Long Non-Coding RNAs in Diffuse Large B-Cell Lymphoma
Abstract
:1. Introduction
2. General Functions of lncRNAs
3. LncRNAs in DLBCL
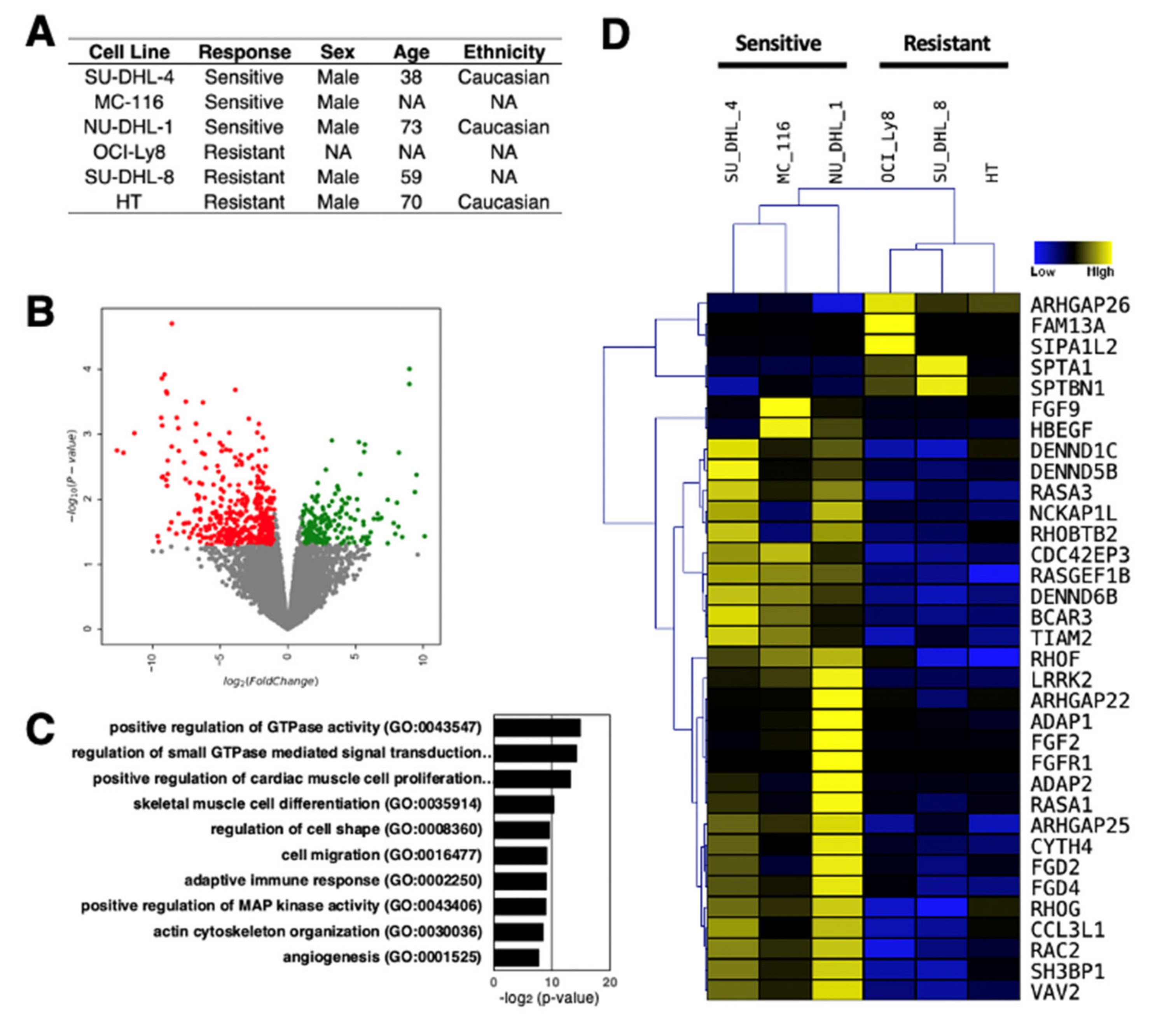
4. Differential Expression of lncRNAs in Rituximab Sensitive and Resistant DLBCL Cell Lines
5. Materials and Methods
5.1. Cell Culture and Treatment
5.2. RNA Isolation and RNA-seq Assay
5.3. Data Analysis
6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uchida, S.; Adams, J.C. Physiological roles of non-coding RNAs. Am. J. Physiol. Cell Physiol. 2019, 317, C1–C2. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Carninci, P. Genome-wide technologies to study RNA-chromatin interactions. Non-Coding RNA 2020, 6. [Google Scholar] [CrossRef]
- Maass, P.G.; Barutcu, A.R.; Rinn, J.L. Interchromosomal interactions: A genomic love story of kissing chromosomes. J. Cell Biol. 2019, 218, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Mishra, K.; Kanduri, C. Understanding long noncoding RNA and chromatin interactions: What we know so far. Non-Coding RNA 2019, 5. [Google Scholar] [CrossRef] [Green Version]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef] [Green Version]
- Ang, C.E.; Trevino, A.E.; Chang, H.Y. Diverse lncRNA mechanisms in brain development and disease. Curr. Opin. Genet. Dev. 2020, 65, 42–46. [Google Scholar] [CrossRef]
- Bar, C.; Chatterjee, S.; Falcao Pires, I.; Rodrigues, P.; Sluijter, J.P.G.; Boon, R.A.; Nevado, R.M.; Andres, V.; Sansonetti, M.; de Windt, L.; et al. Non-coding RNAs: Update on mechanisms and therapeutic targets from the ESC working groups of myocardial function and cellular biology of the heart. Cardiovasc. Res. 2020, 116, 1805–1819. [Google Scholar] [CrossRef]
- Dominguez-Andres, J.; Fanucchi, S.; Joosten, L.A.B.; Mhlanga, M.M.; Netea, M.G. Advances in understanding molecular regulation of innate immune memory. Curr. Opin. Cell. Biol. 2020, 63, 68–75. [Google Scholar] [CrossRef]
- Pereira Fernandes, D.; Bitar, M.; Jacobs, F.M.J.; Barry, G. Long Non-Coding RNAs in Neuronal Aging. Non-Coding RNA 2018, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmeron-Barcenas, E.G.; Mendoza-Catalan, M.A.; Illades-Aguiar, B.; Peralta-Arrieta, I.; Alquisiras-Burgos, I.; Ortiz-Ortiz, J.; Navarro-Tito, N.; Zacapala-Gomez, A.E. Long non-coding RNAs as new players in cervical carcinogenesis: An update. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8314–8328. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Kauppinen, S. Long non-coding RNAs in liver cancer and nonalcoholic steatohepatitis. Non-Coding RNA 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Ginn, L.; Shi, L.; Montagna, M.; Garofalo, M. LncRNAs in non-small-cell lung cancer. Non-Coding RNA 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.A.; Prinz, F.; Calin, G.A.; Pichler, M. Current concepts of non-coding RNA regulation of immune checkpoints in cancer. Mol. Aspects Med. 2019, 70, 117–126. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, J.; Yu, J.; Liu, X.; Yu, C.; Hu, J.; Shi, H.; Ma, X. Long non-coding RNAs: Emerging roles in the Immunosuppressive tumor microenvironment. Front. Oncol. 2020, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Qu, J.; Kamal, M.A.; Yuan, C. The regulatory roles of long non-coding RNA in the chemoresistance process of ovarian cancer. Curr. Pharm. Des. 2019, 25, 856–861. [Google Scholar] [CrossRef]
- Bai, H.; Lei, K.; Huang, F.; Jiang, Z.; Zhou, X. Exo-circRNAs: A new paradigm for anticancer therapy. Mol. Cancer 2019, 18, 56. [Google Scholar] [CrossRef]
- Kong, Y.; Hsieh, C.H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Xie, Z.; Lei, X.; Gan, R. Long non-coding RNA GAS5 in human cancer. Oncol. Lett. 2020, 20, 2587–2594. [Google Scholar] [CrossRef]
- Yu, Y.; Hann, S.S. Novel tumor suppressor lncRNA growth arrest-specific 5 (GAS5) in human cancer. Onco. Targets Ther. 2019, 12, 8421–8436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Shen, A.; Liu, A. Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull. Cancer 2019, 106, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Tietze, L.; Kessler, S.M. The good, the bad, the question-H19 in hepatocellular carcinoma. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Di Bonito, M.; Cerrone, M.; Collina, F.; De Laurentiis, M.; Botti, G. Long non-coding RNA HOTAIR in breast cancer therapy. Cancers 2020, 12. [Google Scholar] [CrossRef]
- Qu, X.; Alsager, S.; Zhuo, Y.; Shan, B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019, 454, 90–97. [Google Scholar] [CrossRef]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [Green Version]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 long non-coding RNA: Functional implications. Non-Coding RNA 2020, 6. [Google Scholar] [CrossRef]
- Chen, Y.K.; Yen, Y. The ambivalent role of lncRNA Xist in carcinogenesis. Stem Cell Rev. Rep. 2019, 15, 314–323. [Google Scholar] [CrossRef]
- Yang, Z.; Jiang, X.; Jiang, X.; Zhao, H. X-inactive-specific transcript: A long noncoding RNA with complex roles in human cancers. Gene 2018, 679, 28–35. [Google Scholar] [CrossRef]
- Ansell, S.M. Hodgkin lymphoma: A 2020 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2020, 95, 978–989. [Google Scholar] [CrossRef]
- Shanbhag, S.; Ambinder, R.F. Hodgkin lymphoma: A review and update on recent progress. CA Cancer J. Clin. 2018, 68, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Armitage, J.O.; Gascoyne, R.D.; Lunning, M.A.; Cavalli, F. Non-Hodgkin lymphoma. Lancet 2017, 390, 298–310. [Google Scholar] [CrossRef]
- Types of B-cell Lymphoma. Available online: https://www.cancer.org/cancer/non-hodgkin-lymphoma/about/b-cell-lymphoma.html (accessed on 22 October 2020).
- Salles, G.; Barrett, M.; Foa, R.; Maurer, J.; O’Brien, S.; Valente, N.; Wenger, M.; Maloney, D.G. Rituximab in B-cell hematologic malignancies: A review of 20 years of clinical experience. Adv. Ther. 2017, 34, 2232–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2017. Available online: https://seer.cancer.gov/csr/1975_2017/ (accessed on 14 September 2020).
- Shaw, J.; Harvey, C.; Richards, C.; Kim, C. Temporal trends in treatment and survival of older adult diffuse large B-cell lymphoma patients in the SEER-medicare linked database. Leuk. Lymphoma 2019, 60, 3235–3243. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, P.; Grillo-Lopez, A.J.; Link, B.K.; Levy, R.; Czuczman, M.S.; Williams, M.E.; Heyman, M.R.; Bence-Bruckler, I.; White, C.A.; Cabanillas, F.; et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: Half of patients respond to a four-dose treatment program. J. Clin. Oncol. 1998, 16, 2825–2833. [Google Scholar] [CrossRef]
- Sehn, L.H.; Donaldson, J.; Chhanabhai, M.; Fitzgerald, C.; Gill, K.; Klasa, R.; MacPherson, N.; O’Reilly, S.; Spinelli, J.J.; Sutherland, J.; et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J. Clin. Oncol. 2005, 23, 5027–5033. [Google Scholar] [CrossRef]
- Vose, J.M.; Link, B.K.; Grossbard, M.L.; Czuczman, M.; Grillo-Lopez, A.; Gilman, P.; Lowe, A.; Kunkel, L.A.; Fisher, R.I. Phase II study of rituximab in combination with chop chemotherapy in patients with previously untreated, aggressive non-Hodgkin’s lymphoma. J. Clin. Oncol. 2001, 19, 389–397. [Google Scholar] [CrossRef]
- Crump, M.; Neelapu, S.S.; Farooq, U.; Van Den Neste, E.; Kuruvilla, J.; Westin, J.; Link, B.K.; Hay, A.; Cerhan, J.R.; Zhu, L.; et al. Outcomes in refractory diffuse large B-cell lymphoma: Results from the international SCHOLAR-1 study. Blood 2017, 130, 1800–1808. [Google Scholar] [CrossRef]
- Rovira, J.; Valera, A.; Colomo, L.; Setoain, X.; Rodriguez, S.; Martinez-Trillos, A.; Gine, E.; Dlouhy, I.; Magnano, L.; Gaya, A.; et al. Prognosis of patients with diffuse large B cell lymphoma not reaching complete response or relapsing after frontline chemotherapy or immunochemotherapy. Ann. Hematol. 2015, 94, 803–812. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Zhang, E.B.; Yin, D.D.; Sun, M.; Kong, R.; Liu, X.H.; You, L.H.; Han, L.; Xia, R.; Wang, K.M.; Yang, J.S.; et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014, 5, e1243. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; de la Cruz, C.C.; Otte, A.P.; Panning, B.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Paucek, R.D.; Gooding, A.R.; Brown, Z.Z.; Ge, E.J.; Muir, T.W.; Cech, T.R. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 2017, 24, 1028–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Goodrich, K.J.; Gooding, A.R.; Naeem, H.; Archer, S.; Paucek, R.D.; Youmans, D.T.; Cech, T.R.; Davidovich, C. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell 2017, 65, 1056–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portoso, M.; Ragazzini, R.; Brencic, Z.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017, 36, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Wang, X.; Cifuentes-Rojas, C.; Goodrich, K.J.; Gooding, A.R.; Lee, J.T.; Cech, T.R. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 2015, 57, 552–558. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes-Rojas, C.; Hernandez, A.J.; Sarma, K.; Lee, J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 2014, 55, 171–185. [Google Scholar] [CrossRef] [Green Version]
- Davidovich, C.; Zheng, L.; Goodrich, K.J.; Cech, T.R. Promiscuous RNA binding by polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013, 20, 1250–1257. [Google Scholar] [CrossRef] [Green Version]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Xie, J.; Shen, C.; Cheng, D.; Shi, Y.; Wu, Z.; Deng, X.; Chen, H.; Shen, B.; Peng, C.; et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015, 75, 3181–3191. [Google Scholar] [CrossRef] [Green Version]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Senergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Muller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA locus handsdown regulates cardiac gene programs and is essential for early mouse development. Dev. Cell 2019, 50, 644–657. [Google Scholar] [CrossRef]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 2011, 17, 792–798. [Google Scholar] [CrossRef] [Green Version]
- Grander, D.; Johnsson, P. Pseudogene-expressed RNAs: Emerging roles in gene regulation and disease. Curr. Top. Microbiol. Immunol. 2016, 394, 111–126. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2020, 21, 10–1038. [Google Scholar] [CrossRef]
- Li, F.; Yang, Q.; He, A.T.; Yang, B.B. Circular RNAs in cancer: Limitations in functional studies and diagnostic potential. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Lei, B.; Tian, Z.; Fan, W.; Ni, B. Circular RNA: A novel biomarker and therapeutic target for human cancers. Int. J. Med. Sci. 2019, 16, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Yang, T.; Xiao, J. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine 2018, 34, 267–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, P.; Ramirez-Martinez, A.; Li, H.; Cannavino, J.; McAnally, J.R.; Shelton, J.M.; Sanchez-Ortiz, E.; Bassel-Duby, R.; Olson, E.N. Control of muscle formation by the fusogenic micropeptide myomixer. Science 2017, 356, 323–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; He, Y.; Zhang, H.; Wu, Z.; Li, D.; Zheng, C. Comprehensive analysis of differentially expressed profiles of lncRNAs and mRNAs reveals ceRNA networks in the transformation of diffuse large B-cell lymphoma. Oncol. Lett. 2018, 16, 882–890. [Google Scholar] [CrossRef] [PubMed]
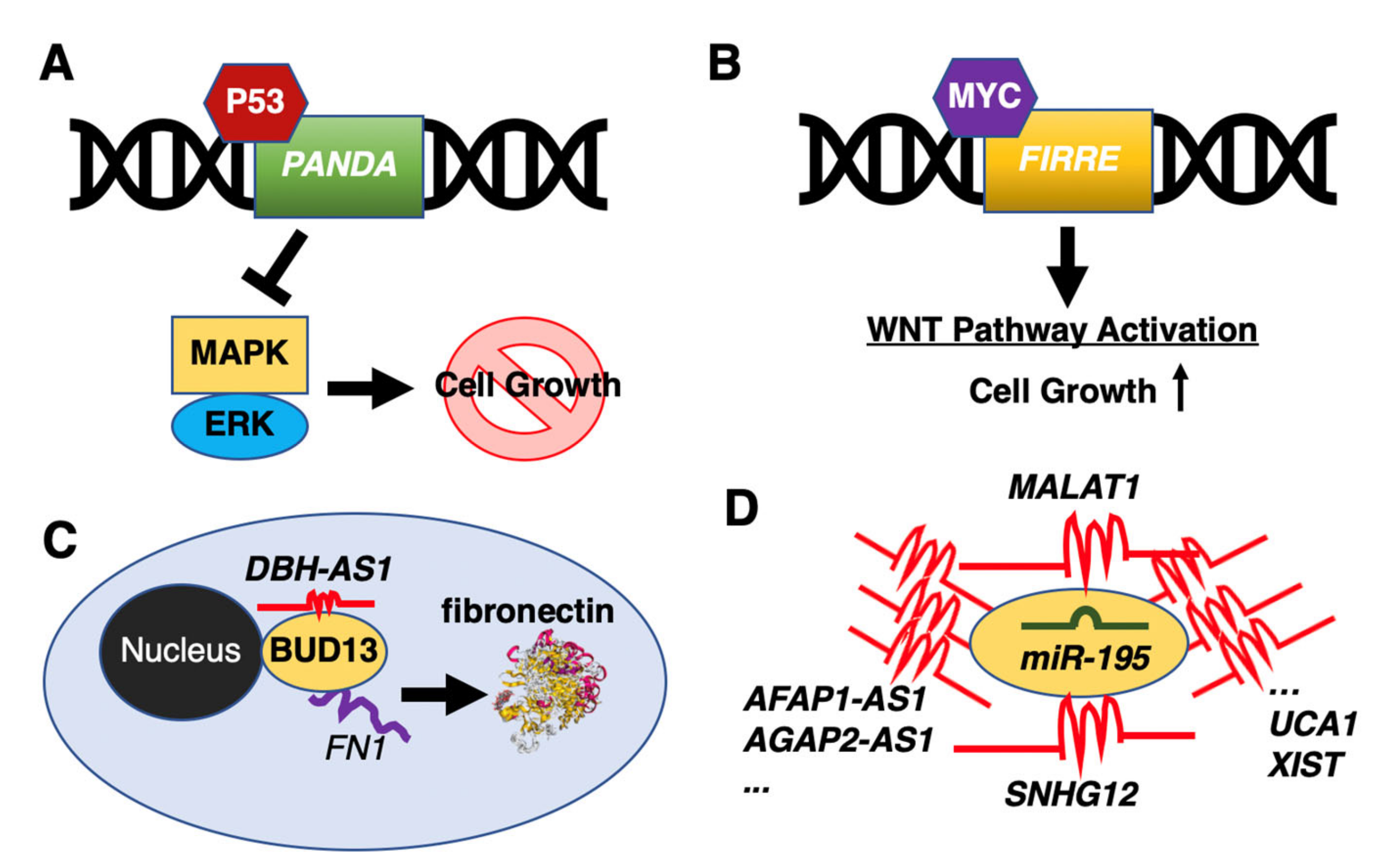
- Wang, Y.; Zhang, M.; Xu, H.; Wang, Y.; Li, Z.; Chang, Y.; Wang, X.; Fu, X.; Zhou, Z.; Yang, S.; et al. Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget 2017, 8, 72182–72196. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.Y.; Wu, B.; Yan, W.; Gong, Z.M.; Sun, Q.; Wang, H.H.; Yang, W. Microarray expression profiles of long non-coding RNAs in germinal center-like diffuse large B-cell lymphoma. Oncol Rep. 2017, 38, 1363–1372. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Fang, C.; Li, X.; Geng, Y.; Li, R.; Wu, C.; Jiang, J.; Wu, C. Predictive analysis of long non-coding RNA expression profiles in diffuse large B-cell lymphoma. Oncotarget 2017, 8, 23228–23236. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Cui, Z.; Liu, X.; Wu, S.; Wu, Y.; Fang, F.; Zhao, H. LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2019, 510, 594–600. [Google Scholar] [CrossRef]
- Song, Y.; Gao, F.; Peng, Y.; Yang, X. Long non-coding RNA DBH-AS1 promotes cancer progression in diffuse large B-cell lymphoma by targeting FN1 via RNA-binding protein BUD13. Cell. Biol. Int. 2020, 44, 1331–1340. [Google Scholar] [CrossRef]
- Sehgal, L.; Mathur, R.; Braun, F.K.; Wise, J.F.; Berkova, Z.; Neelapu, S.; Kwak, L.W.; Samaniego, F. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 2014, 28, 2376–2387. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Yan, Z.; Wang, X.; Cao, J.; Chen, W.; Qi, K.; Zhou, D.; Xia, J.; Qi, N.; Li, Z.; et al. Downregulation of long non-coding RNA TUG1 suppresses tumor growth by promoting ubiquitination of MET in diffuse large B-cell lymphoma. Mol. Cell Biochem. 2019, 461, 47–56. [Google Scholar] [CrossRef]
- Qian, C.S.; Li, L.J.; Huang, H.W.; Yang, H.F.; Wu, D.P. MYC-regulated lncRNA NEAT1 promotes B cell proliferation and lymphomagenesis via the miR-34b-5p-GLI1 pathway in diffuse large B-cell lymphoma. Cancer Cell Int. 2020, 20, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Su, L.; Jiang, J. Long non-coding RNA paternally expressed imprinted gene 10 (PEG10) elevates diffuse large B-cell lymphoma progression by regulating kinesin family member 2A (KIF2A) via Targeting MiR-101-3p. Med. Sci. Monit. 2020, 26, e922810. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.C.; Jiao, Y.; Zhang, Y.Y.; Ning, J.; Zhang, Y.R.; Xu, J.; Wei, W.; Kang-Sheng, G. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/ β -catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Ojha, S.; Malla, S.; Lyons, S.M. snoRNPs: Functions in ribosome biogenesis. Biomolecules 2020, 10, 783. [Google Scholar] [CrossRef]
- Yang, H.; Jiang, Z.; Wang, S.; Zhao, Y.; Song, X.; Xiao, Y.; Yang, S. Long non-coding small nucleolar RNA host genes in digestive cancers. Cancer Med. 2019, 8, 7693–7704. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Y.; Zhang, X.M.; Han, B.Q.; Dai, H.B. Long noncoding RNA SNHG12 indicates the prognosis and accelerates tumorigenesis of diffuse large B-cell lymphoma through sponging microR-195. Onco. Targets Ther. 2020, 13, 5563–5574. [Google Scholar] [CrossRef]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.B.M.; Osman, M.A. The regulatory role of microRNAs in breast cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A review of its role in cancers. Onco. Targets Ther. 2018, 11, 7109–7123. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.M.; Lian, G.Y.; Song, Y.; Huang, Y.F.; Gong, Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019, 231, 116335. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Wu, A.; Yu, H.; Yu, Q.; Zheng, B.; Yang, W.; Tian, D.; Li, P.; Gao, Y. Involvement of the lncRNA AFAP1-AS1/microRNA-195/E2F3 axis in proliferation and migration of enteric neural crest stem cells of Hirschsprung’s disease. Exp. Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Li, K.; Liu, Y.; Liu, X.; Liu, B.; Ba, Y.; Xing, W. Silencing lncRNA AGAP2-AS1 Upregulates miR-195-5p to repress migration and invasion of EC cells via the decrease of FOSL1 expression. Mol. Ther. Nucleic Acids 2020, 20, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Song, D. LncRNA MEG3 Participates in caerulein-induced inflammatory injury in human pancreatic cells via regulating miR-195-5p/FGFR2 axis and inactivating NF- κB pathway. Inflammation 2020. [Google Scholar] [CrossRef]
- Tang, N.; Dong, Y.; Liu, J.; Zhao, H. Silencing of long non-coding RNA NEAT1 upregulates miR-195a to attenuate intervertebral disk degeneration via the BAX/BAK pathway. Front. Mol. Biosci. 2020, 7, 147. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, T.; Tian, L.; Li, Y. LncRNA OIP5-AS1 promotes the proliferation of hemangioma vascular endothelial cells via regulating miR-195-5p/NOB1 axis. Front. Pharmacol. 2019, 10, 449. [Google Scholar] [CrossRef]
- Li, H.J.; Sun, X.M.; Li, Z.K.; Yin, Q.W.; Pang, H.; Pan, J.J.; Li, X.; Chen, W. LncRNA UCA1 promotes mitochondrial function of bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol. Biochem. 2017, 43, 2548–2561. [Google Scholar] [CrossRef]
- Yin, D.; Fu, C.; Sun, D. Silence of lncRNA UCA1 represses the growth and tube formation of human microvascular endothelial cells through miR-195. Cell Physiol. Biochem. 2018, 49, 1499–1511. [Google Scholar] [CrossRef]
- Yang, C.; Wu, K.; Wang, S.; Wei, G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J. Cell Biochem. 2018, 119, 5646–5656. [Google Scholar] [CrossRef]
- Chang, K.C.; Chang, W.C.; Chang, Y.; Hung, L.Y.; Lai, C.H.; Yeh, Y.M.; Chou, Y.W.; Chen, C.H. Ran GTPase-activating protein 1 is a therapeutic target in diffuse large B-cell lymphoma. PLoS ONE 2013, 8, e79863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouw, L.G.; Reading, N.S.; Jenson, S.D.; Lim, M.S.; Elenitoba-Johnson, K.S. Expression of the Rho-family GTPase gene RHOF in lymphocyte subsets and malignant lymphomas. Br. J. Haematol. 2005, 129, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.B.; Reinholdt, L.; Schonherz, A.A.; Due, H.; Jespersen, D.S.; Grubach, L.; Ettrup, M.S.; Roge, R.; Falgreen, S.; Sorensen, S.; et al. High CXCR4 expression impairs rituximab response and the prognosis of R-CHOP-treated diffuse large B-cell lymphoma patients. Oncotarget 2019, 10, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Due, H.; Brondum, R.F.; Young, K.H.; Bogsted, M.; Dybkaer, K. MicroRNAs associated to single drug components of R-CHOP identifies diffuse large B-cell lymphoma patients with poor outcome and adds prognostic value to the international prognostic index. BMC Cancer 2020, 20, 237. [Google Scholar] [CrossRef]
- Whitlock, N.C.; Trostel, S.Y.; Wilkinson, S.; Terrigino, N.T.; Hennigan, S.T.; Lake, R.; Carrabba, N.V.; Atway, R.; Walton, E.D.; Gryder, B.E.; et al. MEIS1 down-regulation by MYC mediates prostate cancer development through elevated HOXB13 expression and AR activity. Oncogene 2020, 39, 5663–5674. [Google Scholar] [CrossRef]
- Farrugia, M.K.; Sharma, S.B.; Lin, C.C.; McLaughlin, S.L.; Vanderbilt, D.B.; Ammer, A.G.; Salkeni, M.A.; Stoilov, P.; Agazie, Y.M.; Creighton, C.J.; et al. Regulation of anti-apoptotic signaling by Kruppel-like factors 4 and 5 mediates lapatinib resistance in breast cancer. Cell Death Dis. 2015, 6, e1699. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- HafezQorani, S.; Houdjedj, A.; Arici, M.; Said, A.; Kazan, H. RBPSponge: Genome-wide identification of lncRNAs that sponge RBPs. Bioinformatics 2019, 35, 4760–4763. [Google Scholar] [CrossRef]
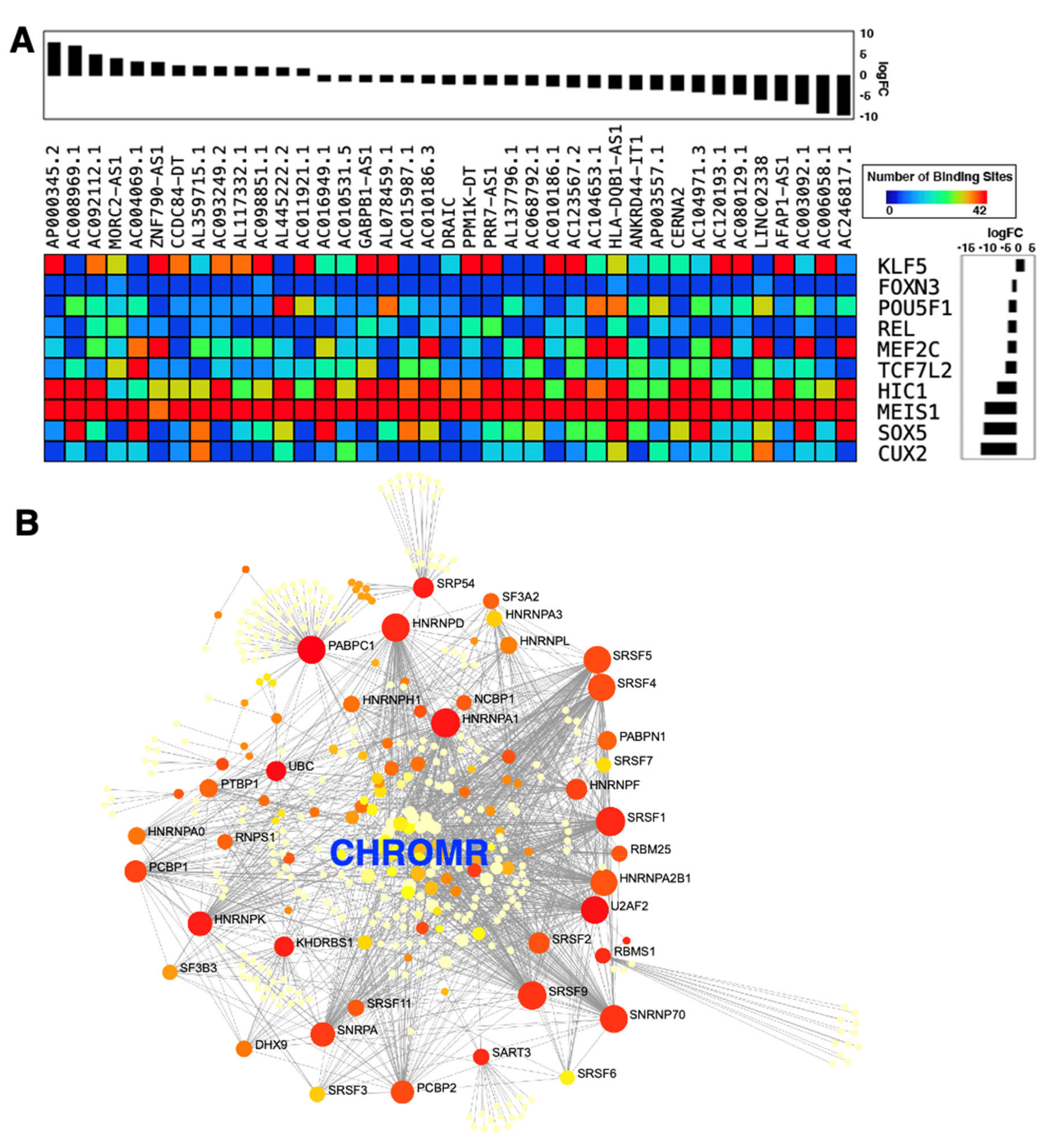
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Huang, B.; Song, B.L.; Xu, C. Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2020, 2, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Rink, J.S.; Yang, S.; Cen, O.; Taxter, T.; McMahon, K.M.; Misener, S.; Behdad, A.; Longnecker, R.; Gordon, L.I.; Thaxton, C.S. rational targeting of cellular cholesterol in diffuse large B-cell lymphoma (DLBCL) enabled by functional lipoprotein nanoparticles: A therapeutic strategy dependent on cell of origin. Mol. Pharm. 2017, 14, 4042–4051. [Google Scholar] [CrossRef] [PubMed]
- Dybkaer, K.; Bogsted, M.; Falgreen, S.; Bodker, J.S.; Kjeldsen, M.K.; Schmitz, A.; Bilgrau, A.E.; Xu-Monette, Z.Y.; Li, L.; Bergkvist, K.S.; et al. Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J. Clin. Oncol. 2015, 33, 1379–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedre, R. reneshbedre/bioinfokit: Bioinformatics Data Analysis and Visualization Toolkit. Available online: https://zenodo.org/record/3747737#.X-SnxthKhPY (accessed on 13 September 2020).
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gearing, L.J.; Cumming, H.E.; Chapman, R.; Finkel, A.M.; Woodhouse, I.B.; Luu, K.; Gould, J.A.; Forster, S.C.; Hertzog, P.J. CiiiDER: A tool for predicting and analysing transcription factor binding sites. PLoS ONE 2019, 14, e0215495. [Google Scholar] [CrossRef] [Green Version]
- Benoit Bouvrette, L.P.; Bovaird, S.; Blanchette, M.; Lecuyer, E. oRNAment: A database of putative RNA binding protein target sites in the transcriptomes of model species. Nucleic Acids Res. 2020, 48, D166–D173. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Cheng, L.; Shi, H.; Zhang, Z.; Zhao, H.; Wang, Z.; Zhou, M. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci. Rep. 2016, 6, 27842. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Jiang, Y.; Du, W.; Fairchild, L.; Melnick, A.; Elemento, O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015, 7, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Zhao, H.; Xu, W.; Bao, S.; Cheng, L.; Sun, J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 2017, 16, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karstensen, K.T.; Schein, A.; Petri, A.; Bøgsted, M.; Dybkær, K.; Uchida, S.; Kauppinen, S. Long Non-Coding RNAs in Diffuse Large B-Cell Lymphoma. Non-Coding RNA 2021, 7, 1. https://doi.org/10.3390/ncrna7010001
Karstensen KT, Schein A, Petri A, Bøgsted M, Dybkær K, Uchida S, Kauppinen S. Long Non-Coding RNAs in Diffuse Large B-Cell Lymphoma. Non-Coding RNA. 2021; 7(1):1. https://doi.org/10.3390/ncrna7010001
Chicago/Turabian StyleKarstensen, Kasper Thystrup, Aleks Schein, Andreas Petri, Martin Bøgsted, Karen Dybkær, Shizuka Uchida, and Sakari Kauppinen. 2021. "Long Non-Coding RNAs in Diffuse Large B-Cell Lymphoma" Non-Coding RNA 7, no. 1: 1. https://doi.org/10.3390/ncrna7010001
APA StyleKarstensen, K. T., Schein, A., Petri, A., Bøgsted, M., Dybkær, K., Uchida, S., & Kauppinen, S. (2021). Long Non-Coding RNAs in Diffuse Large B-Cell Lymphoma. Non-Coding RNA, 7(1), 1. https://doi.org/10.3390/ncrna7010001






