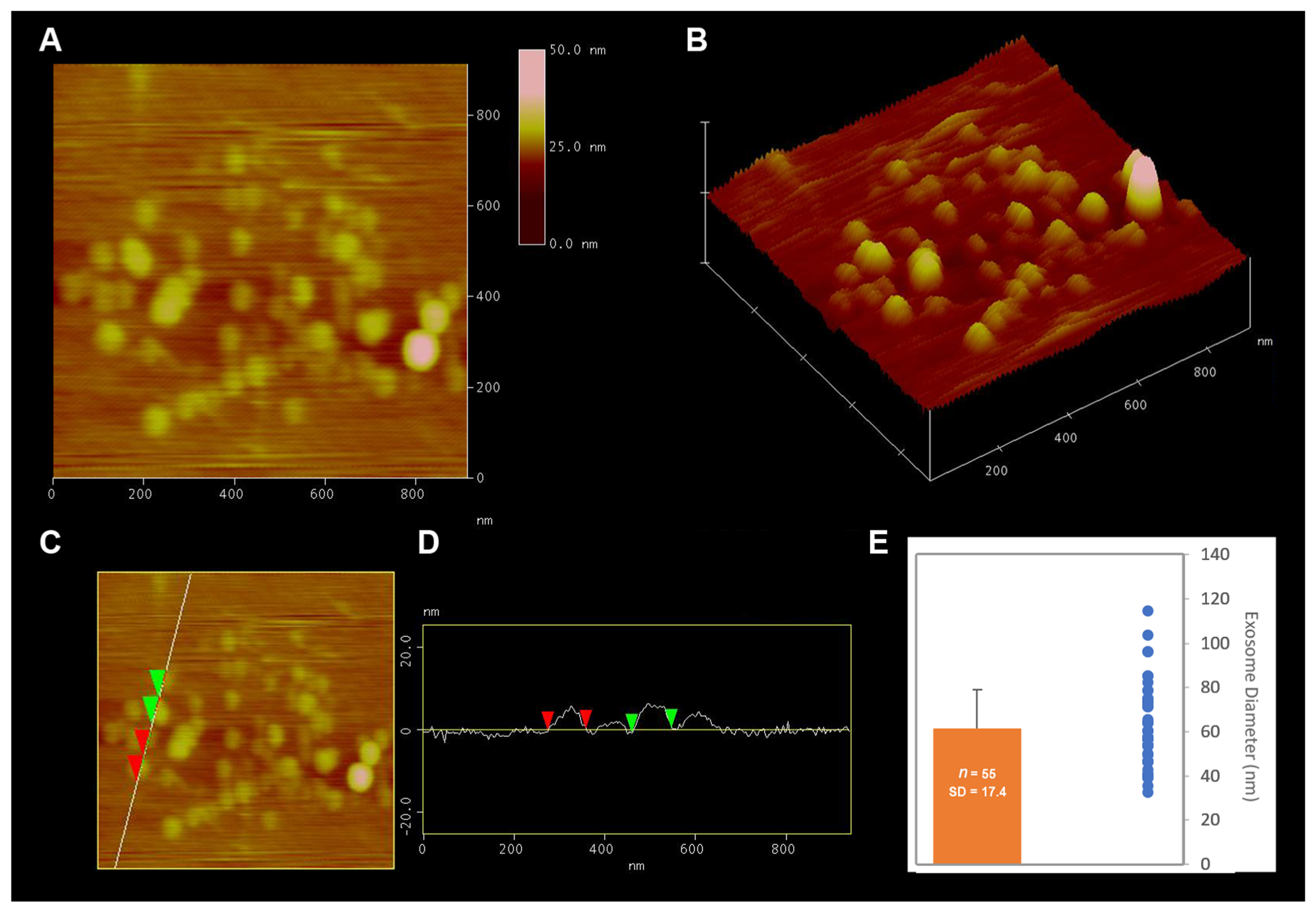
2.1. Isolation and Characterization of Exosome Enriched Fractions of Extracellular Vesicles (EV) from Brain Tissue
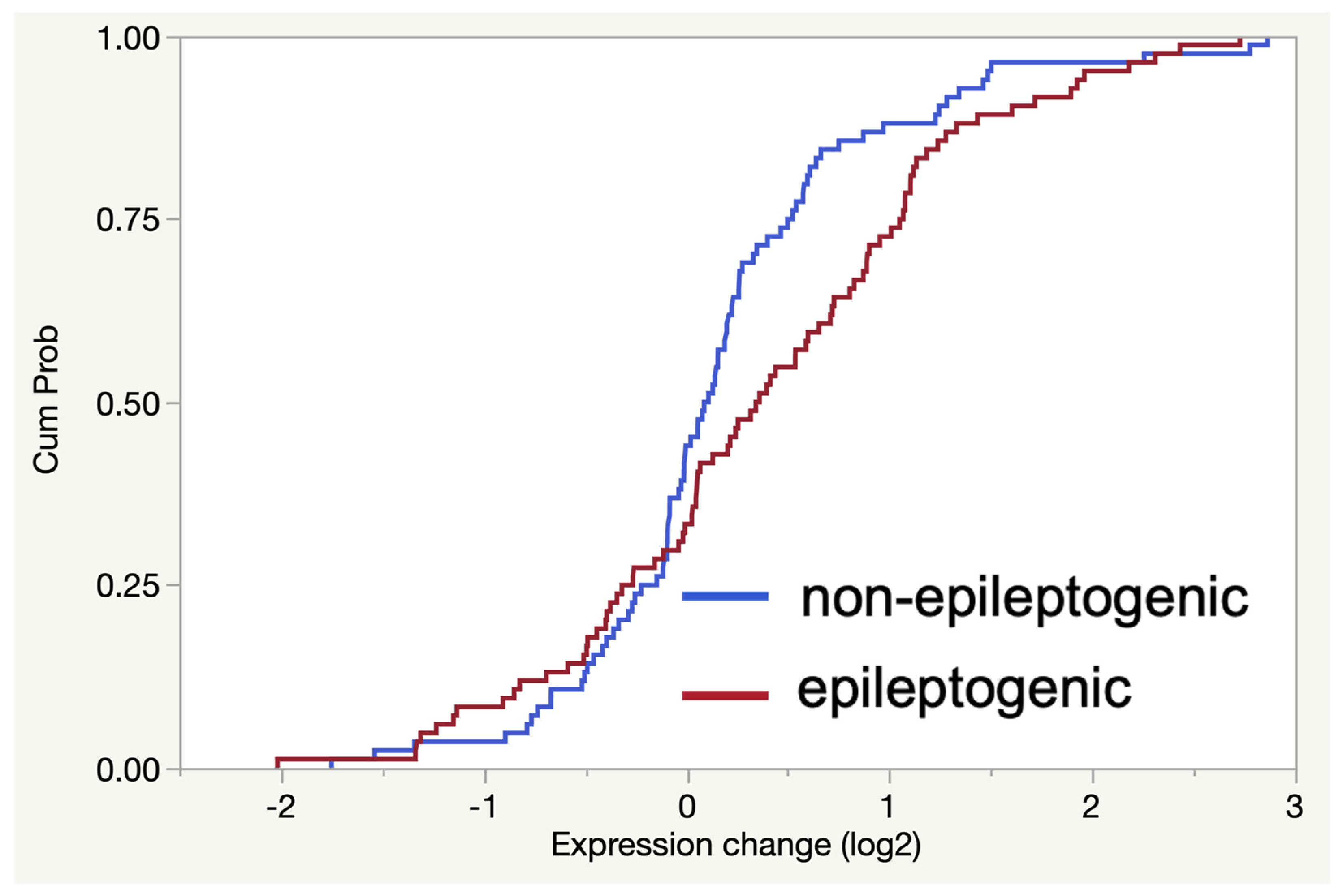
2.2. RNA-Seq Analysis Reveals Enrichment of TLR7/8-Activating MicroRNAs in Epileptogenic Exosomes
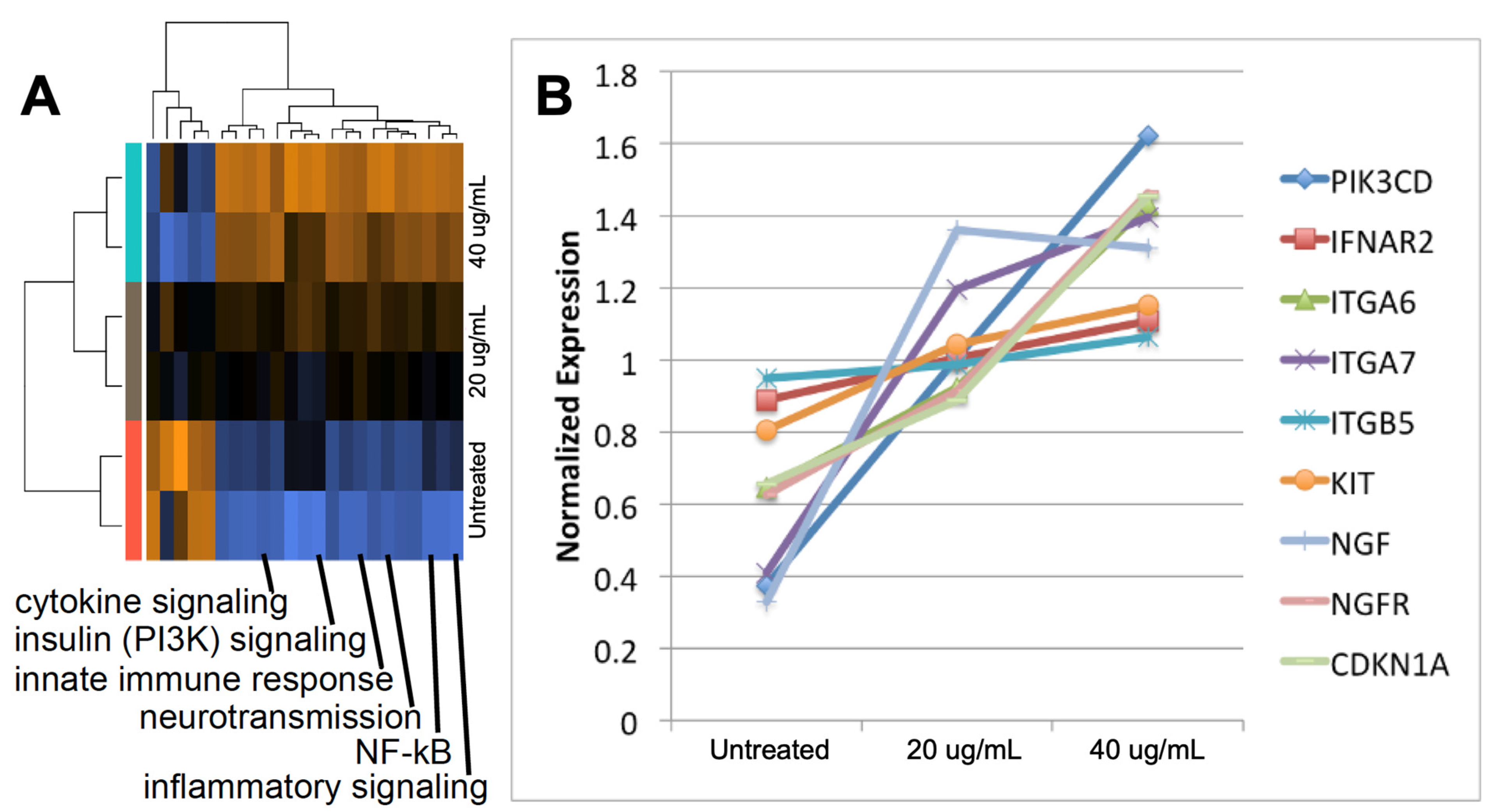
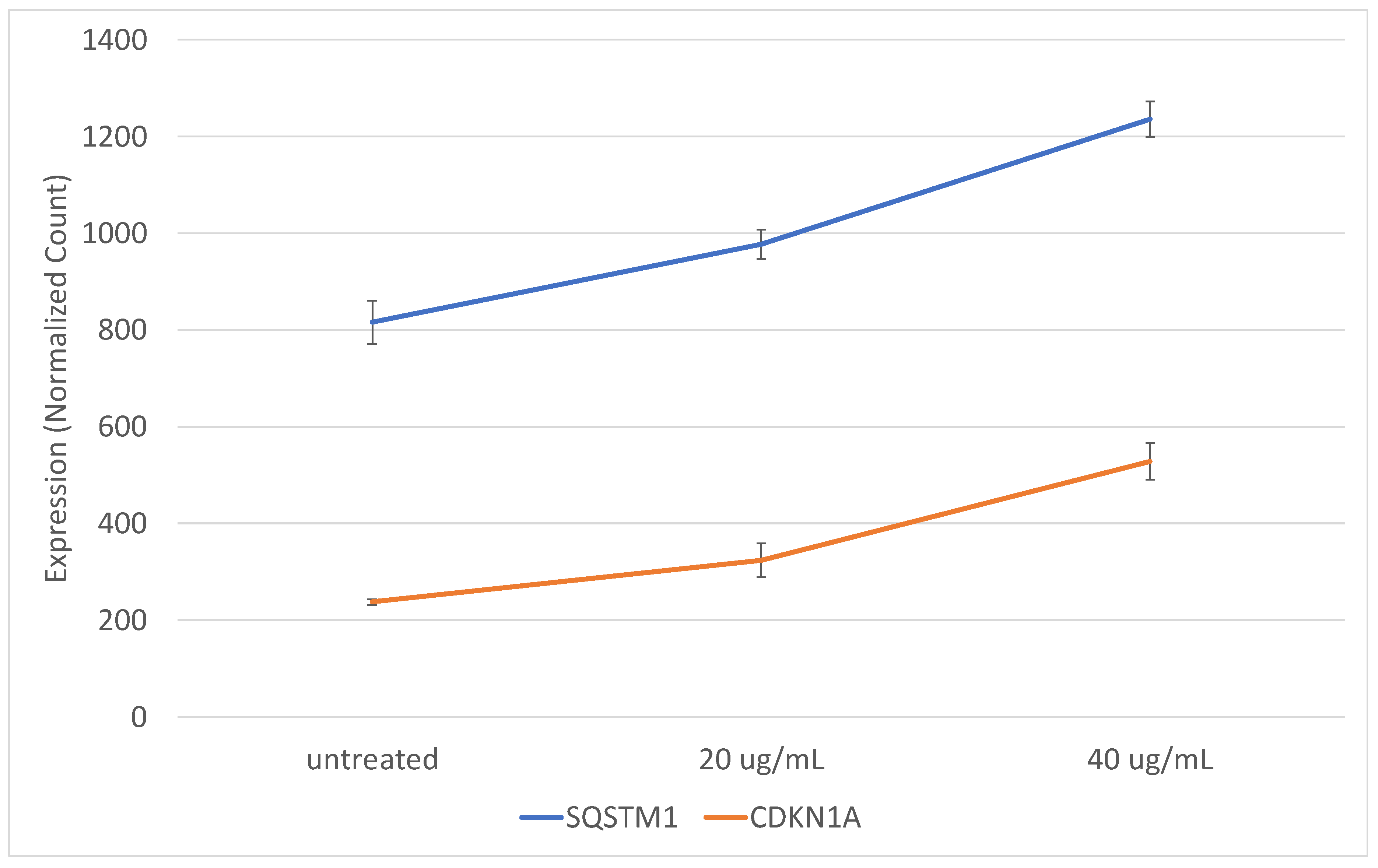
2.3. Exosomes from Epileptogenic Tissue Induce Key Signaling Pathways in Recipient Cells
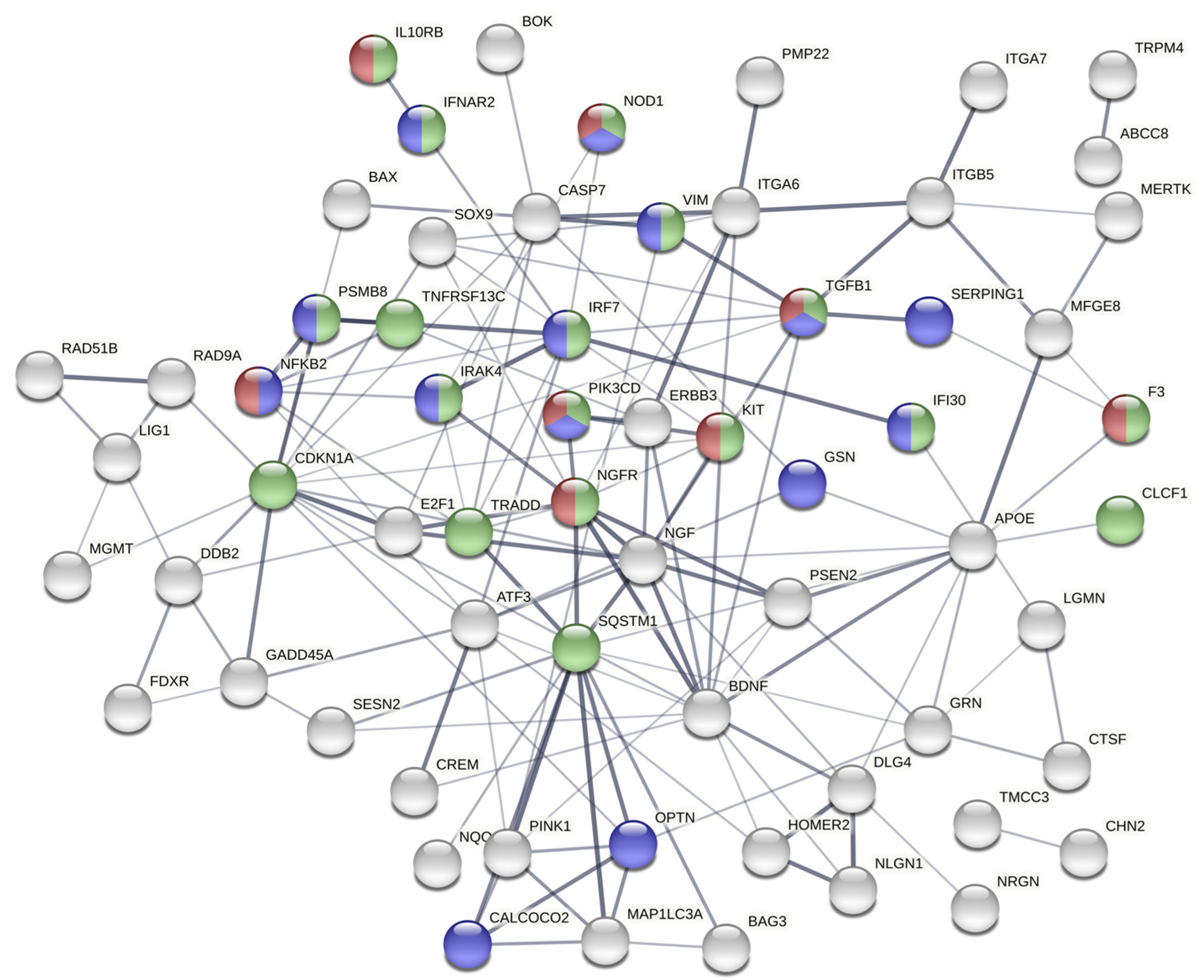
2.4. The Gene Expression Signature Induced In Vitro by Epileptogenic Exosomes Is Also Found in Epileptogenic Tissue
3. Discussion
4. Materials and Methods
4.1. Brain Tissue Used for Isolation of Extracellular Vesicles
4.2. Isolation of Extracellular Vesicles from Brain Tissue
4.3. Size and Morphology Analysis of Extracellular Vesicles Using Atomic Force Microscopy
4.4. Characterization of EV Proteins Using LC-MS/MS Proteomics
4.5. RNA Extraction from Extracellular Vesicles and Tissue
4.6. RNA-Seq of Exosomal RNA Cargo
4.7. In Vitro Analysis of SH-SY5Y Cell Response to Exosomes
4.8. Gene Expression Analysis of SH-SY5Y Cells and TSC Tissue Specimens
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reschke, C.R.; Henshall, D.C. microRNA and Epilepsy. Adv. Exp. Med. Biol. 2015, 888, 41–70. [Google Scholar] [CrossRef]
- Bagla, S.; Cukovic, D.; Asano, E.; Sood, S.; Luat, A.; Chugani, H.T.; Chugani, D.C.; Dombkowski, A.A. A distinct microRNA expression profile is associated with α[11C]-methyl-L-tryptophan (AMT) PET uptake in epileptogenic cortical tubers resected from patients with tuberous sclerosis complex. Neurobiol. Dis. 2018, 109 Pt A, 76–87. [Google Scholar] [CrossRef]
- Vezzani, A.; Aronica, E.; Mazarati, A.; Pittman, Q. Epilepsy and brain inflammation. Exp. Neurol. 2013, 244, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Bagla, S.; Dombkowski, A.A. Neuroinflammatory Nexus of Pediatric Epilepsy. J. Pediatr. Epilepsy 2018, 7, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zou, L.; Yan, F.; Chen, H.; Xu, G.; Jian, W.; Cui, P.; Chao, W. Extracellular MicroRNAs Induce Potent Innate Immune Responses via TLR7/MyD88-Dependent Mechanisms. J. Immunol. 2017, 199, 2106–2117. [Google Scholar] [CrossRef] [PubMed]
- Dombkowski, A.A.; Cukovic, D.; Bagla, S.; Jones, M.; Caruso, J.A.; Chugani, H.T.; Chugani, D.C. TLR7 activation in epilepsy of tuberous sclerosis complex. Inflamm. Res. 2019, 68, 993–998. [Google Scholar] [CrossRef]
- Bayraktar, R.; Bertilaccio, M.T.S.; Calin, G.A. The Interaction Between Two Worlds: MicroRNAs and Toll-Like Receptors. Front. Immunol. 2019, 10, 1053. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Croce, C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013, 10, 169–174. [Google Scholar] [CrossRef]
- Lehnardt, S.; Wallach, T.; Gres, V.; Henneke, P. Guardians of neuroimmunity—Toll-like receptors and their RNA ligands. Neuroforum 2019, 25, 185–193. [Google Scholar] [CrossRef]
- Wallach, T.; Wetzel, M.; Dembny, P.; Staszewski, O.; Krüger, C.; Buonfiglioli, A.; Prinz, M.; Lehnardt, S. Identification of CNS Injury-Related microRNAs as Novel Toll-Like Receptor 7/8 Signaling Activators by Small RNA Sequencing. Cells 2020, 9, 186. [Google Scholar] [CrossRef]
- Chiavegato, A.; Zurolo, E.; Losi, G.; Aronica, E.; Carmignoto, G. The inflammatory molecules IL-1beta and HMGB1 can rapidly enhance focal seizure generation in a brain slice model of temporal lobe epilepsy. Front. Cell Neurosci. 2014, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Takemiya, T.; Sugiura, H.; Yamagata, K. Role of Inflammatory Mediators in the Pathogenesis of Epilepsy. Mediat. Inflamm. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gallart-Palau, X.; Serra, A.; Sze, S.K. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol. Neurodegener. 2016, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhuang, X.; Zhang, S.; Deng, Z.-B.; Grizzle, W.; Miller, D.; Zhang, H.-G. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv. Drug Deliv. Rev. 2013, 65, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.W.; Bond, V.C.; Borràs, F.E.; Breakefield, X.O.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Forsbach, A.; Nemorin, J.-G.; Montino, C.; Müller, C.; Samulowitz, U.; Vicari, A.P.; Jurk, M.; Mutwiri, G.K.; Krieg, A.M.; Lipford, G.B.; et al. Identification of RNA Sequence Motifs Stimulating Sequence-Specific TLR8-Dependent Immune Responses. J. Immunol. 2008, 180, 3729–3738. [Google Scholar] [CrossRef]
- Rivals, I.; Personnaz, L.; Taing, L.; Potier, M.-C. Enrichment or depletion of a GO category within a class of genes: Which test? Bioinformatics 2006, 23, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Xicoy, H.; Wieringa, B.; Martens, G.J.M. The SH-SY5Y cell line in Parkinson’s disease research: A systematic review. Mol. Neurodegener. 2017, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, C.J.; Crews, F.T. Ethanol, TLR3, and TLR4 Agonists Have Unique Innate Immune Responses in Neuron-Like SH-SY5Y and Microglia-Like BV2. Alcohol. Clin. Exp. Res. 2017, 41, 939–954. [Google Scholar] [CrossRef]
- Lawrimore, C.J.; Coleman, L.G.; Crews, F.T. Ethanol induces interferon expression in neurons via TRAIL: Role of astrocyte-to-neuron signaling. Psychopharmacology 2019, 236, 2881–2897. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.M. Digital Multiplexed Gene Expression Analysis Using the NanoString nCounter System. Curr. Protoc. Mol. Biol. 2011, 94, 1–17. [Google Scholar] [CrossRef]
- Guiducci, C.; Gong, M.; Cepika, A.-M.; Xu, Z.; Tripodo, C.; Bennett, L.; Crain, C.; Quartier, P.; Cush, J.J.; Pascual, V.; et al. RNA recognition by human TLR8 can lead to autoimmune inflammation. J. Exp. Med. 2013, 210, 2903–2919. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Cantley, L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015, 25, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Butchi, N.B.; Du, M.; Peterson, K.E. Interactions between TLR7 and TLR9 agonists and receptors regulate innate immune responses by astrocytes and microglia. Glia 2009, 58, 650–664. [Google Scholar] [CrossRef]
- Sanchez-Martin, P.; Saito, T.; Komatsu, M. p62/SQSTM1: ’Jack of all trades’ in health and cancer. FEBS J. 2019, 286, 8–23. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [PubMed]
- Yelamanchili, S.V.; Lamberty, B.G.; Rennard, D.A.; Morsey, B.M.; Hochfelder, C.G.; Meays, B.M.; Fox, H.S. MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease. PLoS Pathog. 2015, 11, e1005032. [Google Scholar]
- Boer, K.; Crino, P.; Gorter, J.; Nellist, M.; Jansen, F.; Spliet, W.; Van Rijen, P.; Wittink, F.R.; Breit, T.; Troost, D.; et al. Gene Expression Analysis of Tuberous Sclerosis Complex Cortical Tubers Reveals Increased Expression of Adhesion and Inflammatory Factors. Brain Pathol. 2009, 20, 704–719. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, G.; Du, H.; Yi, M.; Hengling, C. Integrative analysis of gene expression associated with epilepsy in human epilepsy and animal models. Mol. Med. Rep. 2016, 13, 4920–4926. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Duran, A.; Amanchy, R.; Linares, J.F.; Joshi, J.; Abu-Baker, S.; Porollo, A.; Hansen, M.; Moscat, J.; Diaz-Meco, M.T. p62 Is a Key Regulator of Nutrient Sensing in the mTORC1 Pathway. Mol. Cell 2011, 44, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.A.; Horswell, S.; Ness, T.; Lumsdon, J.; Tooze, S.A.; Kirkham, N.; Armstrong, J.L.; Lovat, P.E. Prognostic Impact of p62 Expression in Cutaneous Malignant Melanoma. J. Investig. Dermatol. 2014, 134, 1476–1478. [Google Scholar] [CrossRef] [PubMed]
- Parkhitko, A.; Myachina, F.; Morrison, T.A.; Hindi, K.M.; Auricchio, N.; Karbowniczek, M.; Wu, J.J.; Finkel, T.; Kwiatkowski, D.J.; Yu, J.J.; et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc. Natl. Acad. Sci. USA 2011, 108, 12455–12460. [Google Scholar] [CrossRef]
- Iyer, A.; Prabowo, A.; Anink, J.; Spliet, W.G.M.; Van Rijen, P.C.; Aronica, E. Cell injury and Premature Neurodegeneration in Focal Malformations of Cortical Development. Brain Pathol. 2013, 24, 1–17. [Google Scholar] [CrossRef]
- Yasin, S.A.; Ali, A.M.; Tata, M.; Picker, S.R.; Anderson, G.W.; Latimer-Bowman, E.; Jacques, T.S. mTOR-dependent abnormalities in autophagy characterize human malformations of cortical development: Evidence from focal cortical dysplasia and tuberous sclerosis. Acta Neuropathol. 2013, 126, 207–218. [Google Scholar] [CrossRef]
- Sanchez-Garrido, J.; Shenoy, A.R. Regulation and repurposing of nutrient sensing and autophagy in innate immunity. Autophagy 2020, 1–21. [Google Scholar] [CrossRef]
- Sipka, A.S.; Chandler, T.L.; Behling-Kelly, E.L.; Overton, T.R.; Mann, S. The effect of ex vivo lipopolysaccharide stimulation and nutrient availability on transition cow innate immune cell AKT/mTOR pathway responsiveness. J. Dairy Sci. 2020, 103, 1956–1968. [Google Scholar] [CrossRef]
- Dombkowski, A.A.; Batista, C.E.; Cukovic, D.; Carruthers, N.J.; Ranganathan, R.; Shukla, U.; Stemmer, P.M.; Chugani, H.T.; Chugani, D.C. Cortical Tubers: Windows into Dysregulation of Epilepsy Risk and Synaptic Signaling Genes by MicroRNAs. Cereb. Cortex 2016, 26, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Manning, B.D. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 2009, 37, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Slota, J.; Booth, S.A. MicroRNAs in Neuroinflammation: Implications in Disease Pathogenesis, Biomarker Discovery and Therapeutic Applications. Non-Coding RNA 2019, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A.; Sharma, P.; Bullock, K.M.; Hansen, K.M.; Ludwig, N.; Whiteside, T.L. Transport of Extracellular Vesicles across the Blood-Brain Barrier: Brain Pharmacokinetics and Effects of Inflammation. Int. J. Mol. Sci. 2020, 21, 4407. [Google Scholar] [CrossRef] [PubMed]
- Northrup, H.; Krueger, D.A.; International Tuberous Sclerosis Complex Consensus Group. Tuberous sclerosis complex diagnostic criteria update: Recommendations of the 2012 International Tuberous Sclerosis Complex Consensus Conference. Pediatr. Neurol. 2013, 49, 243–254. [Google Scholar] [CrossRef]
- Chugani, H.T.; Luat, A.F.; Kumar, A.; Govindan, R.; Pawlik, K.; Asano, E. α-[11C]-Methyl-L-tryptophan-PET in 191 patients with tuberous sclerosis complex. Neurology 2013, 81, 674–680. [Google Scholar] [CrossRef]
- Asano, E.; Juhász, C.; Shah, A.; Sood, S.; Chugani, H.T. Role of subdural electrocorticography in prediction of long-term seizure outcome in epilepsy surgery. Brain 2009, 132, 1038–1047. [Google Scholar] [CrossRef]
- Jena, B.P.; Schneider, S.W.; Geibel, J.P.; Webster, P.; Oberleithner, H.; Sritharan, K.C. Gi regulation of secretory vesicle swelling examined by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1997, 94, 13317–13322. [Google Scholar] [CrossRef]
- Jeremic, A.; Quinn, A.S.; Cho, W.J.; Taatjes, D.J.; Jena, B.P. Energy-Dependent Disassembly of Self-Assembled SNARE Complex: Observation at Nanometer Resolution Using Atosmic Force Microscopy. J. Am. Chem. Soc. 2006, 128, 26–27. [Google Scholar] [CrossRef]
- Ziemann, M.; Kaspi, A.; El-Osta, A. Evaluation of microRNA alignment techniques. RNA 2016, 22, 1120–1138. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of Endogenous Controls for Analyzing Serum Exosomal miRNA in Patients with Hepatitis B or Hepatocellular Carcinoma. Dis. Markers 2015, 2015, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, G.-M.; Liu, L.-L.; Liu, C.; Liu, F.; Jiang, D.-N.; Pu, X.-Y. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol. Med. Rep. 2015, 12, 4683–4691. [Google Scholar] [CrossRef] [PubMed]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; de Couto, G.; Walravens, A.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef]
- Huangda, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
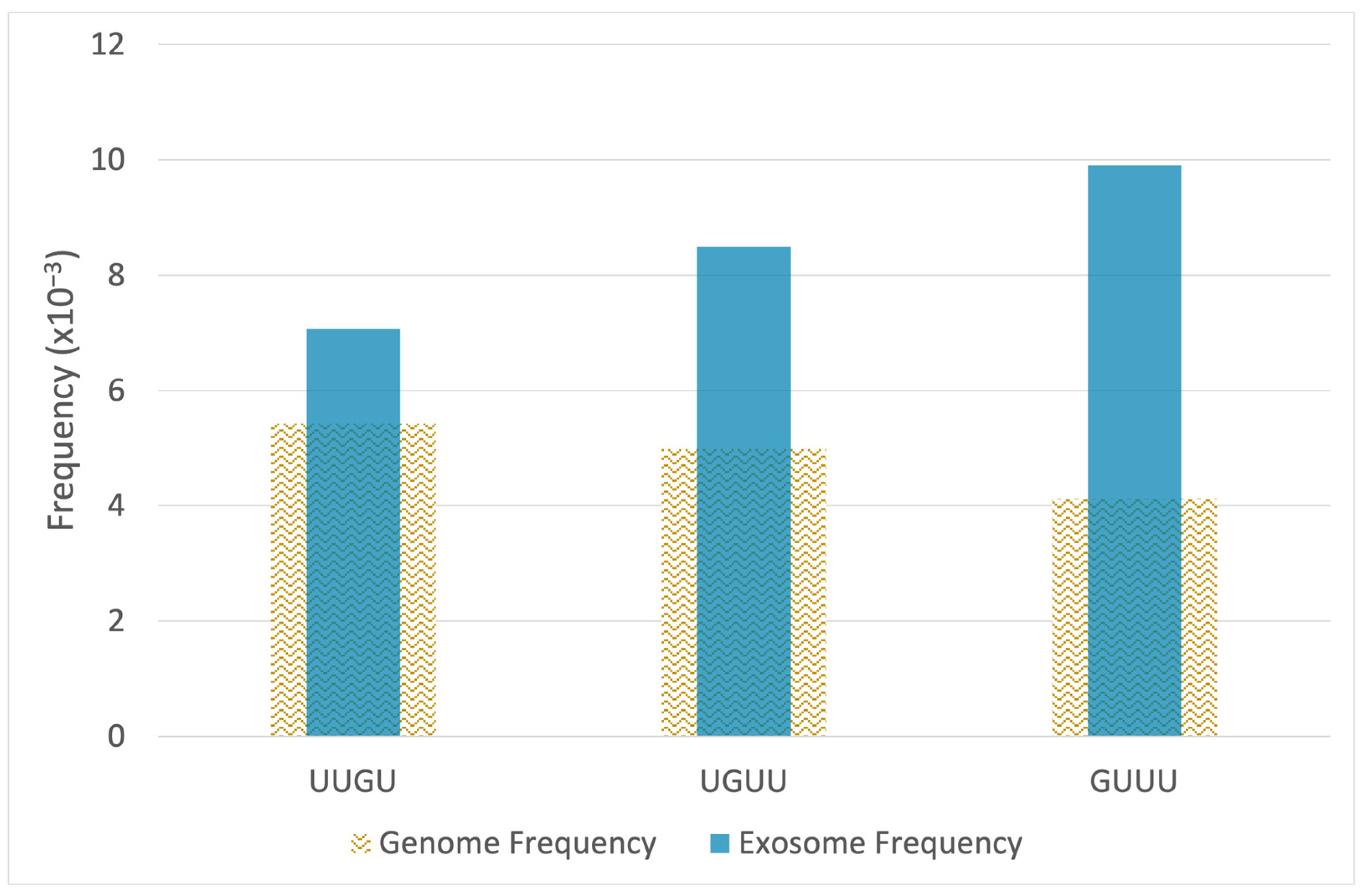
| microRNA | p-Value | FC | Sequence | UUGU | GUUU | UGUU | Total Motifs |
|---|---|---|---|---|---|---|---|
| hsa-miR-27a-5p | 0.04 | 17.41 | AGGGCUUAGCUGCUUGUGAGCA | 1 | 0 | 0 | 1 |
| hsa-miR-744-3p | 0.03 | 5.49 | CUGUUGCCACUAACCUCAACCU | 0 | 0 | 1 | 1 |
| hsa-miR-26a-2-3p | 0.02 | 3.54 | CCUAUUCUUGAUUACUUGUUUC | 1 | 1 | 1 | 3 |
| hsa-miR-652-3p | 0.04 | 3.54 | AAUGGCGCCACUAGGGUUGUG | 1 | 0 | 0 | 1 |
| hsa-miR-21-5p | 0.05 | 3.48 | UAGCUUAUCAGACUGAUGUUGA | 0 | 0 | 1 | 1 |
| hsa-miR-142-3p | 0.05 | 3.21 | UGUAGUGUUUCCUACUUUAUGGA | 0 | 1 | 1 | 2 |
| hsa-miR-29b-1-5p | 0.01 | 2.74 | GCUGGUUUCAUAUGGUGGUUUAGA | 0 | 2 | 0 | 2 |
| hsa-miR-629-5p | 0.03 | 2.48 | UGGGUUUACGUUGGGAGAACU | 0 | 1 | 0 | 1 |
| hsa-miR-28-3p | 0.05 | 2.33 | CACUAGAUUGUGAGCUCCUGGA | 1 | 0 | 0 | 1 |
| hsa-miR-3605-3p | 0.01 | 1.90 | CCUCCGUGUUACCUGUCCUCUAG | 0 | 0 | 1 | 1 |
| hsa-miR-223-3p | 0.04 | 1.75 | UGUCAGUUUGUCAAAUACCCCA | 1 | 1 | 0 | 2 |
| hsa-miR-30a-3p | 0.04 | 1.73 | CUUUCAGUCGGAUGUUUGCAGC | 0 | 1 | 1 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cukovic, D.; Bagla, S.; Ukasik, D.; Stemmer, P.M.; Jena, B.P.; Naik, A.R.; Sood, S.; Asano, E.; Luat, A.; Chugani, D.C.; et al. Exosomes in Epilepsy of Tuberous Sclerosis Complex: Carriers of Pro-Inflammatory MicroRNAs. Non-Coding RNA 2021, 7, 40. https://doi.org/10.3390/ncrna7030040
Cukovic D, Bagla S, Ukasik D, Stemmer PM, Jena BP, Naik AR, Sood S, Asano E, Luat A, Chugani DC, et al. Exosomes in Epilepsy of Tuberous Sclerosis Complex: Carriers of Pro-Inflammatory MicroRNAs. Non-Coding RNA. 2021; 7(3):40. https://doi.org/10.3390/ncrna7030040
Chicago/Turabian StyleCukovic, Daniela, Shruti Bagla, Dylan Ukasik, Paul M. Stemmer, Bhanu P. Jena, Akshata R. Naik, Sandeep Sood, Eishi Asano, Aimee Luat, Diane C. Chugani, and et al. 2021. "Exosomes in Epilepsy of Tuberous Sclerosis Complex: Carriers of Pro-Inflammatory MicroRNAs" Non-Coding RNA 7, no. 3: 40. https://doi.org/10.3390/ncrna7030040
APA StyleCukovic, D., Bagla, S., Ukasik, D., Stemmer, P. M., Jena, B. P., Naik, A. R., Sood, S., Asano, E., Luat, A., Chugani, D. C., & Dombkowski, A. A. (2021). Exosomes in Epilepsy of Tuberous Sclerosis Complex: Carriers of Pro-Inflammatory MicroRNAs. Non-Coding RNA, 7(3), 40. https://doi.org/10.3390/ncrna7030040








